Abstract
Transcription is an essential step in gene expression and its understanding has been one of the major interests in molecular and cellular biology. By precisely tuning gene expression, transcriptional regulation determines the molecular machinery for developmental plasticity, homeostasis and adaptation. In this review, we transmit the main ideas or concepts behind regulation by transcription factors and give just enough examples to sustain these main ideas, thus avoiding a classical ennumeration of facts. We review recent concepts and developments: cis elements and trans regulatory factors, chromosome organization and structure, transcriptional regulatory networks (TRNs) and transcriptomics. We also summarize new important discoveries that will probably affect the direction of research in gene regulation: epigenetics and stochasticity in transcriptional regulation, synthetic circuits and plasticity and evolution of TRNs. Many of the new discoveries in gene regulation are not extensively tested with wetlab approaches. Consequently, we review this broad area in Inference of TRNs and Dynamical Models of TRNs. Finally, we have stepped backwards to trace the origins of these modern concepts, synthesizing their history in a timeline schema.
Keywords: regulatory network inference, regulatory network plasticity, chromosome structure, dynamical models of regulatory networks, regulatory network
Introduction: cis elements and trans regulatory factors
Transcriptional regulation emerges from the interaction between trans factors (Latin for ‘far side of’) that bind to cis-regulatory elements (Latin for ‘this side of’) in the context of a particular chromatin/chromosome structure. Taking the doubled-stranded DNA molecule as a reference, cis elements are all those DNA regions – encoded in a plasmid or in a chromosome – in the vicinity of a gene. In complement, all the diffusible cellular molecules that are able to bind to the DNA are the trans factors. The coactivity of these molecular entities composes the minimal transcriptional regulatory system in all living organisms. In bacterial chromosomes, a transcription unit (TU) is the ordered assembly of the following genetic entities: a regulatory region, a transcription start site, one or more ORFs and a transcription termination site. When a TU comprises more than one ORF, the transcribed mRNA is called polycistronic; otherwise, it is called monocistronic. It is not uncommon for genes to be transcribed by several promoters; thus, TUs overlap. The collection of overlapping TUs constitutes an operon. Historically defined as a polycistronic TU, it has been observed that operons always contain a promoter that transcribes the whole set of genes conforming its TUs. The regulatory region contains cis elements such as the promoter – where the RNA polymerase initially binds – and transcription factor-binding sites (TFBS) – where transcription factors (TFs) bind to modulate the binding of the RNA polymerase (Browning & Busby, 2004). In prokaryotes, these regions occupy up to 400 base pairs (bp) (Collado-Vides et al., 1991).
Transcription initiation in bacteria requires proteins known as sigma factors (σ). These factors – with even dozens of different types per genome – are essential for proper promoter recognition by RNA polymerase (Maeda et al., 2000; Helmann, 2002; Paget & Helmann, 2003; Kazmierczak et al., 2005). In bacteria, σ factors are divided into two main phylogenetic families: σ70 and σ54. The σ70 family includes the housekeeping σ that contributes with most of the gene transcription under normal conditions. One subgroup of factors from this family comprises a varying number of proteins known as extracytoplasmatic factors (ECF) activated in response to environmental stress. Usually, every bacterium has one protein member from the σ54 family. RNA polymerase associated with a member of this family recognizes promoters that are different from those exclusively recognized when associated with a member of σ70. However, there are exceptions where two different σ factors bind to the same promoter (Weber et al., 2005; Wade et al., 2006; Typas et al., 2007). Most σ factors have one anti-σ protein that binds to their σ cognate, inhibiting its action. The σ activity depends on σ/anti-σ ratios and the mechanisms to dissociate σ/anti-σ complexes are diverse (Hughes & Mathee, 1998). Also, there are post-translational mechanisms that modulate the activity of TFs and σ factors such as proteins of transport systems that sequester the factors, releasing them only when special conditions are encountered (Martinez-Antonio & Collado-Vides, 2008).
TFs are classified in several families based on at least two domains, which allow them to function as regulatory switches (Jacob, 1970). One domain functions as a signal sensor by ligand-binding or protein–protein interaction. In many cases, the ligand is a metabolite or a physicochemical signal that conduits the endogenous or environmental information (Ptashne & Gaan, 2002; Martinez-Antonio et al., 2006). The other domain is the responsive element of the switch that directly interacts with a target DNA sequence or TFBS. In bacteria, the helix–turn–helix domain is the most common (Madan Babu & Teichmann, 2003a; Seshasayee et al., 2006). Also, in bacteria, most of these domains are present in one single protein, except for two-component systems (Ulrich et al., 2005). Classically, in these systems, when the sensor protein – usually localized in the cell periplasm – senses an exogenous condition, it phosphorylates itself and its cytoplasmic partner, which has a transcriptional regulatory activity (Mascher et al., 2006). These two-component systems work as a unit: evidences from Escherichia coli show that 26 of the 29 pairs are encoded in the same operon (Janga et al., 2007a).
In general, negative regulators bind to the promoter, interfering directly with RNA polymerase; in contrast, positive regulators bind to the promoter's upstream region, helping to recruit the polymerase and start transcription (Collado-Vides et al., 1991; Madan Babu & Teichmann, 2003b). TFs usually work as homodimers, tetramers, hexamers and even, in a few cases, as heterodimers (Goulian, 2004). TFs work in concert and a regulatory region can be occupied by several TFs. One of the causes of this crowding of the DNA by TFs in some regulatory regions is the degeneracy of TF–TFBS interaction, i.e. there are different sites that are able to recruit the same TF and different TFs that can recognize similar sites. For example, overlapping regulons like E. coli's SoxS, MarA and Rob arise because of TF–TFBS degeneracy (Martin & Rosner, 2002). The regulatory effect depends on the TF concentration and TF–TFBS affinity: to function, weak sites require high concentrations of TFs; in contrast, strong sites work with a lower amount (Alon, 2007a, b). Also, compared with local TFs that tend to have high-affinity sites, global TFs are less specific, bind to a larger collection of sites and must be expressed at higher levels (Lozada-Chavez et al., 2008; Martínez-Antonio et al., 2008). Furthermore, there are TFs with a dual regulatory role, being activators and repressors at the same time. One simple example are TFs that bind to a single site in the intergenic region between divergently transcribed units, regulating each one of them in a different manner. This is a common theme in sugar catabolism loci where a structural operon is activated, whereas the gene that codes for the TF itself is repressed. An alternative process by which dual regulation works is by the interplay between TF concentration and binding site strength: imagine two TFBSs for the same TF, a weak negative site inside a promoter and a strong positive site next to it. When the TF concentration is low, the strong positive site recruits the TF and transcription is promoted. As the TF concentration increases, the strong site saturates and the weak site begins to be occupied, thus preventing the union of the polymerase to the promoter. The transcriptional regulator factor for inversion stimulation (Fis) has a dual function over some TUs using the previous strategy (Weinstein-Fischer & Altuvia, 2007).
It is not yet possible to predict the regions of DNA binding from protein structure and experimental mapping is necessary. In general, the number of genes encoding TFs increases with the number of total genes. In particular, in bacterial genomes this increment is proportional to the squared number of genes, suggesting that the increase in genome size is followed by a greater regulatory complexity (Cases et al., 2003; van Nimwegen, 2003; Aravind et al., 2005; Molina & van Nimwegen, 2008). Also, genes in small genomes are relatively more clustered in operons compared with genes in larger genomes (Moreno-Hagelsieb, 2006). However, recent evidences support the idea that the average number of TFBS per regulatory region is independent of genome size (Molina & van Nimwegen, 2008). (Box 1).
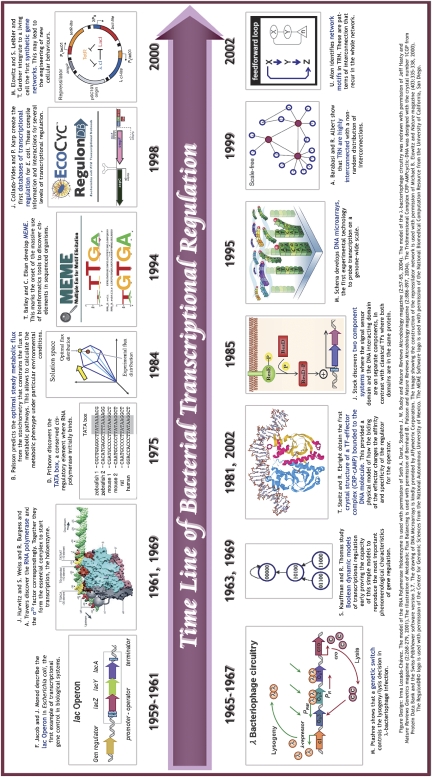
Box 1. Timeline
Transcription is regulated. This was realized currently with two classical examples: the induction of the lac operon (Jacob & Monod, 1959, 1961) and the control of the lytic-lysogenic decision in λ-phage infection (Ptashne, 1965). The circuits controlling these processes are canonical examples that present almost all properties ubiquitous to all gene regulation. It did not take too much time to realize that all cellular functional states, for example cell types, could be codified in a genetic network. This hypothesis gave rise to the first theoretical studies on gene networks that showed that stable genetic patterns indeed arise on very simple models (Kauffman, 1969, 1995; Thomas & D'Ari, 1990). RNA polymerase is essential for transcription (Hurwitz, 1959) and many factors concur to modulate gene expression through the regulation of the binding to DNA of this molecular machine. For example, σ factors and anti-σ factors coordinate the rapid response of many processes in the face of environmental changes and are essential for proper transcription (Burgess et al., 1969; Stevens & Rhoton, 1975). Also, factors can be inherited across multiple cell generations, giving rise to epigenetic phenomena that are not always determined by the DNA sequence (Luger et al., 1997; Bao et al., 2007). Much of the knowledge on transcriptional regulation was discovered with many clever experiments. Nonetheless, direct evidence of metabolite–TF–DNA interaction was not available until the first crystallographic structures were obtained (McKay & Steitz, 1981; Weber & Steitz, 1987; Benoff et al., 2002). The continuous accumulation of experimental facts showed that there are generalities on how cells sense the external environment and couple that change to gene regulation: repressible/inducible systems and two-component systems (Savageau, 1974; Stock et al., 1985). However, the disperse increase of these data did not allow a genome-wide analysis. The solution to this problem began with the appearance of comprehensive structured compendia of transcriptional data with authoritative editing (Wingender, 1988; Huerta et al., 1998). Modularity is present in transcriptional regulation, and one of the first evidences was the discovery of the TATA box motif (Pribnow, 1975), a modular element of transcription initiation. The necessity to automatize motif searching was evident, and many computational sequence-searching algorithms emerged out, among them MEME (Bailey & Elkan, 1994). Genes are highly interrelated and this was clear when the pictures of the first – albeit incomplete – transcription networks (Barabasi & Albert, 1999; Guelzim et al., 2002) and high-throughput experiments appeared, i.e. microarrays and ChIP-chip experiments (Schena et al., 1995; DeRisi et al., 1996; Lockhart et al., 1996; Ren et al., 2000). One of the peculiarities of gene networks is that they have an over-representation of network motifs, a signature of evolutionary and structural constraints (Milo et al., 2002; Shen-Orr et al., 2002). Transcriptional regulation controls the presence/absence of cellular components, allowing, for example, to metabolize available nutrients. Even though these networks are highly intricate, the metabolic fluxes of bacterial colonies can be predicted (Palsson & Lightfoot, 1984; Palsson et al., 1984). Many technologies and knowledge on gene regulation have converged to synthesize the first gene circuits (Elowitz & Leibler, 2000; Gardner et al., 2000). With the aid of new technological applications, transcription in single cells has been detected, showing that promoter activity is stochastic, producing bursts of proteins when messenger is transcribed (Yu et al., 2006). Furthermore, single-molecule detection in individual cells reports that 90% of the time the LacI repressor is bound unspecifically to DNA, wandering along it until it encounters its operator (Elf et al., 2007) (Fig. 1).
Fig. 1.
Timeline of bacterial transcription regulation.
Chromosome organization and structure
Chromosome compactness might represent a physical constraint to transcription initiation (Willenbrock & Ussery, 2004; Marr et al., 2008). Recent studies suggest that the E. coli chromosome is arranged in structural domains with a loop-like conformation, with sizes that range from 10 to 117 kb (Postow et al., 2004; Gitai et al., 2005). The packing of some regions depends on the activity of nucleoid-associated proteins: in bacteria, these are DNA-bending [integration host factor (IHF), HU and Fis] and DNA-bridging proteins [histone-like protein (H-NS)]. The expression of these proteins depends on the growth phase, suggesting a correlation between growth and nucleoid structure (Ali Azam et al., 1999; Luijsterburg et al., 2006; Zimmerman, 2006). In addition, DNA isomerases, DNA chaperones and accessory proteins also regulate DNA access, coiling, bending and packing. Fis recognizes specific TFBSs and in some DNA regions (100–200 bp) clusters of high-affinity Fis sites can be found. However, Fis may also bind nonspecifically to stabilize DNA loops (Skoko et al., 2006). As opposed to Fis-induced bending, H–NS is a condensing agent of the DNA. However, surprisingly, some experiments have shown that it can also have the opposite relaxing effect (Dorman, 2004). It has been suggested that one of the functions of H–NS is to silence horizontally acquired genes, especially those of low GC content (Navarre et al., 2006).
Chromosome size in bacteria ranges from c. 0.5 mbp (intracellular pathogens and endosymbionts) to c. 9 mbp (free-living bacteria) (Cordero & Hogeweg, 2007; Vinuelas et al., 2007). A chromosome contains from hundreds to thousands of genes that are encoded in both leading and lagging DNA strands. There is a preference for essential and highly expressed genes (such as those for ribosomal proteins) to be localized in the leading strand near the origin of replication (Rocha, 2004). The strategic orientation of these genes has been explained as an advantage for efficient transcription, for example to avoid head-on collisions between the transcription and the replication machinery (Brewer et al., 1992; Mirkin et al., 2006). The G+C content differs among genomes, although regulatory regions have a rich A+T content, an observation related to the access of the transcriptional machinery (Dekhtyar et al., 2008).
Epigenetics in transcriptional regulation
Inherited stable changes in cell functioning that cannot be explained as the result of mutations or modifications in the DNA sequence are considered as epigenetic (Bird, 2007). Specific molecular mechanisms are responsible for the transmission of particular acquired characteristics in a nongenetic manner: biochemical modifications in DNA or DNA-binding proteins can act as epigenetic markers. Bacterial DNA can be methylated in several ways, resulting in N4-methyl-cytosine (m4C), N6-methyl-adenine (m6A) and N5-methyl-cytosine (m5C). Among these three chemical markers, m4C has been clearly related to epigenetic transcriptional regulation besides its relation to other cellular processes (Casadesus & Low, 2006). Epigenetic markers are conserved through bacterial generations thanks to the capacity of methyltransferases to recognize preferentially hemimethylated DNA. This covalent modification can alter the interactions of restriction enzymes or regulatory proteins with DNA by a direct steric effect. In E. coli, many genes such as dnaA and trp can be regulated by Dam methyltransferase (Low et al., 2001). A well-studied specific example of epigenetic inheritance by DNA methylation is the switching of the pap operon in the uropathogenic E. coli. The operon is regulated by the interplay of two leucine-responsive protein (Lrp)-binding sites. In the repressed state, Lrp binds the proximal site interfering with transcription and Dam methylates the distal site blocking Lrp binding. The operon is derepressed when PapI dimerizes with Lrp. The PapI–Lrp complex has a higher affinity for the distal site, thus freeing the proximal site from Lrp. Dam methylates the proximal site and transcription begins (Hernday et al., 2002). Any of the two states of the pap operon is passed on to daughter cells using the methylation signal.
It is not always necessary to have molecular markers for epigenetic inheritance. One commonly unnoticed – and misconceived as a trivial – example is the transmission, to the daughter cells, of the cellular components in the mother's cytoplasm in every cell division cycle. The cytoplasm contains specific factors that prime the daughter's transcription in order to recover the transcription state of the mother cell. For example, it is known that low levels of the gratuitous inducer isopropyl β-d-1-thiogalactopyranoside (IPTG) do not derepress the lac operon. However, once high IPTG concentrations have induced the transcription of the operon, it is possible to lower the IPTG concentration to noninducing levels and maintain induced a colony previously induced with high IPTG concentrations. This is because daughters of preinduced mothers have a high level of β-galactoside permease in their membranes. This allows them to import, even at low concentrations, IPTG and maintain the lac operon derepressed (Casadesus & D'Ari, 2002).
Transcriptional regulatory networks
The direct influence of TFs over the transcription activity of different target genes (TG) is customarily drawn in a network of causal relationships known as a transcriptional regulatory network (TRN) (McAdams & Arkin, 1998; Thieffry & Thomas, 1998; Lee et al., 2002a). The network representation unveils the global organization of transcriptional regulation such as its modular and hierarchical structure (Thieffry & Romero, 1999; Ihmels et al., 2002; Segal et al., 2003; Wolf & Arkin, 2003; Barabasi & Oltvai, 2004; Resendis-Antonio et al., 2005; Yu & Gerstein, 2006; Martínez-Antonio et al., 2008) or the fact that on average every TG is controlled by two TFs (Albert, 2005; Aldana et al., 2007). One natural unit in TRNs is the regulon: a set of TGs coregulated by the same set of TFs; this concept was originally defined as the group of genes subject to the exclusive regulation of one TF (Maas, 1964). Regulons are divided into simple or complex if regulated by a single or by multiple TFs, correspondingly. The majority of regulons in bacteria correspond to the last category (Gutierrez-Rios et al., 2003).
The E. coli TRN seems to be dominated by probably <10 global TFs (Martinez-Antonio & Collado-Vides, 2003). Local TFs usually act in concert with global TFs and are also regulated by them, forming a feedforward loop motif (Alon, 2007a, b). In E. coli, most of the local TFs tend to be encoded in close chromosomal proximity with one of their regulated genes (Janga et al., 2007a). In addition to simple horizontal cotransfer, a biophysical explanation for local TFs and TGs colocalization is that, because the number of local TF molecules is low, they must be close to their regulated target in order to quickly reach their binding site by jumping and sliding along the DNA molecule (Kolesov et al., 2007; Wunderlich & Mirny, 2008). As a rule, global TFs do not regulate each other directly, a phenomenon known as ‘hubs repulsion’ or disassortativity (Song et al., 2006; Takemoto & Oosawa, 2007). As a general observation, the promiscuity of a TF for binding sites diminishes as its local character augments (Lozada-Chavez et al., 2008), and global and local regulators tend to coordinate jointly a general and a particular condition (Balaji et al., 2007; Janga et al., 2007b). Global TFs and some recently duplicated TF pairs can coregulate some TUs, forming a network motif named bifan (Shen-Orr et al., 2002). In fact, this motif is a particular class of the complex regulons coordinated by only two TFs. Escherichia coli, for instance, has regulons with as many as four to six TFs mutually affecting expression of their TGs. The transcriptional response concentrating regulatory changes – triggered by environmental signals – is partitioned by global TFs as well as by sigma promoter subsets. For example, this is evident when considering E. coli's σ interactions, giving a very clear separation of gene subsets participating coordinately in heat shock, σ32 (Nonaka et al., 2006), stress response σE (Johansen et al., 2006), and stationary-phase σS (Typas et al., 2007), etc.
Local regulators and nucleoid-associated factors (many of them global TFs) affect the transcription rate of TGs in drastically distinct ways. Evidence shows that nucleoid-associated TFs and DNA-supercoiling induce continuous changes in the transcription rate, whereas local TFs induce discrete changes (i.e. On/Off transcription states). These two aspects have been compared with the analog and digital components of electronic devices (Blot et al., 2006; Marr et al., 2008).
Plasticity and evolution of TRN
Thanks to the availability of hundreds of sequenced bacterial genomes, one can consider the following evolutionary question: in bacteria, to what extent are TRNs conserved? Recent studies show that TFs evolve much faster than their TGs, suggesting that TRNs in bacteria are highly flexible and dynamic (Lozada-Chavez et al., 2006; Madan Babu et al., 2006). Several reports that analyze different components of TRNs strongly support their plasticity. For example, multiple evidences show that nonorthologous TFs control equivalent pathways, for example the nonorthologous NagC, NagR and NagQ regulate the utilization of N-acetylglucosamine and chitin in various groups of proteobacteria (Meibom et al., 2004; Yang et al., 2006). In contrast and to a lesser extent, orthologous regulators may control distinct pathways in different species, for example the orthologous Fur (Alpha-, Beta-, Gammaproteobacteria, bacilli and cyanobacteria) and Mur (alphaproteobacterial rhizobial species Rhizobium leguminosarum and Sinorhizobium meliloti) regulate iron homeostasis and manganese uptake, respectively (Rodionov et al., 2006). Also, even global TFs do not necessarily regulate similar metabolic responses in different organisms (Friedberg et al., 2001; Suh et al., 2002; Derouaux et al., 2004; Moreno-Campuzano et al., 2006). Likewise, as phylogenetic distances decrease, TFBS conservation increases (Makarova et al., 2001; Mazon et al., 2004). However, there are some exceptions to this rule: TFBSs of BirA (regulation of biotin biosynthesis) are highly conserved in Bacteria and Archaea (Rodionov et al., 2002), while TFBSs of ArgR/AhrC (control of arginine regulon) and NrdR (ribonucleotide reductase regulon) are strongly conserved in Bacteria (Makarova et al., 2001; Rodionov & Gelfand, 2005). This suggests that biotin, arginine and ribonucleotide reductase regulatory sites may be ancient. In addition, bacterial species that live in ever changing environments have a tendency to increase the number of encoded stress-responsive TFs and σ ECF; this may be a simple effect of a larger number of regulators encoded in larger genomes (Helmann, 2002). Finally, studies in E. coli show that some parts of its TRN are more conserved if they are involved in basic processes (Cosentino Lagomarsino et al., 2007; Salgado et al., 2007).
Several evolutionary processes, such as duplication and horizontal gene transfer (HGT), must be studied to understand TRN flexibility. For example, loss and duplication of TFs and TFBS may result in regulon expansion, shrinkage, fusions, fissions and even creation and destruction. It is possible to see the contribution of gene duplication at all levels of TRNs (Teichmann & Babu, 2004), although it seems to be more frequent at the bottom layers (Cosentino Lagomarsino et al., 2007; Lozada-Chavez et al., 2008). There are coordinated TF–TG duplications in bacterial TRN. These events account for 38% of the regulatory interactions in E. coli's TRN and 45% in S. cerevisiae's TRN (Teichmann & Babu, 2004; Zhang et al., 2005). The percentages were obtained considering only paralogy within each species; this can mask a convergent evolution within paralogs. For E. coli, the previous percentage contrast with the 8% obtained when HGT events are eliminated from the regulatory interactions arose within the E. coli lineage (Price et al., 2008).
Although most TFs have paralogs, they seem to have arisen by HGT rather than by gene duplication within the E. coli lineage (Price et al., 2008). Moreover, it seems that, in horizontal transfer events, local regulators flow more easily within near phylogenetic distances than global regulators (Lercher & Pal, 2008; Price et al., 2008). Therefore, global regulators are gained and lost more slowly and are even prone to undergoing a slower sequence evolution than other regulators within a bacterial lineage (Rajewsky et al., 2002; Price et al., 2008). This fact does not ensure the maintenance of their global functional role (Friedberg et al., 2001) because the property of global regulation depends on several evolutionary forces and on TF's particular molecular properties (Lozada-Chavez et al., 2008). In addition, genes recently transferred have low expression levels; probably this is a sign of slow but steady integration of transferred genes into the existing regulatory circuits (Taoka et al., 2004; Price et al., 2008). In E. coli, the evolutionary rate of TFBSs of horizontal transferred TGs is fast but gradually decelerates with the age of horizontal transfer (Lercher & Pal, 2008). These facts show that TFs and their TFBSs can evolve largely independently, allowing genes to join or leave regulons and allowing regulatory regions to increase their complexity by augmenting the quantity and type of cis-regulatory interactions. HGT, complex gene duplication events and an accelerated sequence divergence may mask the discovery of orthologs, making comparative studies of TRN a particularly difficult task; see Box 2.
Box 2. Inference of TRNs
Before any biological question about TRNs can be asked, the technical problem of obtaining a reliable network must be solved. There are essentially three methodologically different ways of doing this: (i) by the compilation of different facts reported in research articles whose main interest could have not been to obtain a network, (ii) by ChIP-chip or ChIP-Seq and (iii) by computational methods with DNA sequences, microarrays or scientific articles as input data. We provide a short description of each one of these methods.
Databases of compiled isolated experiments
Interactions derived from the literature are the standard to validate any computational or high-throughput experimental inference (Jacques et al., 2005; Munch et al., 2005; Baumbach et al., 2006; Kazakov et al., 2007; Gama-Castro et al., 2008; Sierro et al., 2008). However, not all the annotated regulatory interactions are equally well supported by experimental facts, and subtleties arise. The experience in RegulonDB has dictated that evidences of TF–TG interaction must be divided into at least two categories: strong and weak. Evidence is strong if, for example, it comes from footprinting or EMSA plus change in expression or binding site mutation plus change in expression. An example of weak evidence is: expression change detected in a microarray plus existence of a binding site – for a certain TF – detected ‘by eye’ by the researcher. Because of its nature, ‘weak’ interactions may become ‘strong’ interactions or may disappear depending on new evidence.
ChIP-chip and ChIP-Seq
These high-throughput experimental techniques are designed to locate, in vivo and at a genome-wide scale, regions in the DNA where specific proteins bind, in particular TFs. Both techniques start with chromatin immunoprecipitation: cells are treated with a reagent that crosslinks proteins and DNA. Then, cells are lysed and DNA is digested. By immunoprecipitation, all DNA fragments bound to a TF are recovered. The fragments are denatured and amplified. At this point, if ChIP-Seq technology is used, the amplified fragments are sequenced in an ultrahigh-throughput sequencing machine. Detection of binding sites is performed mapping back the sequenced fragments to the genome (Fields, 2007). If ChIP-chip technology is used, fragments are labeled with fluorescent tags to subsequently be hybridized in a special microarray. The microarray may contain only intergenic regions or may be a high-density tiling array (Grainger et al., 2005a; Wade et al., 2005; Cho et al., 2008). All regions that contain a binding site for the TF will have a signal above background in the microarray. The ChIP-chip technique has been used to infer the component of the TRN of S. cerevisiae that is under the control of 106 TFs (Lee et al., 2002b; Grainger et al., 2005b, 2006; MacIsaac et al., 2006).
Computational approaches
There is a plethora of computational approaches (Albert, 2007; Margolin & Califano, 2007). Here, we enlist some of the core ideas/techniques behind many inference algorithms and give some of the most representative examples in the literature. It is worth mentioning that all these approaches have a high false-positive rate; they are sometimes unable to discern between direct and indirect regulations and some of them do not detect regulatory feedback loops. However, their informative guidance must not be underestimated. For example, consider the detection of regulatory candidates for some arbitrary gene in E. coli. Without any type of previous information, the candidates would be c. 4500 genes, i.e. every gene in the E. coli's genome. Using for example Mutual information, the set would be reduced to a dozen of putative regulators – a tractable set size.
Bayesian networks
A Bayesian approach solves the following problem: given a set of genes and their expression patterns, find the network that explains the observed patterns with the maximum of probabilities (Pearl, 1988; Heckerman, 1999; Neapolitan, 2003). To discriminate among the different possible networks, a score function – known as the Bayesian–Dirichlet metric – is evaluated. This inference method has been applied to propose a TRN in S. cerevisiae (Segal et al., 2003).
Mutual information
This is the most general way to detect dependence between two variables. The method is used to estimate, from a group of genes and their expression patterns, whether there exists dependence between all possible pairs. An interaction between a pair is proposed if their mutual information is significantly different from that of the same pair but with the expression patterns randomized (Steuer et al., 2002). This is the core idea behind the inference of the TRN of E. coli (Faith et al., 2007).
Discovery of TF-binding sites
From a collection of TFBSs for a specific TF, it is possible to obtain an estimate of the binding energy between the TF and any arbitrary site. To construct a network, the estimated binding energy between the TF and every possible site in intergenic regions is obtained. The sites with the highest binding energies are proposed as targets, thus inferring an interaction between the TF and the gene in the surroundings of the binding site (McCue et al., 2002; Aerts et al., 2003; Tompa et al., 2005; Chang et al., 2006; Rodionov, 2007; van Nimwegen, 2007).
Orthology-based algorithms
From a model organism, where some regulatory interactions are known, the evolutionary and functional relationships between the components of transcriptional regulation can be studied using phylogenetic trees or bidirectional best blast hits, BBHs. With these tools, a search for orthologous counterparts of TF–TG pairs in the model organism is performed in closely related species. When orthologous pairs are found, new regulatory interactions are proposed (Yu et al., 2004). This has been used to show that networks of transcriptional regulation are highly evolvable in Bacteria (Lozada-Chavez et al., 2006; Madan Babu et al., 2006; Price et al., 2007). Some experimental works have supported the orthology predictions and their regulatory extrapolation based on this approach. This is the case of the Lrp regulon within the E. coli lineage (Lintner et al., 2008) and of the σB regulon in Gram-positive bacteria (van Schaik et al., 2007).
Natural language processing
First, a lexicon (e.g. a set of gene names) related to transcriptional regulation is compiled. These nouns are concatenated with verbs – to regulate, to inhibit, to promote, etc. – to, depending on certain grammatical rules, discover regulatory interactions in a collection of related scientific articles. Using this method, from 200 000 E. coli's article abstracts, it was possible to recover 395 regulatory interactions with 85% accuracy (Saric et al., 2006).
The regulatory network of E. coli can be perturbed globally, rewiring it to a great extent; this might be a consequence of the inherent plasticity of TRNs. For example, Isalan et al. (2008) reconnect some global and local regulators and σ factors also by transforming wild-type strains with constructs of almost all possible combinations of these genes with their different promoters. They rewire the network in 600 different ways, every time adding up to five new interactions. Remarkably, in a wild-type genome background, bacterial colonies are viable in 95% of the cases. Another example of network perturbation in a wild-type background shows that mutations in the housekeeping σ factor induce global rewiring (Alper & Stephanopoulos, 2007). The authors show how this rewiring more efficiently solves several problems of metabolic optimization thanks to the interplay of many changes in gene expression that make possible the exploration of complex phenotypes. These results must be confronted with metabolic networks where enzymes have great specificity for their substrates and many catabolic and anabolic pathways are highly conserved. In this respect, metabolic networks appear to be stiff; in contrast, TRN seem to be loose. TFs bind to a broad spectrum of binding sites with different affinity and change targets widely among species. In the light of the previous facts, the rapid adaptation of bacterial organisms to almost every niche on earth is greatly explained thanks to the plasticity of transcriptional regulation.
Stochasticity in transcriptional regulation
In transcription, all the time TFs are binding to or unbinding from different sequences in the DNA. The greater the affinity, the greater the time they remain bound. If the sequence is regulatory, there is a likelihood that the rest of the transcription machinery assemblies begin transcription before the TF tears off from DNA by thermal fluctuations. In this picture, there is no natural threshold in affinity above which TFs undoubtedly induce transcription. In general, there are a variety of binding sites and for every one of them a TF will have a different affinity, inducing, with some probability, transcription. When promoters are strong and TFs abound, transcription is certain and has a well-defined rate (Elowitz et al., 2002). However, when promoter strength is weak or TF numbers oscillate around the dozens, stochastic fluctuations in the mean TF numbers are very large and transcription becomes ‘noisy’.
In transcription, variability in the number of messages arises from two sources of noise: one intrinsic and the other extrinsic. In a hypothetical cell with two identical genes, intrinsic noise would cause differences in their number of transcripts. This effect is analogous to the tossing of two identical coins that do not generate the same sequence of heads and tails. Extrinsic noise originates from the cell-to-cell variation of cellular components, for example the exact number of polymerase molecules. Elowitz et al. (2002) measured the individual contribution of the two components of noise by the ingenious construct of two fluorescent proteins of different colors in the same plasmid that were subjected – every one of them separately – to the control of a promoter with the same sequence. Transformed with this construct, individuals of ‘noisy’ strains appear under the microscope with any of the two possible colors (intrinsic noise high). In quiet strains, every individual appears with the same color obtained when combining equal quantities of the two fluorescent proteins (intrinsic noise low). Extrinsic noise is obtained when comparing the fluorescence intensity among cells of the same strain.
One fact with profound consequences in the cell fate decision is the metastable gene expression patterns originating from the random fluctuations of the expression of individual genes. The metastability is attained thanks to TRNs that amplify random fluctuations of gene expression and then sustain stable patterns over biological relevant lapses of time. This causes growing isogenetic colonies of microorganisms to differentiate in subcolonies of specialized ‘cell types’spontaneously (Maamar et al., 2007; Suel et al., 2007; Chai et al., 2008). Any single cell from an original isogenetic colony can give rise, in turn, to descendants that differentiate in subcolonies that are in the same proportion as the ones in the original colony.
Transcriptomics
At present, there are basically two options to probe the transcriptional state of the cell: microarrays and ultra-high-throughput sequencing. In the first technology, different single-stranded DNA probes are designed and arrayed to monitor the mRNA expression of different genes. These transcriptional products, isolated from a culture sample, are tagged with fluorescent proteins and then hybridized in the microarray against their complementary sequences. The intensity of the fluorescence, in the different locations of the array, gives an estimate of the abundance of the different probed transcripts. Microarray technology has been refined since its first appearance in the mid 1990s when they detected exclusively annotated ORFs (Schena et al., 1995). Today, state-of-the-art microarray technology is represented by high-density whole-genome tiling arrays. In this implementation, the arrayed set of probes is richer, containing, for example, DNA probes for both intragenic and intergeneic regions. This improvement allows for the identification of complex transcript structures – such as genes in operons – as well as novel short transcripts – such as small RNAs – that would be missed by previous low-density arrays (Reppas et al., 2006). The raw data generated from microarrays must be transformed in two steps: correction for background noise and normalization. The first transformation attempts to eliminate the contribution from unspecific hybridization; the second transformation intends to make gene intensities from different experiments comparable (Quackenbush, 2002). The widespread use of this technology has led to the appearance of useful databases with collections of hundreds of arrays of different bacterial organisms under diverse experimental conditions (Demeter et al., 2007; Faith et al., 2008; Kanehisa et al., 2008).
There are particular problems that are inherent to microarray technology. For example, prior selection of probes in the arrays biases the possible set of transcripts that can be detected; unspecific hybridization cannot be completely eliminated; the differential efficiency of probes makes it impossible to compare the expression of different genes in the same sample, etc. It appears that the solution to these problems is to use the sheer brute force of massive sequencing with the new ultra-high-throughput sequencing technologies (Bennett et al., 2005; Margulies et al., 2005). The idea is simple: sequence all the transcripts that the cell expresses under a particular condition and then map these sequences back to their corresponding regions on the genome to detect presence or absence (Nagalakshmi et al., 2008). Note that the detection of transcripts is not conditioned on a possibly biased set of probes nor on the resolution of the array. This translates into the possible discovery of new gene products. Also, the effect of unspecific hybridization is not present in the sequencing, and comparison between gene transcript levels is possible because the number of sequenced transcripts is directly counted. At least one study has compared microarray and sequencing technology, showing that data in the latter are highly replicable and that the sequencing technology can detect differentially expressed genes between two samples at a higher positive discovery rate (Marioni et al., 2008).
The processing of transcription data and the rationale behind that same processing is as important as the technology to probe transcription. The traditional data workflow screens for differentially expressed genes; this proceeding has been described, pejoratively, as fishing expeditions (Gibson, 2003). This criticism indirectly points to the fact that the community lacks methods to synthesize gene expression data and methods to analyze this synthesis at higher levels of description, for example gene expression data organized coherently in TRN or genes of related function sorted out in functional classes. One way to amend this situation is the use of a clustering method known as Self-Organizing Maps. This clustering reorganizes transcription data in such a way that genes with similar expression levels are contiguously located in a squared lattice, generating an image of the state of the transcriptome. Surprisingly, with this reordering, it is possible to sort out different cellular functional states just by seeing the image, a gestalt analysis (Guo et al., 2006). Another method of higher level analysis is to take advantage of the decades of molecular knowledge and organize transcriptional data into sets of genes that together perform a cellular process (Subramanian et al., 2005). This gene set analysis has a higher statistical power to discriminate changes at the gene set level that would be unnoticed at the single-gene level. (Box 3).
Box 3. Dynamical models of TRNS
Dynamical models of TRN present different degrees of granularity that are appropriate to particular aspects of and questions in cell regulation. In deciding the proper model and its coarseness – perhaps the most important step in modeling – all prior available knowledge is important: number of genes, available physicochemical parameters, TF affinities for TFBSs, kinetic parameters, etc. In general, there are three classes of models with particular levels of granularity: Boolean, stochastic and continuous models (Smolen et al., 2000; Bower & Bolouri, 2001; Christensen et al., 2007). It is also possible to combine any of them to produce hybrid models.
Boolean models
The sigmoidal induction/repression of gene expression by different factors is well approximated by step functions with two states: On/Off (Thomas & D'Ari, 1990). Using this simplification, we can define a TRN model: genes with two states (inhibited or induced), interacting through logical rules (ORs, ANDs or Boolean tables in general) in discrete time steps. Model networks with these modest characteristics are Boolean and they present – remarkably – much of the higher order phenomena sustained by gene networks (Kauffman, 1969; Thomas & D'Ari, 1990). To give but one example: only a few gene expression patterns in Boolean models are stable and have a direct correspondence with gene expression patterns of real cell types (Mendoza & Alvarez-Buylla, 1998; Albert & Othmer, 2003; Huang & Ingber, 2007). Also, recent investigations using the Boolean abstraction of real networks provide a first explanation of how – paradoxically – robustness and adaptability coexist in living organisms (Aldana et al., 2007; Balleza et al., 2008; Nykter et al., 2008).
Stochastic models
Stochasticity inevitably emerges when molecular components are present at low cellular concentrations (McAdams & Arkin, 1997; Kierzek et al., 2001). This physical phenomenon generates noise in synthetic and natural circuits (Paulsson, 2004; Mettetal et al., 2006), and its consequences over the phenotype are starting to be explored (Suel et al., 2006). For example, noise constitutes the driving force behind differentiation in isogenetic colonies (Colman-Lerner et al., 2005). Biological and theoretical studies have aided to delineate the regulatory mechanism by which the cell handles noise efficiently and effectively to carry out its biological functions (Gardner & Collins, 2000; Orrell & Bolouri, 2004; Raser & O'Shea, 2005). From a theoretical point of view, stochastic models are the most challenging but also the most realistic ones: there is a precise counting of how, through individual chemical reactions, the populations of every chemical species change. The milestone to simulate stochastic processes is the Gillespie algorithm (Gillespie, 1992). Because of their analytical and computational complexity, the present models do not surpass a handful of chemical species. Two immediate problems must be solved to model systems with several dozens of genetic components: the systematic determination of kinetic constants and the efficient computation of thousands of chemical stochastic equations (Kuwahara et al., 2006; Sanchez & Kondev, 2008).
Continuous models
Contrary to stochastic models, continuous modeling assumes that the number of components in TRNs is sufficiently high to assume that concentrations are continuous variables. This framework allows taking into account realistic effects such as complex TF interactions, spatial diffusion of molecules and the gradual variation of mRNA expression, to mention just a few (Bower & Bolouri, 2001; de Jong, 2002). The continuous description has been useful for the design of genetic circuits in synthetic biology (Atkinson et al., 2003; Kaern et al., 2003). Remarkably, this sort of approach allows one to understand the biological function of network motifs. For instance, dynamical analyses of feed forward loops show how this circuit controls the slow activation and rapid deactivation of the regulated gene. Also, the analysis of feedback loops evidence their role as the units behind memory (Alon, 2007a, b). As in stochastic models, the continuous approach is useful to quantitatively analyze the dynamics of TRN only when the topology, the regulatory type of the interactions and the kinetic constants are known (Shea & Ackers, 1985; Kobiler et al., 2005). When kinetic constants are not available, plausible values can be used to obtain the possible dynamical responses of TRNs.
Hybrid models
Experimental evidence shows that different levels of cell functioning are carried out at different time scales and at different concentrations of their components – seconds and thousands of molecules in metabolism, minutes and hundreds of molecules in transcription. How these levels can be combined to be consistent between them in a single model constitutes an active field of research (Puchalka & Kierzek, 2004; English et al., 2006; Samoilov & Arkin, 2006; Covert et al., 2008). One solution is hybrid models; these combine any of the above approaches to integrating different cues of cell functioning (Bower & Bolouri, 2001). For example, there are hybrid models that take into account the continuous character of the concentration of some transcripts and their abrupt discrete change in transcription during regulation (Glass & Kauffman, 1973; Thomas & D'Ari, 1990). Another hybrid model is one in which a noise function is added to the continuous concentration of transcripts to introduce stochastic fluctuations (Ozbudak et al., 2002). Even though these models are more complex than purely discrete ones, they provide a more approximate picture to transcriptional regulation, making it easier, for example, to relate and compare the models with real transcriptome data.
Synthetic transcriptional regulatory circuits
The previous sections show the detailed knowledge we have accumulated on transcriptional regulation by TFs. The synthesis of TRNs attempts to go from this understanding to a rational transcriptional network design. It aspires to integrate new complex functions into cell behavior; not just the addition of stationary properties such as the constitutive expression of exogenous proteins but the addition of the dynamically controlled expression of complete gene programs. There are several first examples in this direction that show the feasibility of program integration into cell behavior: rational design of memory circuits (Ajo-Franklin et al., 2007), insertion of complete regulated metabolic pathways (Pfleger et al., 2006), toggle switches (Gardner et al., 2000), oscillators (Elowitz & Leibler, 2000) and the creation of new ways of cell–cell communication (Bulter et al., 2004). In all these cases, small gene circuits compute their output based on the external/internal input signal sensed. Promoters controlling the expression of the genes in the circuit are the essential piece to accomplish the required computations, for it is in this element where signals – transduced by TFs – converge and are integrated. The particular importance of promoters has naturally led to an interest in their characterization and synthesis. For example, with respect to their characterization, it has been shown that, more often than not, the activity of different promoters controlled by two regulators is not a simple OR/AND function (Cox et al., 2007; Kaplan et al., 2008). With regard to their synthesis, now we have available complete characterized libraries of synthetic promoters with different strengths; this last fact was verified indirectly by measuring the specific β-galactosidase activity. Remarkably, more than six orders of magnitude in β-galactosidase activity can be covered using different promoters (Mijakovic et al., 2005). It is also possible to create libraries of regulated promoters by combinatorial synthesis (Cox et al., 2007). This consists of the combinatorial ligation of previously created promoter regions, i.e. sequences that correspond to the distal region (upstream the −35 box), to the core region (between the −35 and −10 boxes) and to the proximal region (downstream the −10 box). These regions contain one operator site for any of the following regulators: LacI, AraC, LuxR or TetR. Using this strategy, thousands of promoters with different regulated strengths can be generated.
Supposing complete characterized libraries of different promoters exist, the main challenge in synthetic circuits still remains: to integrate these small networks into the cell environment without killing the cell, for example without overproducing toxic intermediates or causing metabolic bottlenecks that would inhibit growth. The problem is to find the exact promoter strengths with the correct regulatory region to balance and coordinate the expression of multiple genes. One promising solution is to generate a library of networks and then select the best-performing one under a given criterion. This is the same strategy followed in the directed evolution of proteins, where a library of mutant protein sequences is created and then screened for the best variation of the protein. The difference in the library of networks lies in the fact that the mutations are in the noncoding regions that regulate transcription and translation. One example of this approach is the combinatorial synthesis of intergenic regions in operons to tune the translation of polycistronic transcripts (Pfleger et al., 2006). The approach, without a specific design, generates transcripts with slight variations in intergenic regions that change RNAase cleavage sites, ribosomal binding sites sequestering sequences and mRNA secondary structures. With this technique, it was possible to introduce in E. coli a heterologous mevalonate biosynthetic pathway by tuning the expression of three genes in an operon. In one last example of combinatorial synthesis, a collection of 125 different networks was produced from these units: five different promoters regulated by three different TFs (LacI, TetR and λ cI) (Guet et al., 2002). Among the networks, it is possible to find positive and negative feedback loops, oscillators and toggle switches. It must be stressed that all these different network functions can be encoded with the same set of genes, the difference residing only in the interaction graph of the constituent genes.
Concluding remarks: the need for integrative schemes
Even though recent progress to unravel the underlying mechanisms of transcriptional regulation has been spectacular, the community lacks an integrative framework to direct new advances. In this respect, systems biology in bacteria has the challenge to show its promised capabilities of new levels of integration and understanding combining modeling and experiments of the whole network and cell behavior. To achieve this, there are two complementary procedures: bottom-up and top-down schemes (Beer & Tavazoie, 2004; Bonneau et al., 2007). The former traces its origins to the systems sciences, whose essence is to explore the collective phenomena emerging when integrating its building parts (Bruggeman & Westerhoff, 2007). Bottom-up schemes constitute the base to develop mechanistic models that are useful to discern the transcriptional organization by which the cell faces a genetic or an environmental perturbation at a genome scale (Segre et al., 2002; Covert et al., 2004; Resendis-Antonio et al., 2007). On the other hand, top-down procedures require deductive methods, whose main interest is to identify causal interactions between the individual genes measured by high-throughput technologies (Wagner, 2001; de la Fuente et al., 2002). Successful integration of top-down and bottom-up schemes is not a trivial activity; it requires permanent comparison between types of modeling and its experimental verification to reconstruct a coherent explanation of cell activity. The navigation towards progress here depends on how simplified models can capture the essentials to predict, and the fact that biological systems can be engineered in synthetic approaches, even if they are also extremely interconnected.
This review has focused on regulation by TFs. However, there are other layers of cellular regulation that ultimately influence regulation by TFs. This situation creates feedback loops that transmit information from almost any regulatory layer to any other one in order to maintain cellular homeostasis. This is in clear contrast with the isolated picture of TRN, where a cellular hierarchical decision-making structure is emphasized. Thus, a major conceptual challenge is to change our way of thinking about causality in a complex system with an important connectivity and an important amount of circularity, i.e. feedback loops, in the ‘decision’ network of gene regulation at the whole-cell level.
Acknowledgments
We thank S. Gama-Castro for useful comments. We also thank anonymous referee no. 2 for detailed observations. Y.I.B.-M. acknowledges PDCB-UNAM and CONACyT-México for a PhD scholarship; E.B. also thanks CCG-UNAM and CIC-UNAM for a postdoctoral scholarship. We acknowledge support by NIH grant no. R01 GM071962-05 and PAPPIT IN214905.
Statement
Reuse of this article is permitted in accordance with the Creative Commons Deed, Attribution 2.5, which does not permit commercial exploitation.
Authors' contribution
L.N.L.-B., A.M.-A., O.R.-A. and I.L.-C. contributed equally to this article.
References
- Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R. Scale-free networks in cell biology. J Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- Albert R. Network inference, analysis, and modeling in systems biology. Plant Cell. 2007;19:3327–3338. doi: 10.1105/tpc.107.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldana M, Balleza E, Kauffman S, Resendiz O. Robustness and evolvability in genetic regulatory networks. J Theor Biol. 2007;245:433–448. doi: 10.1016/j.jtbi.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. London: Chapman & Hall/CRC; 2007a. [Google Scholar]
- Alon U. Network motifs: theory and experimental apporaches. Nat Rev Genet. 2007b;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- Balaji S, Babu MM, Aravind L. Interplay between network structures, regulatory modes and sensing mechanisms of transcription factors in the transcriptional regulatory network of E. coli. J Mol Biol. 2007;372:1108–1122. doi: 10.1016/j.jmb.2007.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Alvarez-Buylla ER, Chaos A, Kauffman S, Shmulevich I, Aldana M. Critical dynamics in genetic regulatory networks: examples from four kingdoms. PLoS ONE. 2008;3:e2456. doi: 10.1371/journal.pone.0002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Chen H, Liu Y, Yan J, Droge P, Davey CA. A divalent metal-mediated switch controlling protein-induced DNA bending. J Mol Biol. 2007;367:731–740. doi: 10.1016/j.jmb.2006.09.082. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Baumbach J, Brinkrolf K, Czaja LF, Rahmann S, Tauch A. CoryneRegNet: an ontology-based data warehouse of corynebacterial transcription factors and regulatory networks. BMC Genomics. 2006;7:24. doi: 10.1186/1471-2164-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH. Structural basis of transcription activation: the CAP-alpha CTD–DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Facciotti MT, Reiss DJ, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- Bower JM, Bolouri H. Computational Modeling of Genetic and Biochemical Networks. Cambridge, MA: MIT Press; 2001. [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol. 2007;15:45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Bulter T, Lee SG, Wong WW, Fung E, Connor MR, Liao JC. Design of artificial cell–cell communication using gene and metabolic networks. P Natl Acad Sci USA. 2004;101:2299–2304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Casadesus J, D'Ari R. Memory in bacteria and phage. Bioessays. 2002;24:512–518. doi: 10.1002/bies.10102. [DOI] [PubMed] [Google Scholar]
- Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol R. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I, de Lorenzo V, Ouzounis CA. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003;11:248–253. doi: 10.1016/s0966-842x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD. A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C, Thakar J, Albert R. Systems-level insights into cellular regulation: inferring, analysing, and modelling intracellular networks. IET Syst Biol. 2007;1:61–77. doi: 10.1049/iet-syb:20060071. [DOI] [PubMed] [Google Scholar]
- Collado-Vides J, Magasanik B, Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. Large changes in regulome size herald the main prokaryotic lineages. Trends Genet. 2007;23:488–493. doi: 10.1016/j.tig.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Cosentino Lagomarsino M, Jona P, Bassetti B, Isambert H. Hierarchy and feedback in the evolution of the Escherichia coli transcription network. P Natl Acad Sci USA. 2007;104:5516–5520. doi: 10.1073/pnas.0609023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- Covert MW, Xiao N, Chen TJ, Karr JR. Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics. 2008;24:2044–2050. doi: 10.1093/bioinformatics/btn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RS, III, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- Dekhtyar M, Morin A, Sakanyan V. Triad pattern algorithm for predicting strong promoter candidates in bacterial genomes. BMC Bioinformatics. 2008;9:233. doi: 10.1186/1471-2105-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A, Brazhnik P, Mendes P. Linking the genes: inferring quantitative gene networks from microarray data. Trends Genet. 2002;18:395–398. doi: 10.1016/s0168-9525(02)02692-6. [DOI] [PubMed] [Google Scholar]
- Demeter J, Beauheim C, Gollub J, et al. The Stanford microarray database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Derouaux A, Dehareng D, Lecocq E, et al. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem Biophys Res Co. 2004;325:983–990. doi: 10.1016/j.bbrc.2004.10.143. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. H–NS: a universal regulator for a dynamic genome. Nat Rev. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Ever-fluctuating single enzyme molecules: Michaelis–Menten equation revisited. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, Schneider SJ, Gardner TS. Many microbe microarrays database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 2008;36:D866–D870. doi: 10.1093/nar/gkm815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. Molecular biology. Site-seeing by sequencing. Science. 2007;316:1441–1442. doi: 10.1126/science.1144479. [DOI] [PubMed] [Google Scholar]
- Friedberg D, Midkiff M, Calvo JM. Global versus local regulatory roles for Lrp-related proteins: Haemophilus influenzae as a case study. J Bacteriol. 2001;183:4004–4011. doi: 10.1128/JB.183.13.4004-4011.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Collins JJ. Neutralizing noise in gene networks. Nature. 2000;405:520–521. doi: 10.1038/35014708. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gibson G. Microarray analysis: genome-scale hypothesis scanning. PLoS Biol. 2003;1:E15. doi: 10.1371/journal.pbio.0000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT. Markov Processes: An Introduction for Physical Scientists. Boston, MA: Academic Press; 1992. [Google Scholar]
- Gitai Z, Thanbichler M, Shapiro L. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 2005;13:221–228. doi: 10.1016/j.tim.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Glass L, Kauffman SA. The logical analysis of continuous, non-linear biochemical control networks. J Theor Biol. 1973;39:103–129. doi: 10.1016/0022-5193(73)90208-7. [DOI] [PubMed] [Google Scholar]
- Goulian M. Robust control in bacterial regulatory circuits. Curr Opin Microbiol. 2004;7:198–202. doi: 10.1016/j.mib.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. P Natl Acad Sci USA. 2005a;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. P Natl Acad Sci USA. 2005b;102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJW. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucliec Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelzim N, Bottani S, Bourgine P, Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- Guo Y, Eichler GS, Feng Y, Ingber DE, Huang S. Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. J Biomed Biotechnol. 2006;2006:69141. doi: 10.1155/JBB/2006/69141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Rios RM, Rosenblueth DA, Loza JA, Huerta AM, Glasner JD, Blattner FR, Collado-Vides J. Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckerman D. A tutorial on learning with Bayesian networks. In: Jordan MI, editor. Learning in Graphical Models. Cambridge, MA: MIT Press; 1999. pp. 301–354. [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Hernday A, Krabbe M, Braaten B, Low D. Self-perpetuating epigenetic pili switches in bacteria. P Natl Acad Sci USA. 2002;99(suppl 4):16470–16476. doi: 10.1073/pnas.182427199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ingber DE. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis. 2007;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- Huerta AM, Salgado H, Thieffry D, Collado-Vides J. RegulonDB: a database on transcriptional regulation in Escherichia coli. Nucleic Acids Res. 1998;26:55–59. doi: 10.1093/nar/26.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- Hurwitz J. The enzymatic incorporation of ribonucleotides into polydeoxynucleotide material. J Biol Chem. 1959;234:2351–2358. [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, Raineri E, Garriga-Canut M, Serrano L. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–845. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. La Logique du Vivant, Une Histoire de L'Hérédité. Paris: Gallimard; 1970. [Google Scholar]
- Jacob F, Monod J. Genes of structure and genes of regulation in the biosynthesis of proteins. C R Hebd Seances Acad Sci. 1959;249:1282–1284. [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jacques PE, Gervais AL, Cantin M, Lucier JF, Dallaire G, Drouin G, Gaudreau L, Goulet J, Brzezinski R. MtbRegList, a database dedicated to the analysis of transcriptional regulation in Mycobacterium tuberculosis. Bioinformatics. 2005;21:2563–2565. doi: 10.1093/bioinformatics/bti321. [DOI] [PubMed] [Google Scholar]
- Janga SC, Salgado H, Collado-Vides J, Martinez-Antonio A. Internal versus external effector and transcription factor gene pairs differ in their relative chromosomal position in Escherichia coli. J Mol Biol. 2007a;368:263–272. doi: 10.1016/j.jmb.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Janga SC, Salgado H, Martinez-Antonio A, Collado-Vides J. Coordination logic of the sensing machinery in the transcriptional regulatory network of Escherichia coli. Nucleic Acids Res. 2007b;35:6963–6972. doi: 10.1093/nar/gkm743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu Rev Biomed Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan S, Bren A, Zaslaver A, Dekel E, Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol Cell. 2008;29:786–792. doi: 10.1016/j.molcel.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. At Home in the Universe: The Search for Laws of Self-Organization and Complexity. New York: Oxford University Press; 1995. p. viii. p. 321pp. [Google Scholar]
- Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, Mironov AA, Gelfand MS, Dubchak I. RegTransBase – a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007;35:D407–D412. doi: 10.1093/nar/gkl865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol R. 2005;69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek AM, Zaim J, Zielenkiewicz P. The effect of transcription and translation initiation frequencies on the stochastic fluctuations in prokaryotic gene expression. J Biol Chem. 2001;276:8165–8172. doi: 10.1074/jbc.M006264200. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Rokney A, Friedman N, Court DL, Stavans J, Oppenheim AB. Quantitative kinetic analysis of the bacteriophage lambda genetic network. P Natl Acad Sci USA. 2005;102:4470–4475. doi: 10.1073/pnas.0500670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesov G, Wunderlich Z, Laikova ON, Gelfand MS, Mirny LA. How gene order is influenced by the biophysics of transcription regulation. P Natl Acad Sci USA. 2007;104:13948–13953. doi: 10.1073/pnas.0700672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara H, Myers CJ, Samoilov MS, Barker NA, Arkin AP. Automated abstraction methodology for genetic regulatory networks. In: Plotkin G, editor. Transactions on Computational Systems Biology VI. Berlin: Springer; 2006. pp. 150–175. [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002a;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002b;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol. 2008;25:559–567. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- Lintner RE, Mishra PK, Srivastava P, Martinez-Vaz BM, Khodursky AB, Blumenthal RM. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 2008;8:60. doi: 10.1186/1471-2180-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J Mol Biol. 2008;379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas WK. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. Ii. dominance of repressibility in diploids. J Mol Biol. 1964;8:365–370. doi: 10.1016/s0022-2836(64)80200-x. [DOI] [PubMed] [Google Scholar]
- MacIsaac K, Wang T, Gordon DB, Gifford D, Stormo G, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003a;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003b;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Mironov AA, Gelfand MS. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2001;2841 doi: 10.1186/gb-2001-2-4-research0013. research 0013.1–0013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Califano A. Theory and limitations of genetic network inference from microarray data. Ann NY Acad Sci. 2007;1115:51–72. doi: 10.1196/annals.1407.019. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr C, Geertz M, Hutt MT, Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Martinez-Antonio A, Collado-Vides J. Comparative Mechanisms for Transcription and Regulatory Signals in Archaea and Bacteria. Chapter 8. Computational Methods for Understanding Archaeal and Bacterial Genomes. London: Imperial College Press; 2008. pp. 1–24. [Google Scholar]
- Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Martínez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. J Mol Biol. 2008;381:238–247. doi: 10.1016/j.jmb.2008.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol R. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Erill I, Campoy S, Cortes P, Forano E, Barbe J. Reconstruction of the evolutionary history of the LexA-binding sequence. Microbiology. 2004;150:3783–3795. doi: 10.1099/mic.0.27315-0. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. P Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Simulation of prokaryotic genetic circuits. Annu Rev Bioph Biom. 1998;27:199–224. doi: 10.1146/annurev.biophys.27.1.199. [DOI] [PubMed] [Google Scholar]
- McCue LA, Thompson W, Carmack CS, Lawrence CE. Factors influencing the identification of transcription factor binding sites by cross-species comparison. Genome Res. 2002;12:1523–1532. doi: 10.1101/gr.323602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. P Natl Acad Sci USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza L, Alvarez-Buylla ER. Dynamics of the genetic regulatory network for Arabidopsis thaliana flower morphogenesis. J Theor Biol. 1998;193:307–319. doi: 10.1006/jtbi.1998.0701. [DOI] [PubMed] [Google Scholar]
- Mettetal JT, Muzzey D, Pedraza JM, Ozbudak EM, van Oudenaarden A. Predicting stochastic gene expression dynamics in single cells. P Natl Acad Sci USA. 2006;103:7304–7309. doi: 10.1073/pnas.0509874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Petranovic D, Jensen PR. Tunable promoters in systems biology. Curr Opin Biotech. 2005;16:329–335. doi: 10.1016/j.copbio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Castro Roa D, Nudler E, Mirkin SM. Transcription regulatory elements are punctuation marks for DNA replication. P Natl Acad Sci USA. 2006;103:7276–7281. doi: 10.1073/pnas.0601127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina N, van Nimwegen E. Universal patterns of purifying selection at noncoding positions in bacteria. Genome Res. 2008;18:148–160. doi: 10.1101/gr.6759507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Campuzano S, Janga SC, Perez-Rueda E. Identification and analysis of DNA-binding transcription factors in Bacillus subtilis and other Firmicutes – a genomic approach. BMC Genomics. 2006;7:147. doi: 10.1186/1471-2164-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G. Operons across prokaryotes: genomic analyses and predictions 300+ genomes later. Curr Genomics. 2006;7:163–170. [Google Scholar]
- Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Neapolitan RE. Learning Bayesian Networks. Harlow: Prentice Hall; 2003. p. xv. p. 674pp. [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykter M, Price ND, Aldana M, Ramsey SA, Kauffman SA, Hood LE, Yli-Harja O, Shmulevich I. Gene expression dynamics in the macrophage exhibit criticality. P Natl Acad Sci USA. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrell D, Bolouri H. Control of internal and external noise in genetic regulatory networks. J Theor Biol. 2004;230:301–312. doi: 10.1016/j.jtbi.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson BO, Lightfoot EN. Mathematical modelling of dynamics and control in metabolic networks. I. On Michaelis–Menten kinetics. J Theor Biol. 1984;111:273–302. doi: 10.1016/s0022-5193(84)80211-8. [DOI] [PubMed] [Google Scholar]
- Palsson BO, Jamier R, Lightfoot EN. Mathematical modelling of dynamics and control in metabolic networks. II. Simple dimeric enzymes. J Theor Biol. 1984;111:303–321. doi: 10.1016/s0022-5193(84)80212-x. [DOI] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. San Mateo, CA: Morgan Kaufmann Publishers; 1988. [Google Scholar]
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Gene Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. P Natl Acad Sci USA. 1975;72:784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. The detachment and maturation of conserved lambda prophage DNA. J Mol Biol. 1965;11:90–96. doi: 10.1016/s0022-2836(65)80174-7. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gaan A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Puchalka J, Kierzek AM. Bridging the gap between stochastic and deterministic regimes in the kinetic simulations of the biochemical reaction networks. Biophys J. 2004;86:1357–1372. doi: 10.1016/S0006-3495(04)74207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Rajewsky N, Socci ND, Zapotocky M, Siggia ED. The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res. 2002;12:298–308. doi: 10.1101/gr.207502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Resendis-Antonio O, Freyre-Gonzalez JA, Menchaca-Mendez R, Gutierrez-Rios RM, Martinez-Antonio A, Avila-Sanchez C, Collado-Vides J. Modular analysis of the transcriptional regulatory network of E. coli. Trends Genet. 2005;21:16–20. doi: 10.1016/j.tig.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Resendis-Antonio O, Reed JL, Encarnacion S, Collado-Vides J, Palsson BO. Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLoS Comput Biol. 2007;3:1887–1895. doi: 10.1371/journal.pcbi.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev. 2007;107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 2005;21:385–389. doi: 10.1016/j.tig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comput Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado H, Martinez-Antonio A, Janga SC. Conservation of transcriptional sensing systems in prokaryotes: a perspective from Escherichia coli. FEBS Lett. 2007;581:3499–3506. doi: 10.1016/j.febslet.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoilov MS, Arkin AP. Deviant effects in molecular reaction pathways. Nat Biotechnol. 2006;24:1235–1240. doi: 10.1038/nbt1253. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Kondev J. Transcriptional control of noise in gene expression. P Natl Acad Sci USA. 2008;105:5081–5086. doi: 10.1073/pnas.0707904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22:645–650. doi: 10.1093/bioinformatics/bti597. [DOI] [PubMed] [Google Scholar]
- Savageau MA. Comparison of classical and autogenous systems of regulation in inducible operons. Nature. 1974;252:546–549. doi: 10.1038/252546a0. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. P Natl Acad Sci USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshasayee AS, Bertone P, Fraser GM, Luscombe NM. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr Opin Microbiol. 2006;9:511–519. doi: 10.1016/j.mib.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical–chemical model for gene regulation. J Mol Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucliec Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoko D, Yoo D, Bai H, Schnurr B, Yan J, McLeod SM, Marko JF, Johnson RC. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P, Baxter DA, Byrne JH. Modeling transcriptional control in gene networks – methods, recent results, and future directions. B Math Biol. 2000;62:247–292. doi: 10.1006/bulm.1999.0155. [DOI] [PubMed] [Google Scholar]
- Song C, Havlin S, Makse HA. Origins of fractality in the growth of complex networks. Nat Phys. 2006;2:275–281. [Google Scholar]
- Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(suppl 2):S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- Stevens A, Rhoton JC. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry. 1975;14:5074–5079. doi: 10.1021/bi00694a007. [DOI] [PubMed] [Google Scholar]
- Stock A, Koshland DE, Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. P Natl Acad Sci USA. 1985;82:7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- Suh SJ, Runyen-Janecky LJ, Maleniak TC, Hager P, MacGregor CH, Zielinski-Mozny NA, Phibbs PV, Jr, West SE. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology. 2002;148:1561–1569. doi: 10.1099/00221287-148-5-1561. [DOI] [PubMed] [Google Scholar]
- Takemoto K, Oosawa C. Modeling for evolving biological networks with scale-free connectivity, hierarchical modularity, and disassortativity. Math Biosci. 2007;208:454–468. doi: 10.1016/j.mbs.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Taoka M, Yamauchi Y, Shinkawa T, Kaji H, Motohashi W, Nakayama H, Takahashi N, Isobe T. Only a small subset of the horizontally transferred chromosomal genes in Escherichia coli are translated into proteins. Mol Cell Proteomics. 2004;3:780–787. doi: 10.1074/mcp.M400030-MCP200. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Thieffry D, Romero D. The modularity of biological regulatory networks. Biosystems. 1999;50:49–59. doi: 10.1016/s0303-2647(98)00087-2. [DOI] [PubMed] [Google Scholar]
- Thieffry D, Thomas R. Qualitative analysis of gene networks. Pac Symp Biocomput. 1998:77–88. [PubMed] [Google Scholar]
- Thomas R, D'Ari R. Biological Feedback. Boca Raton: CRC Press; 1990. 316pp. [Google Scholar]
- Tompa M, Li N, Bailey TL, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Typas A, Becker G, Hengge R. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- van Nimwegen E. Finding regulatory elements and regulatory motifs: a general probabilistic framework. BMC Bioinformatics. 2007;8(suppl 6):S4. doi: 10.1186/1471-2105-8-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, van der Voort M, Molenaar D, Moezelaar R, de Vos WM, Abee T. Identification of the sigmaB regulon of Bacillus cereus and conservation of sigmaB-regulated genes in low-GC-content gram-positive bacteria. J Bacteriol. 2007;189:4384–4390. doi: 10.1128/JB.00313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuelas J, Calevro F, Remond D, Bernillon J, Rahbe Y, Febvay G, Fayard JM, Charles H. Conservation of the links between gene transcription and chromosomal organization in the highly reduced genome of Buchnera aphidicola. BMC Genomics. 2007;8:143. doi: 10.1186/1471-2164-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJW, Struhl K, Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- Wagner A. How to reconstruct a large genetic network from n gene perturbations in fewer than n(2) easy steps. Bioinformatics. 2001;17:1183–1197. doi: 10.1093/bioinformatics/17.12.1183. [DOI] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: {sigma}S-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Weinstein-Fischer D, Altuvia S. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol. 2007;63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Ussery DW. Chromatin architecture and gene expression in Escherichia coli. Genome Biol. 2004;5:252. doi: 10.1186/gb-2004-5-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988;16:1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Arkin AP. Motifs, modules and games in bacteria. Curr Opin Microbiol. 2003;6:125–134. doi: 10.1016/s1369-5274(03)00033-x. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z, Mirny LA. Spatial effects on the speed and reliability of protein-DNA search. Nucleic Acids Res. 2008;36:3570–3578. doi: 10.1093/nar/gkn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J Biol Chem. 2006;281:29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. P Natl Acad Sci USA. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: protein–protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Zhang LV, King OD, Wong SL, Goldberg DS, Tong AH, Lesage G, Andrews B, Bussey H, Boone C, Roth FP. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J Biol. 2005;4:6. doi: 10.1186/jbiol23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB. Shape and compaction of Escherichia coli nucleoids. J Struct Biol. 2006;156:255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]