Abstract
Objective
To define the molecular basis of TGF-β1 function in cervical carcinogenesis, we explored the expression and mutational status of TGF-β1, TGF-β1 receptors, and Smads, the regulators of the TGF-β1 signaling pathway, in human cervical cancers.
Methods
Expression of TGF-β1, TGF-β1 receptors, and Smads transcripts were determined by quantitative reverse transcription-polymerase chain reaction (RT-PCR), and sequence alteration was analyzed using RT-PCR-single-strand conformation polymorphism (SSCP) analysis. Genomic levels of TGF-β1, TGF-β1 receptors and Smads was also measured by quantitative genomic PCR.
Results
Abnormal overexpression of TGF-β1 and abnormal reduction of type II TGF-β1 receptor were identified in 36% (18 of 50) and 20% (10 of 50) of cervical cancer tissues, respectively. 22% (11 of 50) in Smad2 and 14% (7 of 50) in Smad4 revealed tumor specific mRNA reduction less than a half of normal means. In addition, no evidence for sequence alterations of the gene was found by RT-PCR-SSCP analysis.
Conclusion
Our study demonstrates that disruption of TGF-β/Smad signaling pathway exist in human cervical cancer, suggesting that abnormal expressions of the member of TGF-β/Smad signaling pathway might contribute to the malignant progression of human cervical tumors via suppressing the tumor suppression function of TGF-β1 1's tumor suppression function.
Keywords: TGF-β/Smads, Cervical cancer, Expression
INTRODUCTION
Uterine cervical cancer is the most common neoplasm of female genital tract accounting for 9.1% of total malignancies in Korean women in 2002.1 It is well known that the human papilloma virus (HPV) is the major causative agent.2 However, many patients who suffer from HPV infection will remain stable.
A particularly interesting biological characteristic associated with carcinogenesis of cervical epithelial cells is their progressive loss of responsiveness to transforming growth factor-β (TGF-β).3,4 TGF-β is a member of a multifunctional family of growth factors that regulate basic cellular functions, such as proliferation, apoptosis, differentiation, extracellular matrix turnover and immunosuppression.5 One of the most widely studied roles of TGF-β is its function as a tumor suppressor.
There are three isoforms of TGF-β: TGF-β1, TGF-β2, and TGF-β3 and each isoform is encoded by a distinct gene.6 TGF-β1 is expressed in endothelial, hematopoietic, and connective tissue cells, and TGF-β2 is expressed in epithelial and neuronal cells, while TGF-β3 is expressed primarily in mesenchymal cells.7
The biological effects of TGF-β1 are primarily mediated by a heteromeric complex of two transmembrane serine/threonine kinases, the type I and type II TGF-β1 receptors (TGF-β RI and TGF-βRII).5,8,9 TGF-β/Smad cascade is composed of a ternary signaling complex, activated when TGF-β1 binds to TGF-βRII, TGF-βRII interacts with and phosphorylates TGF-βRI, which in turn phosphorylates cytoplasmic mediators, Smad2 and/or Smad3, and a heterotrimeric complex is formed with Smad4 that translocates into the nucleus, binds a consensus sequence, and directly or indirectly regulates gene transcription.5,10 Many cancer cells display mutations in the TGF-β/Smad signaling pathway, with over 15% of colon carcinomas displaying mutations of the TGF-βRII. Furthermore, 25% of ovarian cancers have a mutation of the TGF-β RII and 33% of the TGF-βRI.11 Mutations in the TGF-βRI are also found, albeit with lesser frequency, in cancers of the lung, breast, and others.12,13 Furthermore, approximately half of all pancreatic and 15% of colorectal cancers have a mutation in Smad4, and mutations in Smad2 have been noted in some lung and colon cancers.9,14
The aim of this investigation was to determine if members of the TGF-β/Smad are abnormally expressed in human cervical cancers and, if so, whether such abnormal expression is associated with mutation. We also analyzed the clinical significance of TGF-β/Smad signaling pathway in human cervical cancers.
MATERIALS AND METHODS
1. Tumor specimens
From January 1999 to December 2004, 50 cervical cancers tissue specimens were obtained by surgical resection at the Kyung Hee University hospital. Tissue specimens were snap-frozen in liquid N2 and stored at -70℃ until used. The study group included 38 patients with the International Federation of Gynecology and Obstetrics (FIGO) stage IB, and 12 patients with FIGO stage II disease. The normal cervical tissues from 10 patients who underwent hysterectomy for benign indications were also collected as controls. Clinical outcomes were recorded through retrospective chart review.
2. Quantitative RT-PCR analysis
One µL of total cellular RNA was converted to cDNA by reverse transcription using random hexamer primers and Moloney murine leukemia virus reverse transcriptase (Life Technologies Inc., Gaithersburg, MD, USA). PCR was initially carried out over a range of cycles (22-40 cycles) using 1:4 diluted cDNA (12.5 ng/50 µL PCR reaction), and PCR undergoing 24-34 cycles were observed to be within the logarithmic phase of amplification with all primers used for TGF-β1, TGF-βRI and TGF-βRII, Smad2, Smad3, Smad4, and an endogenous expression standard gene GAPDH. Primer sequences are available upon request. The cDNA was then subjected to 26-32 cycles of PCR at 95℃ for denaturation (1 min), 58-64℃ for annealing (0.5 min), and 72℃ for extension (1 min) in reaction buffer containing 1.5 mM MgCl2 (PCR buffer II; Perkin-Elmer Corp., Norwalk, CT, USA). Quantitation of gene expression levels was performed by laser densitometric measurement of the ethidium bromide-stained gel. Absolute area integrations of the curves representing each specimen were compared after adjustment for the expression level of the housekeeping gene, GAPDH. Integration and analysis were performed using Molecular Analyst software program (Bio-Rad, Richmond, CA, USA). Quantitative PCR was repeated at least three times for each specimen and the mean was obtained. The relative levels of expression were represented as the ratio of target genes/GAPDH. We considered less than 50% expression of normal mean as being abnormally low, and more than double times expression of normal mean as abnormally high.
3. Nonisotopic RT-PCR-SSCP analysis
The entire coding regions of the TGF-β1, TGF-βRI, TGF-βRII, Smad2, Smad3, and Smad4 transcripts were amplified by PCR using several sets of primer for amplification of 200-to 280-bp lengths of fragments optimal for single-strand conformation polymorphism (SSCP) analysis. Twenty µL of these nest-PCR products were mixed with 5 µL of 0.5 N NaOH, 10 mM EDTA, 10 µL of denaturing loading buffer (95% formamide, 20 mM EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol), and 15 ml of double distilled H2O. After heating at 95℃ for 5 min, samples were loaded in gels wells pre-cooled to 4℃. SSCP analysis was performed using 8% nondenaturing acrylamide gels containing 10% glycerol at room temperature.
4. Quantitative genomic DNA-PCR
Genomic DNA was prepared from tissue specimens using QIAGEN genomic-tip system (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The concentration of extracted DNA was determined by a spectrophotometric measurement (Schimadzu Scientific Instruments Inc., Concord, CA, USA). PCR amplification using intron-specific primers was performed with 200 µg genomic DNA as template for 38 cycles at 95℃ (1 min), 58-62℃ (0.5 min), and 72℃ (0.5 min) in 1.5 mmol/L MgCl2-containing reaction buffer (PCR buffer II, Perkin Elmer-Cetus, Concord). Ten µL of the PCR products were electrophoresed on 2% agarose gel. Quantitation was achieved by densitometric scanning and absolute area integrations of the curves representing each specimen were compared after adjustment for GAPDH.
5. Statistical analysis
To compare means the independent samples T test, and to compare frequencies the Chi-square test, or Fisher's exact test when appropriate, was used. A p-value of less than 0.05 was considered to be significant. All analyses were performed using the statistical software SPSS for Windows ver. 12 (SPSS Inc., Chicago, USA).
RESULTS
1. Expression of TGF-β1, TGF-βRI and TGF-βRII (RT-PCR analysis)
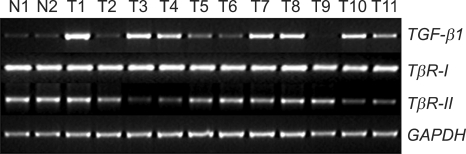
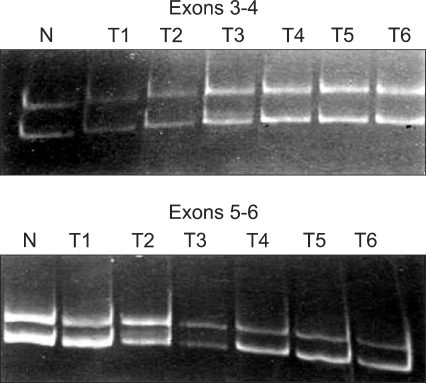
We evaluated the mRNA expression profile of TGF-β1, TGF-βRI, and TGF-βRII in a total of 60 cervical tissue specimens (Fig. 1). Ten specimens were normal cervical tissues from hysterectomies performed for non-malignant conditions, 44 were squamous cell carcinoma, 4 were adenocarcinoma, and 2 were adenosquamous carcinoma.
Fig. 1.
TGF-β1, TGF-βRI and TGF-βRII expression (Quantitative reverse transcription-polymerase chain reaction analysis).
N: normal cervix tissues, T: cervical tumor tissues
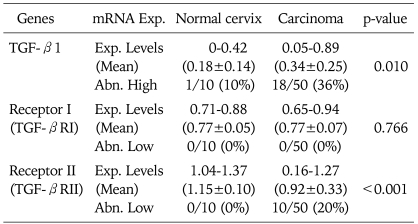
Quantitative RT-PCR was repeated at least three times for each specimen, and the mean expression level of TGF-β1, TGF-βRI, and TGF-βRII in noncancerous tissues was determined as 0.18, 0.77, and 1.15, respectively. Expression levels (Target/GAPDH) of TGF-β1, TGF-βRI, and TGF-βRII in cervical cancer tissues were observed within the range of 0.05-0.89 (mean 0.34), 0.65-0.94 (mean 0.77), and 0.16-1.27 (mean 0.92), respectively.
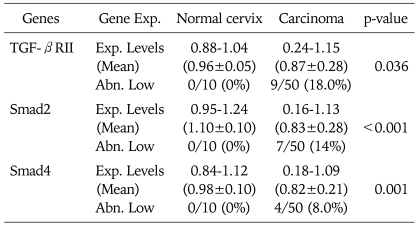
Compared with normal controls, RT-PCR analysis demonstrated an abnormal overexpression of TGF-β1 mRNA and decreased expression of TGF-βRII mRNA expression in the cervical cancer tissues (Table 1). We arbitrarily classified tumors with expression levels of less than a half of normal means as abnormally low expression. Abnormal overexpression of TGF-β1 and abnormal reduction of TGF-βRII were identified in 36% (18 of 50) and 20% (10 of 50) of cervical cancer tissues, respectively.
Table 1.
Expression level of TGF-β1, TGF-βRI, and TGF-βRII mRNA
Exp. Levels: expression levels (target gene/GAPDH), Abn. High: abnormally high (>normal mean×2), Abn. Low: abnormally low (<normal mean×1/2)
2. Expression of Smads (RT-PCR analysis)
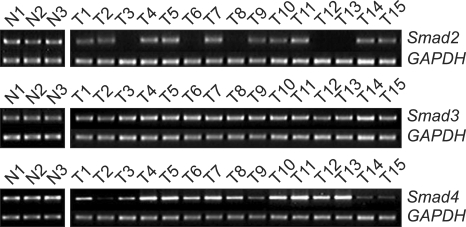
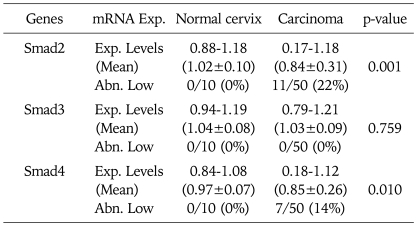
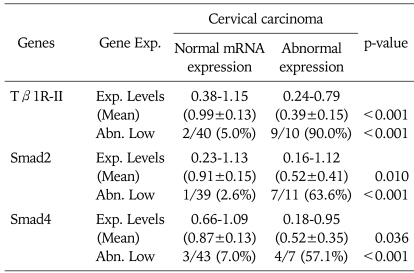
Next, we evaluated Smads expression in 50 cervical cancer tissues and 10 noncancerous tissues (Fig. 2). Our findings indicate that Smad2 and Smad4 mRNA expression is significantly related to the condition of the cervix (normal, cancer). As a result of RT-PCR analysis, cervical cancer tissues showed variable expression of Smad2, Smad3, and Smad4 (0.17-1.18, 0.79-1.21, and 0.18-1.12, respectively), and their levels were markedly reduced in Smad2 and Smad4 (mean; 0.84 and 0.85, respectively) compared to that of noncancerous controls (mean; 1.02 and 0.97, respectively). According to our arbitrary classification, 22% (11 of 50) in Smad2 and 14% (7 of 50) in Smad4 revealed tumor specific mRNA reduction of less than a half of normal means (Table 2)
Fig. 2.
Smads expression (Quantitative reverse transcription-polymerase chain reaction analysis).
N: normal cervix tissues, T: cervical tumor tissues
Table 2.
Expression level of Smads mRNA
Exp. Levels: expression levels (target gene/GAPDH), Abn. Low: abnormally low (<normal mean×1/2)
3. Genomic levels of abnormally low expressed TGF/Smads (DNA-PCR analysis)
To analyze the allelic status of TGF-βRII, Smad2, and Smad4 in cervical cancers, we first determined the genomic level of TGF-βRII, Smad2, and Smad4 using quantitative PCR. As shown in Table 3, the absence and marked reduction of TGF-βRII, Smad2, and Smad4 were detected in abnormal low expressed cervical cancer tissues which have homozygous deletion or LOH of the gene, while normal levels of TGF-β RII, Smad2, and Smad4 were detected in noncancerous tissues and normally expressed cancer tissues, indicating that genomic level determined by our quantitative PCR is consistent with the allelic status of the gene. Nine of the 10 (90%), 7 of the 11 (63.6%), and 4 of the 7 (57.1%) of cervical cancer tissues showing low mRNA expression represented a detectable reduction of TGF-βRII, Smad2, and Smad4 levels, respectively. In contrast, none of the noncancerous tissues and cervical cancer tissues showing normal mRNA expression cell lines had absence or significantly low frequencies of genomic reduction (Table 4).
Table 3.
Genomic level of TGF-βRII, Smad2, Smad4
Exp. Levels: expression levels (target gene/GAPDH), Abn. Low: abnormally low (<normal mean×1/2)
Table 4.
Comparison of genomic level of TGF-βRII, Smad2, Smad4 in cervical cancer
Exp. Levels: expression levels (target gene/GAPDH), Abn. Low: abnormally low (<normal mean×1/2)
4. Mutational analysis (RT-PCR-SSCP)
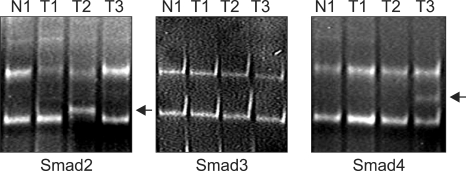
To explore the presence of mutational alteration of TGF-β RII and Smads, we performed RT-PCR-SSCP analysis for TGF-βRII and Smads transcripts expressed from 50 primary tumor tissues. The entire coding region for TGF-βRII and Smads transcript was amplified using seven sets of exon specific primer sets, and the RT-PCR products were subjected to SSCP analysis. To improve mutation detection sensitivity, the same PCR products were digested using different restriction endonuclease(s), and SSCP was carried out under two different running conditions. However, we failed to detect any types of mutation leading to amino-acid substitutions or frameshifts of TGF-βRII and Smad3 in cervical cancer tissues, whereas Smad2 and Smad4 mutations were detected in 2 of 50 (4%) and 1 of 50 (2%), respectively (Figs. 3 and 4).
Fig. 3.
Mutational analysis of Tβ1R-II transcripts (reverse transcription-polymerase chain reaction-single-strand-conformation-polymorphism analysis).
Fig. 4.
Mutational analysis of Smad transcripts (reverse transcription-polymerase chain reaction-single-strand-conformation-polymorphism analysis). Abnormal migration shift of ssDNA (mutational sequence alteration, arrow). Smad2: 2 of 50 (4%) cervical cancers, Smad3: 0 of 50 (0%) cervical cancers, Smad4: 1 of 50 (2%) cervical cancers.
N: normal cervix tissues, T: cervical tumor tissues
DISCUSSION
TGF-β1 is characterized as an antiproliferative cytokine, especially in the early stage of cancer development.15,16 It is clear that the TGF-β/Smad pathway is an important tumor suppressor mechanism and in several cancer types, and there is a strong correlation between malignant progression and loss of sensitivity to the antiproliferative effects of TGF-β, which is frequently associated with reduced expression or mutational inactivation of TGF-β receptors.17,18
One characteristic associated with malignant progression of cervical epithelial cells is their progressive loss of responsiveness to TGF-β1.3,4 Furthermore, resistance to TGF-β1 that is acquired by several cell lines correlates with HPV tumorigenic potential. Such studies indicate that after HPV infection, additional cellular or molecular changes might participate in the loss of TGF-β responsiveness, which then promotes malignant transformation. Our results agree with these observations, and suggest that one significant mechanism is the lack of TGF-βRII expression.
Thus, the TGF-β1 restriction point is an important obstacle many cells must overcome in order to evolve into a malignancy and achieve unrestricted proliferation. This can be accomplished in multiple ways, including reduced activation of TGF-β1 from its latent form, decreased expression of the TGF-β1 receptors, mutation of individual TGF-β1 receptor subunits, and mutation of Smad2, 3, or 4 which propagate the TGF-β signal.
In this report, we demonstrated that disruption of TGF-β/Smad signaling pathway exist in human cervical cancers. Such alterations in members of TGF-β/Smad signaling might lead to loss of TGF-β sensitivity in cervical cancer tissues, and ultimately to escape from growth regulatory signals imposed by TGF-β. Of note, we observed a correlation between reduced or absent phosphorylation of TGF-βRII, Smad2, 4 and high TGF-β1 expression. Sample size also limited our ability to provide a meaningful analysis of clinical outcomes.
Our data suggest that disruption of the TGF-β/Smad signaling pathway by alteration of TGF-βRII, Smad2, and Smad4 may play an important role in development and progression of cervical carcinomas. This escape from growth control in cervical cancer seems to be linked to allelic deletion and mutation of members of TGF-β/Smad signaling pathway.
Together, our study demonstrates that activation of TGF-β signaling associated with overexpression of TGF-β1 coexists in cervical cancer compared to the normal cervix, suggesting that elevated TGF-β1 activity might contribute to the malignant progression of human cervical tumors at some special stage of carcinogenesis.
Overproduction of TGF-β1 has been associated with tumors of many histologic types including those of breast, prostate, lung, liver, and colon.6 These high TGF-β1 levels in tumor tissues, including cervical cancer, correlate with markers of higher metastatic phenotype and/or poor patient outcome,19 and many tumor cells exhibit increased invasiveness in response to TGF-β.20
Recent studies have provided us insight into how aberrant negative regulation in TGF-β/Smad signaling is linked to human carcinogenesis. Although clinical translation will not be easy considering the multifunctional and contextual nature of TGF-β/Smad signaling, we anticipate that further progress will provide us opportunities for therapeutic intervention.
Footnotes
This study was supported by the Kyung Hee University Research Fund in 2006 (KHU-20060391).
This study was awarded a best poster presentation prize at 23rd annual meeting of Korean Society of Gynecologic Oncology and Colposcopy.
References
- 1.Shin HR, Jung KW, Won YJ, Park JG 139 KCCR-affiliated Hospitals. 2002 annual report of the Korea central cancer registry: based on registered data from 139 hospitals. Cancer Res Treat. 2004;36:103–114. doi: 10.4143/crt.2004.36.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waggoner SE. Cervical cancer. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- 3.Kang SH, Won K, Chung HW, Jong HS, Song YS, Kim SJ, et al. Genetic integrity of transforming growth factor beta (TGF-beta) receptors in cervical carcinoma cell lines: loss of growth sensitivity but conserved transcriptional response to TGF-beta. Int J Cancer. 1998;77:620–625. doi: 10.1002/(sici)1097-0215(19980812)77:4<620::aid-ijc23>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Kim JW, Kim HS, Kim IK, Kim MR, Cho EY, Kim HK, et al. Transforming growth factor-β1 induces apoptosis through down-regulation of c-mycGene and overexpression of p27Kip1 protein in cervical carcinoma. Gynecol Oncol. 1998;69:230–236. doi: 10.1006/gyno.1998.5003. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Massagu J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 6.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 7.Pasche B. Role of transforming growth factor beta in cancer. J Cell Physiol. 2001;186:153–168. doi: 10.1002/1097-4652(200002)186:2<153::AID-JCP1016>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Sun Y, Weinberg RA, Lodish HF. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 10.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 11.Lynch MA, Nakashima R, Song H, DeGroff VL, Wang D, Enomoto T, et al. Mutational analysis of the transforming growth factor beta receptor type II gene in human ovarian carcinoma. Cancer Res. 1998;58:4227–4232. [PubMed] [Google Scholar]
- 12.Kim SJ, Im YH, Markowitz SD, Bang YJ. Molecular mechanisms of inactivation of TGF-beta receptors during carcinogenesis. Cytokine Growth Factor Rev. 2000;11:159–168. doi: 10.1016/s1359-6101(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 13.Chen T, Triplett J, Dehner B, Hurst B, Colligan B, Pemberton J, et al. Transforming growth factor-beta receptor type I gene is frequently mutated in ovarian carcinomas. Cancer Res. 2001;61:4679–4682. [PubMed] [Google Scholar]
- 14.Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- 15.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 16.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 17.Kang SH, Bang YJ, Im YH, Yang HK, Lee DA, Lee HY, et al. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18:7280–7286. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 18.Markowitz SD, Roberts AB. Tumor suppressor activity of the TGF-beta pathway in human cancers. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 19.Hazelbag S, Kenter GG, Gorter A, Dreef EJ, Koopman LA, Violette SM, et al. Overexpression of the alpha v beta 6 integrin in cervical squamous cell carcinoma is a prognostic factor for decreased survival. J Pathol. 2007;212:316–324. doi: 10.1002/path.2168. [DOI] [PubMed] [Google Scholar]
- 20.Welch DR, Fabra A, Nakajima M. Transforming growth factor beta stimulates mammary adenocarcinoma cell invasion and metastatic potential. Proc Natl Acad Sci USA. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]