Abstract
The human β2-adrenergic receptor gene has multiple single-nucleotide polymorphisms (SNPs), but the relevance of chromosomally phased SNPs (haplotypes) is not known. The phylogeny and the in vitro and in vivo consequences of variations in the 5′ upstream and ORF were delineated in a multiethnic reference population and an asthmatic cohort. Thirteen SNPs were found organized into 12 haplotypes out of the theoretically possible 8,192 combinations. Deep divergence in the distribution of some haplotypes was noted in Caucasian, African-American, Asian, and Hispanic-Latino ethnic groups with >20-fold differences among the frequencies of the four major haplotypes. The relevance of the five most common β2-adrenergic receptor haplotype pairs was determined in vivo by assessing the bronchodilator response to β agonist in asthmatics. Mean responses by haplotype pair varied by >2-fold, and response was significantly related to the haplotype pair (P = 0.007) but not to individual SNPs. Expression vectors representing two of the haplotypes differing at eight of the SNP loci and associated with divergent in vivo responsiveness to agonist were used to transfect HEK293 cells. β2-adrenergic receptor mRNA levels and receptor density in cells transfected with the haplotype associated with the greater physiologic response were ≈50% greater than those transfected with the lower response haplotype. The results indicate that the unique interactions of multiple SNPs within a haplotype ultimately can affect biologic and therapeutic phenotype and that individual SNPs may have poor predictive power as pharmacogenetic loci.
The β2-adrenergic receptors (β2ARs) are G protein-coupled receptors that mediate the actions of catecholamines in multiple tissues. Like other members of the superfamily, the β2AR has an extracellular amino terminus, seven transmembrane-spanning domains, three intracellular and three extracellular loops, and an intracellular carboxyl terminus. The receptor is encoded by an intronless gene on chromosome 5q31–32. In the human population, we have identified (1) nonsynonymous single-nucleotide polymorphisms (SNPs) at nucleotides 46, 79, and 491 that result in changes in amino acid residues 16 and 27 of the amino terminus and 164 of the fourth intracellular loop (Table 1 and GenBank accession nos. AF022953, AF022954, and AF022956). In transfected cells, we have shown that one of these polymorphic receptors (Ile164) is markedly dysfunctional, with altered high-affinity binding and decreased coupling to the stimulatory G protein, Gs (2). In transgenic mice overexpressing Thr164 or Ile164 in the heart, a similar functional defect was observed in myocytes and in cardiac function in vivo (3). In cell-based systems, we also have shown that the amino-terminal polymorphisms at amino acid positions 16 and 27 alter cellular trafficking of the receptor such that the magnitude of agonist-promoted down-regulation varies with certain alleles (4, 5). Ligand binding and functional coupling are not altered by these SNPs. The β2AR transcription start site is 5′ to a small ORF (termed the β2AR 5′-leader cistron) that encodes a 19-aa peptide. This peptide, denoted the β2AR upstream peptide (BUP), modulates receptor translation (6). We recently have shown that a human polymorphism at amino acid 19 of BUP alters this modulatory effect, thereby affecting β2AR expression (7). In addition to the above, SNPs in the 5′ promoter region have recently been identified (8).
Table 1.
Localization of SNPs and identification of haplotypes of the β2AR gene
Nucleotide number is based on the first nucleotide of the start codon being +1. Frequencies of the haplotypes were determined in the indicated populations as described in Methods. Allele, the two nucleotide possibilities at each SNP position; Ca, Caucasian; A-A, African-American; As, Asian; H-L, Hispanic-Latinos; 5′, 5′ upstream of β2AR ORF; syn, synonymous SNP.
The β2AR expressed on bronchial smooth muscle acts to relax contracted muscle, resulting in bronchodilation (9). Indeed, β agonists are the most effective acute treatment for reversal of bronchospasm in asthma. The bronchodilating response to β agonists, although, is known to exhibit significant interindividual variation (10). Given the in vitro findings with the polymorphic β2AR, we and others have considered that these SNPs may act as disease modifiers in asthma or may be the basis for the variability in the response to β agonists (11). Indeed, some studies have shown that an individual β2AR SNP correlates with the bronchodilatory response to β agonists (12–14) or the extent of tachyphylaxis to repeated use of these agents (15, 16). Other studies, although, have failed to show any such correlations (17) or have shown discordant results (15, 16).
Of note, the above in vitro studies used constructs designed to assess phenotypic consequences of a given SNP in isolation, without consideration as to potential interactions with other SNPs in the promoter or coding regions. Nor have any of the physiologic studies in humans assessed the relevance of combinations of multiple β2AR SNPs, or specific haplotypes (the array of SNPs on a given chromosome), in these regions for predicting the bronchodilator response to β agonists. And finally, the phylogeny of these haplotypes and their distribution amongst different ethnic groups, which has particular relevance to pharmacogenetics, has not been explored.
In this work, we have delineated SNPs within a continuous region of 5′ untranslated region (UTR) and β2AR coding sequence, analyzed phylogeny, and determined the haplotypes in a defined reference population consisting of four ethnic groups. Then the relationship between the bronchodilatory response to the β2AR agonist albuterol and the common haplotype pairs was explored in asthmatics. And finally, to assign a molecular basis for the haplotype-specific physiologic response that was observed, two haplotypes were studied within the context of the β2AR gene and protein expression in vitro in transient transfection experiments.
Methods
Nomenclature.
The reference sequence for the intronless human β2AR gene is that from GenBank accession no. M15169. For clarity, in this paper we refer to the first nucleotide of the ORF as nucleotide 1 (corresponding to nucleotide 1,588 of M15169), with the 5′ UTR beginning at −1 and proceeding in the negative direction. Sequencing was confined to ≈1,100 bp of 5′ UTR and ≈700 bp of ORF. In the former, eight SNPs were identified as shown in Table 1. Within the ORF, sequence was determined from the start codon through nucleotide 523, which is a synonymous SNP that has been reported to be associated with the response to albuterol (14). As shown, this region also contains all of the known nonsynonymous SNPs that alter the encoded residues at amino acid positions 16, 27, and 164 of the protein (1).
Constructs and Cell Transfections.
PCR products of the β2AR gene derived from human genomic DNA were subcloned into pCR2.1 and the sequence was verified. This vector lacks a eukaryotic-responsive promoter and thus the expression of the β2AR gene in mammalian cells is directed by the included β2AR promoter sequence. The luciferase expression vector driven by a human β actin promoter has been described previously (7). For expression studies, HEK293 cells were transfected by using a liposome method (Gene Therapy Systems, San Diego). Cells were transfected with 10 μg of β2AR plasmid, 2 μg of luciferase plasmid, and 50 μl of liposome reagent. Two days later, cells were harvested for radioligand binding, mRNA studies, and luciferase activity as previously described (7). Briefly, cells were washed three times in PBS and lysed in hypotonic 5 mM Tris/2 mM EDTA (pH 7.40) buffer, and the particulates were centrifuged at 40,000 × g for 10 min. Receptor expression was determined by radioligand binding with 400 pM 125I-cyanopindolol. Nonspecific binding was determined in the presence of 1 μM propranolol. Luciferase activity of cell lysates was determined by using a commercial assay (Promega) and was used to control for minor differences in transfection efficiency from plate to plate. β2AR density thus is expressed as fmol/mg of membrane protein or fmol/light unit (fmol/LU) as previously described (7). mRNA levels were determined by using ribonuclease protection assays with a 563-bp antisense riboprobe corresponding to the most 3′ region of the β2AR ORF as described previously (18). β actin mRNA was simultaneously quantitated, confirming the equivalent loading of the samples.
Genotyping.
Two approaches were used to obtain unphased and phased genotypes. Unphased genotypes from both parental chromosomes were determined by sequencing PCR products obtained by using genomic DNA as template. These products consisted of two PCR reactions with the following two sets of primers (sense/antisense): 5′-CTGTCTTCATGCCTGCAAATTCC-3′/5′-CAAACACGATGGCCAGGACGATGAGAG-3′ and 5′-GGACGAGGTGTGGGTGGTGGGCATG-3′/5′-GGGGGTCTTTAAAAGTAGAAAAACTGC-3′. The 1,241- and 1,187-bp respective products were sequenced by using dye terminator chemistry (Big-Dye, Applied Biosystems, Foster City, CA) and an Applied Biosystems 3700 capillary sequencer. Sequencing primers were designed to provide for overlapping ≈500-bp reads. To delineate phase, a single 3,398-bp PCR product consisting of the β2AR ORF and 5′ and 3′ UTR was cloned into the vector pCR 2.1. Primers for this PCR were 5′-AGTAGCTGGGACTACAGGTACG-3′/5′-AGGCAACAGCACTCCAGTCAAG-3′. After transformation, a single colony representing the sequence from one chromosome was chosen for sequencing. The difference between the genomic sequence and the cloned sequence thus provided for phased genotypes for both chromosomes (termed the “molecular haplotype”).
Haplotype Analysis.
Haplotypes were estimated from unphased genotypes by using an extension of Clark's algorithm (19) in which haplotypes are assigned directly from individuals who are homozygous at all sites or heterozygous at no more than one of the variable sites. This list of haplotypes is then used to deconvolute the unphased genotypes in the remaining (multiply heterozygous) individuals. In our analysis, the list of haplotypes was augmented with haplotypes obtained from three families (two multigeneration Caucasian families and one two-generation African-American family). Phylogenetic analysis of the individual haplotypes used a variation of the minimal-spanning network algorithm (20). An advantage of this algorithm over other methods is that it does not force a strictly bifurcating tree as a result. Thus, actual reticulations in the “tree,” such as those arising from evolutionary recombination among the haplotypes, can be visualized and interpreted. Linkage disequilibrium was quantitated as Δ as described by Hill and colleagues (21, 22).
Subjects.
To delineate the diversity of SNPs of the β2AR gene, sequence was obtained from DNA that was derived from immortalized lymphocytes from an index repository of apparently normal individuals. This repository consisted of 23 Caucasians, 19 African-Americans, 20 Asians, and 15 Hispanic-Latinos. This number of individuals in each ethnic group provides for a >90% probability of detecting an SNP with an allele frequency between 0.05 (Caucasian group) and 0.08 (Hispanic-Latino group). To determine whether haplotypes of the β2AR gene are associated with the bronchodilatory response to the agonist albuterol, 121 Caucasian patients with asthma were enrolled from an outpatient facility as described in detail elsewhere (23). Patients underwent spirometry before and 30 min after inhalation of 180 μg of albuterol delivered by nebulization. Forced expiratory volume in 1 sec (FEV1) and forced vital capacity (FVC) were determined in triplicate. The predicted values for these measurements were calculated based on standard algorithms (24). The change in the percentage predicted FEV1 was considered the primary measure of responsiveness to albuterol (25).
Statistical Analysis.
The association between changes in percentage predicted FEV1 and haplotype pair was assessed by fitting an analysis of covariance model (ANCOVA) with terms for haplotype pair, sex, and baseline severity. P values from pairwise comparisons by haplotype pair were adjusted for multiple comparisons by applying the Holm–Sidak step-down procedure (26). For the analysis of individual SNPs, a similar ANCOVA model was used with a discrete term for SNP genotype and terms for sex and baseline severity. The Holm–Sidak step-down procedure was used to adjust the P values from the individual SNP analyses for the number of tests performed.
Results and Discussion
Thirteen variable sites within a span of 1.6 kb were identified in the β2AR gene (Table 1). Two SNPs, at −709 and −406, have not been previously reported. Of the 213 (=8,192) possible combinations of these SNPs, only 12 haplotypes were found in individuals from the index repository and the asthmatic cohort (Table 1). All SNPs and haplotypes were found to be in conformance with Hardy–Weinberg equilibrium, with the exception of homozygotes for haplotype 1 in Hispanic-Latinos, attributable to the existence of a single homozygote in this population for an otherwise rare haplotype. Four of the observed haplotypes occur in all populations sampled, although at markedly different frequencies. Haplotype 2, the most frequent in Caucasians (48%), is seen only at frequencies of 6%, 10%, and 27% in samples of African-Americans, Asians, and Hispanic-Latinos, respectively. Furthermore, this particular haplotype is by far the most distinctive at the nucleotide level, having unique differences at four sites from all other haplotypes sampled. The distribution of haplotype 1 also indicates population differentiation at this locus with as much as a >20-fold lower frequency in Caucasians as compared with the other groups. Also, haplotype 6 is more common in African-Americans and Asians as compared with the other two groups. In contrast to the above findings, the frequency of haplotype 4 is similar in all groups. Assigning haplotypes from unphased genotype data from 200 individuals by using an extension of standard algorithms (19) gave the same results as molecular haplotyping, except in a single subject because of a discrepancy at one SNP position.
Phylogenetic analysis (Fig. 1) was carried out by the minimum-spanning network algorithm (20). With this approach, every haplotype is connected to the haplotype(s) most similar to itself. This method allows multifurcations (i.e., not just binary branching) and reticulations appropriate for allelic sequence data. Reticulations, in particular, often suggest recombination events or evolutionarily recombinant haplotypes. An analysis of the β2AR haplotypes revealed a deep divergence of haplotype 2 as well as potential evolutionary recombination events. Haplotype 12 appears to be a recombinant between haplotypes 1 and 2. Also, haplotype 8 is best explained as a recombination between the highly frequent haplotypes 4 and 6. Results from estimations of linkage disequilibrium (Δ) for our largest sample, Caucasians, are shown in Fig. 2. Although many of the sites are in strong disequilibrium, it is clear that some pairs of close sites have reduced levels of linkage disequilibrium relative to more distantly spaced pairs of sites. No individual SNP adequately predicted these complex haplotypes. This finding illustrates the hazards inherent in randomly selecting an individual SNP as a surrogate marker for a haplotype.
Figure 1.
Phylogeny of β2AR haplotypes. Each haplotype is represented by a circle whose area represents the overall frequency of that haplotype in the sample. Lines connecting haplotypes are solid black for single-site differences, solid blue for two-site differences, and dashed for more than two differences. Each circle is subdivided to show the proportion of the individual haplotype frequency found in each of the four population groups as represented by the indicated colors.
Figure 2.
Linkage disequilibrium between β2AR SNPs. Genotypes from the Caucasian samples were determined at the 13 loci shown in Table 1 and the degree of linkage disequilibrium (Δ) between SNPs was calculated. Results are color coded as shown. The site at −406 was monomorphic in the Caucasian sample.
Having identified β2AR haplotypes in the population, we next explored the predictive value of haplotypes in the response to albuterol. In the asthmatic cohort, the two rare SNPs at −709 and −406 were not found. (For purposes of consistency, the haplotypes discussed below continue to list these positions although they were invariant in this cohort.) No other differences were found in the frequencies of the haplotypes between the index and the asthmatic population. The haplotypes were assembled as pairs, and the 18 haplotype pairs that were found in the asthmatic cohort are shown in Table 2. The five most common haplotype pairs represent almost 88% of the asthmatic cohort. Haplotypes observed in <1% of the cohort that were single-nucleotide derivatives of another, more frequent haplotype were collapsed into the more frequent haplotype if the single-nucleotide difference was unique to the rare haplotype. And, for purposes of analysis, the final data set excluded haplotype pairs that were observed in <5% of the cohort. The responsiveness to the β agonist albuterol for individuals with the five common haplotype pairs is shown in Fig. 3. Haplotype pair was significantly related to improvements in FEV1 (P = 0.007 from ANCOVA). To delineate which pairs differ from one another, comparisons were made for haplotype pair 4/4 (which had the lowest response) and haplotype 4/6 (which had the highest response) versus the other haplotype pairs. Pairwise tests were made, correcting for multiple comparisons. These results showed that the responses of those with haplotype pairs 4/4 and 4/6 were highly significant (change percentage FEV1 = 8.53 ± 1.78 vs. 19.1 ± 2.79; P = 0.008). Significant differences also were found between those with pairs 2/4 vs. 4/6 and 2/2 vs. 4/4 (P = 0.036 and 0.046, respectively). In contrast to these results with haplotypes, we found no association between the response to albuterol and any individual SNP. For this analysis, a similar ANCOVA model as above was used. The P values for each SNP were all substantially >0.05 (adjusted for multiple comparisons). Based on this in vivo data, it appears that haplotype 4 is associated with depressed responsiveness and haplotype 2 with increased responsiveness. Because 0, 1, or 2 copies of these two haplotypes are present in our population as haplotype pairs 2/2, 2/4, and 4/4, a potential gene-dose effect can be assessed by regression analysis. Such an analysis indeed showed a highly significant relationship between copy number of haplotype 2 (or 4) and the response to albuterol (P = 0.009). It would be of interest to carry out a similar analysis with haplotype 6, however no homozygous individuals (haplotype pair 6/6) were found in our Caucasian cohort. We would expect that these individuals would have an enhanced response as compared to those with 4/6. Interestingly, in regards to haplotype 2, the 2/6 and 2/2 responses were not significantly different.
Table 2.
β2AR haplotype pairs found in the asthmatic cohort
| Haplotype pair | Chromosome A haplotype | Chromosome B haplotype | n | % | |
|---|---|---|---|---|---|
| 2/4 | A C G G C C C C G G G C C | / | G C A C C T T T A C G C C | 37 | 30.6 |
| 2/2 | A C G G C C C C G G C C | / | A C G G C C C C G G G C C | 25 | 20.7 |
| 2/6 | A C G G C C C C G G G C C | / | G C G C C T T T G C A C A | 22 | 18.2 |
| 4/4 | G C A C C T T T A C G C C | / | G C A C C T T T A C G C C | 14 | 11.6 |
| 4/6 | G C A C C T T T A C G C C | / | G C G C C T T T G C A C A | 8 | 6.6 |
| 2/5 | A C G G C C C C G G G C C | / | G C A C C T T T G C G C C | 2 | 1.7 |
| 4/10 | G C A C C T T T A C G C C | / | G C G C C T T T G C A C C | 2 | 1.7 |
| 1/4 | A C G C C T T T A C G C C | / | G C A C C T T T A C G C C | 1 | 0.8 |
| 1/6 | A C G C C T T T A C G C C | / | G C G C C T T T G C A C A | 1 | 0.8 |
| 2/11 | A C G G C C C C G G G C C | / | G C G C C T T T G C G C C | 1 | 0.8 |
| 2/3 | A C G G C C C C G G G C C | / | G A A C C T T T A C G C C | 1 | 0.8 |
| 2/7 | A C G G C C C C G G G C C | / | G C G C C T T T G C A T A | 1 | 0.8 |
| 2/8 | A C G G C C C C G G G C C | / | G C A C C T T T A C A C A | 1 | 0.8 |
| 3/4 | G A A C C T T T A C G C C | / | G C A C C T T T A C G C C | 1 | 0.8 |
| 4/5 | G C A C C T T T A C G C C | / | G C A C C T T T G C G C C | 1 | 0.8 |
| 4/7 | G C A C C T T T A C G C C | / | G C G C C T T T G C A T A | 1 | 0.8 |
| 4/8 | G C A C C T T T A C G C C | / | G C A C C T T T A C A C A | 1 | 0.8 |
| 6/7 | G C G C C T T T G C A C A | / | G C G C C T T T G C A T A | 1 | 0.8 |
Nucleotide positions are omitted for clarity but are the same as in Table 1. Chromosomes A and B are arbitrarily assigned.
Figure 3.
In vivo responses to β2AR agonist depend on β2AR haplotypes. Shown are data from 121 subjects whose FEV1s were measured before and after inhalation of the agonist albuterol as described in Methods. The response was significantly related to β2AR genotype (P = 0.007 by ANCOVA). See text for additional statistical analysis.
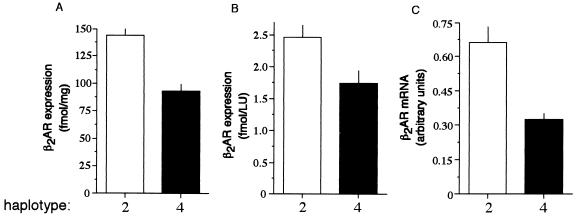
The two homozygous haplotype pairs, 4/4 and 2/2, were common in our population and displayed significant differences in the in vivo response to the agonist albuterol. To determine whether the SNPs within these two haplotypes result in different levels of β2AR mRNA or protein expression, transfection studies were carried out in the human embryonic kidney cell line HEK293. These cells were chosen because of their high transfection efficiency, their human origin, and the fact that they express β2AR (≈10 fmol/mg) and, thus, presumably have the relevant transcription factors for expression of the human gene. The constructs used for transfection lacked the typical eukaryotic promoters, but instead used the two β2AR 5′ UTR haplotypes as the promoter in the same context as is found in the native gene. As might be expected, the levels of expression with the β2AR promoter were significantly less than what we have previously reported with viral promoters (27). Nevertheless, the levels obtained (≈100 fmol/mg receptor by radioligand binding) were clearly above background and, in fact, are similar to β2AR expression in the lung (5). As shown in Fig. 4, the level of β2AR expression was clearly different between haplotypes 2 and 4. When the construct consisting of haplotype 2 was used, β2AR expression was 144 ± 12.8 fmol/mg as compared with 93.6 ± 5.7 fmol/mg when the haplotype 4 construct was used (P < 0.005). When corrected for transfection efficiency by quantitating luciferase activity derived from coexpression of a luciferase construct, the differences in expression of the β2AR (fmol/LU) between the two haplotypes remained (Fig. 4B). β2AR mRNA levels were determined by quantitative RNase protection assays (Fig. 4C). The β2AR mRNA levels for the haplotype 2-transfected cells were consistently higher than those of the haplotype 4-transfected cells (0.663 ± 0.067 vs. 0.320 ± 0.024 arbitrary units; P < 0.005). Of note, in HEK293 cells, increases in β2AR do not result in proportional increases in agonist-stimulated adenylyl cyclase activities (27). Thus, although a 50% increase in expression would not be expected to result in increases in adenylyl cyclase in these cells, such an increase in β2AR is considered highly significant in physiologically relevant cell types (28). The above results for both protein and mRNA expression are entirely consistent with the in vivo findings, where individuals with haplotype pair 2/2 had a ≈50% greater response than did those with haplotype pair 4/4 (Fig. 3).
Figure 4.
β2AR haplotype determines receptor transcript and protein expression. HEK293 cells were transiently transfected with the indicated β2AR haplotypic vector and a luciferase vector (to control for transfection efficiency) as described in Methods. β2AR protein density was determined by 125I-cyanopindolol radioligand binding and normalized to membrane protein (A) or luciferase expression (B). β2AR mRNA levels were determined by quantitative ribonuclease protection assays. Shown are results from four to seven experiments. Haplotype 4 expression was less than haplotype 2 at P < 0.005.
Comparisons of the sequence of haplotypes 2 and 4 reveal eight differences in the 13 SNP positions. These include differences in amino acid 19 of the BUP and in amino acids 16 and 27 of the receptor protein. Each of these, studied in isolation, has been shown to alter expression or trafficking of the receptor (4, 7), but the effects of the various SNP combinations at these loci have not previously been explored. And, indeed, our current results with haplotypes are different from those previously obtained with individual SNPs taken out of context of a verified haplotype. For example, based on our previous work with the BUP SNP (5′-leader cistron, position −47) studied in isolation, we would have predicted that the T (cys) allele would be associated with higher expression. In that study, however, the BUP SNPs were within the context of the Gly16 (G at position 46) and Glu27 (G at position 79) alleles, which, as shown in Table 1, were never found in combination with T (cys) in position −47. This finding emphasizes the importance of studying SNPs in vitro within the context of a validated haplotype. The SNPs at the other five loci that differ between haplotypes 2 and 4 are at positions −1,023, −654, −468, −367, and −20. A database search for transcription factor binding sites (29) shows that these SNPs are located within, or closely flank, a number of potential cis-acting elements. For example, the SNP at −1,023 flanks potential binding sites for AP-4 and C/EBP; the SNPs at −654 and −468 each flank an NF-1 consensus sequence; and the SNP at −367 is within a CP2 consensus sequence. Of note, the synonymous SNP at position 523 that has been associated with altered responsiveness to albuterol in Japanese families (14) was invariant between haplotypes 2 and 4. However, this SNP along with several others distinguishes one common haplotype (haplotype 6) from the others. And haplotype 6 appears to have an effect on response (Fig. 3). Whether a smaller subset of β2AR SNPs defines the cellular expression phenotype cannot be ascertained from the current molecular approach because it would require systematic construction of vectors representing many unique haplotypes. Because a large fraction of these would in fact be rare (or never found) in the human population, we have taken the approach of restricting our examination to the common haplotypes, because ultimately these are most relevant to pharmacogenetics. However, based on the results of the current in vivo responsiveness studies, the cell transfection experiments, and previous studies with isolated SNPs (4, 7), it is likely that the biologic phenotype is directed by an interaction involving transcription, translation, and protein processing that ultimately defines the effect of these haplotypes.
In summary, we have identified 13 SNPs in a contiguous region of the 5′ upstream and coding sequence of the β2AR in humans. Twelve distinct haplotypes were represented in a population of four major ethnic groups. Several relatively recent recombination events seem to be responsible for some haplotypes. A striking divergence in ethnic distribution was found for several haplotypes. Five haplotype pairs were common in asthmatics, and there were clear differences in the in vivo response to a β2AR agonist based on haplotype pair. In contrast, no isolated SNP had any predictive utility. The homozygous haplotypes 2/2 and 4/4 with divergent agonist efficacies were shown to have differential effects on β2AR gene and protein expression in vitro, consistent with the direction and magnitude of the in vivo responses. These findings are a delineation of phylogeny, and in vivo and in vitro relevance, of haplotypic combinations of SNPs. The results indicate that the unique interactions of multiple SNPs within a haplotype ultimately affect biologic and therapeutic phenotype and that individual SNPs may have poor predictive power as pharmacogenetic loci.
Acknowledgments
We thank Jonathan Bernstein for providing some of the patients and Melenie Meyers for data collection. This work was supported by National Institutes of Health Grants HL45967, HL03986, and ES06096. The member who communicated this paper serves on the Science Advisory Board of Genaissance and holds stock in the company.
Abbreviations
- β2AR
β2-adrenergic receptor
- SNP
single-nucleotide polymorphism
- FEV1
forced expiratory volume in 1 sec
- ANCOVA
analysis of covariance
- BUP
β2AR upstream peptide
- UTR
untranslated region
References
- 1.Reihsaus E, Innis M, MacIntyre N, Liggett S B. Am J Respir Cell Mol Biol. 1993;8:334–339. doi: 10.1165/ajrcmb/8.3.334. [DOI] [PubMed] [Google Scholar]
- 2.Green S A, Cole G, Jacinto M, Innis M, Liggett S B. J Biol Chem. 1993;268:23116–23121. [PubMed] [Google Scholar]
- 3.Turki J, Lorenz J N, Green S A, Donnelly E T, Jacinto M, Liggett S B. Proc Natl Acad Sci USA. 1996;93:10483–10488. doi: 10.1073/pnas.93.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green S, Turki J, Innis M, Liggett S B. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 5.Green S A, Turki J, Bejarano P, Hall I P, Liggett S B. Am J Respir Cell Mol Biol. 1995;13:25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 6.Parola A L, Kobilka B K. J Biol Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 7.McGraw D W, Forbes S L, Kramer L A, Liggett S B. J Clin Invest. 1998;102:1927–1932. doi: 10.1172/JCI4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott M G H, Swan C, Wheatley A P, Hall I P. Br J Pharmacol. 1999;126:841–844. doi: 10.1038/sj.bjp.0702385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green S A, Liggett S B. In: The Genetics of Asthma. Liggett S B, Meyers D, editors. New York: Marcel Dekker; 1996. pp. 67–90. [Google Scholar]
- 10.Drazen J M, Israel E, Boushey H A, Chinchilli V M, Fahy J V, Fish J E, Lazarus S C, Lemanske R F, Martin R J, Peters S P, et al. N Engl J Med. 1996;335:841–847. doi: 10.1056/NEJM199609193351202. [DOI] [PubMed] [Google Scholar]
- 11.Liggett S B. In: The Genetics of Asthma. Liggett S B, Meyers D A, editors. New York: Marcel Dekker; 1996. pp. 455–478. [Google Scholar]
- 12.Martinez F D, Graves P E, Baldini M, Solomon S, Erickson R. J Clin Invest. 1997;100:3184–3188. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima J J, Thomason D B, Mohamed M H N, Eberle L V, Self T H, Johnson J A. Clin Pharmacol Ther. 1999;65:519–525. doi: 10.1016/S0009-9236(99)70071-8. [DOI] [PubMed] [Google Scholar]
- 14.Ohe M, Munakata M, Hizawa N, Itoh A, Doi I, Yamaguchi E, Homma Y, Kawakami Y. Thorax. 1995;50:353–359. doi: 10.1136/thx.50.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S, Hall I P, Dewar J, Dow E, Lipworth B. Lancet. 1997;350:995–999. doi: 10.1016/S0140-6736(97)03211-X. [DOI] [PubMed] [Google Scholar]
- 16.Israel E, Drazen J M, Liggett S B, Boushey H A, Cherniack R M, Chinchilli V M, Cooper D M, Fahy J V, Fish J E, Ford J G, et al. Am J Respir Crit Care Med. 2000;162:75–80. doi: 10.1164/ajrccm.162.1.9907092. [DOI] [PubMed] [Google Scholar]
- 17.Lipworth B J, Hall I P, Aziz I, Tan K S, Wheatley A. Clin Sci. 1999;96:253–259. [PubMed] [Google Scholar]
- 18.McGraw D W, Forbes S L, Witte D P, Fortner C N, Paul R J, Liggett S B. J Biol Chem. 1999;274:32241–32247. doi: 10.1074/jbc.274.45.32241. [DOI] [PubMed] [Google Scholar]
- 19.Clark A G. Mol Biol Evol. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- 20.Excoffier L, Smouse P E, Quattro J M. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill W G, Robertson A. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 22.Hill W G, Weir B S. Am J Hum Genet. 1994;54:705–714. [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Galinsky R E, Bernstein J A, Liggett S B, Weinshilboum R M. Pharmacogenetics. 2000;10:261–266. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Morris J F, Koski A, Johnson L C. Am Rev Respir Dis. 1971;103:57–67. doi: 10.1164/arrd.1971.103.1.57. [DOI] [PubMed] [Google Scholar]
- 25.Dales R E, Spitzer W O, Tousignant P, Schechter M, Suissa S. Am Rev Respir Dis. 1988;138:317–320. doi: 10.1164/ajrccm/138.2.317. [DOI] [PubMed] [Google Scholar]
- 26.Ludbrook J. Clin Exp Pharmacol Physiol. 1998;25:1032–1037. doi: 10.1111/j.1440-1681.1998.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 27.Tepe N M, Liggett S B. J Recept Signal Transduction Res. 2000;20:75–85. doi: 10.3109/10799890009150038. [DOI] [PubMed] [Google Scholar]
- 28.Barnes P J. Am J Respir Crit Care Med. 1995;152:838–860. doi: 10.1164/ajrccm.152.3.7663795. [DOI] [PubMed] [Google Scholar]
- 29.Heinemeyer T, Chen X, Karas H, Kel A E, Kel O V, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]