Abstract
Cytosolic NADPH can be directly oxidized by a calcium-dependent NADPH dehydrogenase, NDB1, present in the plant mitochondrial electron transport chain. However, little is known regarding the impact of modified cytosolic NADPH reduction levels on growth and metabolism. Nicotiana sylvestris plants overexpressing potato (Solanum tuberosum) NDB1 displayed early bolting, whereas sense suppression of the same gene led to delayed bolting, with consequential changes in flowering time. The phenotype was dependent on light irradiance but not linked to any change in biomass accumulation. Whereas the leaf NADPH/NADP+ ratio was unaffected, the stem NADPH/NADP+ ratio was altered following the genetic modification and strongly correlated with the bolting phenotype. Metabolic profiling of the stem showed that the NADP(H) change affected relatively few, albeit central, metabolites, including 2-oxoglutarate, glutamate, ascorbate, sugars, and hexose-phosphates. Consistent with the phenotype, the modified NDB1 level also affected the expression of putative floral meristem identity genes of the SQUAMOSA and LEAFY types. Further evidence for involvement of the NADPH redox in stem development was seen in the distinct decrease in the stem apex NADPH/NADP+ ratio during bolting. Additionally, the potato NDB1 protein was specifically detected in mitochondria, and a survey of its abundance in major organs revealed that the highest levels are found in green stems. These results thus strongly suggest that NDB1 in the mitochondrial electron transport chain can, by modifying cell redox levels, specifically affect developmental processes.
Plant mitochondria possess an electron transport chain (ETC) that in known complexity supersedes that of other eukaryotes (Rasmusson et al., 2008). In particular, a setup of non-proton-pumping plant type II NAD(P)H dehydrogenases and alternative oxidases bypasses the standard proton-pumping protein complexes of oxidative phosphorylation and, as a consequence, ATP formation (Vanlerberghe and McIntosh, 1997; Rasmusson et al., 2004). Additionally, the uncoupling protein, which is present in both plants and mammals, can directly bypass the ATP synthase itself (Vercesi et al., 2006).
The type II NAD(P)H dehydrogenases are located on the internal and external sides of the inner mitochondrial membrane and oxidize NADH or NADPH from the mitochondrial matrix or the cytosol. These reactions bypass the first energy conservation step maintained by the proton-pumping complex I, which oxidizes matrix NADH. Among the type II NAD(P)H dehydrogenases, NDB proteins of potato (Solanum tuberosum) and Arabidopsis (Arabidopsis thaliana) are present on the external surface of the inner mitochondrial membrane (Rasmusson et al., 1999; Elhafez et al., 2006). The NDB proteins characteristically possess an inserted domain carrying more or less conserved EF-hand motifs for calcium binding (Michalecka et al., 2003). Upon analyses of cytoplasmic membranes of Escherichia coli producing Arabidopsis NDB fusion proteins, AtNDB2 and AtNDB4 oxidized NADH but not NADPH, and calcium ions bound to and stimulated AtNDB2 but not AtNDB4 (Geisler et al., 2007). This was consistent with the generally observed partial calcium dependence of external NADH oxidation (Rasmusson et al., 2004). In contrast, potato StNDB1 expressed in Nicotiana sylvestris catalyzed calcium-dependent NADPH oxidation (Michalecka et al., 2004). Consistently, the E. coli-expressed AtNDB1 was shown to be specific for NADPH and to bind calcium via its EF-hand domain as a prerequisite for its activity (Geisler et al., 2007).
External NADPH-specific type II dehydrogenases have to date been reported in the ETC of plants (Michalecka et al., 2004) and the filamentous fungus Neurospora crassa (Melo et al., 2001) but are absent in yeasts and animals. This indicates that the enzyme has specific physiological roles, which are not required in all eukaryotes. Given that it is not directly linked to energy conservation, NDB1, like other type II NAD(P)H dehydrogenases, may be able to modulate cellular NADP(H) redox levels, irrespective of the cellular ATP phosphorylation status. In N. sylvestris overexpressing StNDB1 and thus having elevated levels of mitochondrial NADPH dehydrogenase, a decrease in NADPH/NADP+ ratio independent of the NADH/NAD+ ratio was observed in leaves during the day. This provided a physiological demonstration in which the NDB1 enzyme was active and able to specifically modify the cellular NADP(H) pools (Liu et al., 2008). However, under the growth conditions used, no visual phenotype was observed, and it is hitherto not known what the physiological importance of mitochondrial NADPH oxidation is.
Here, we report that when grown under higher light intensities, N. sylvestris plants overexpressing StNDB1 exhibit an earlier transition from rosette stage to bolting, whereas a line suppressing both StNDB1 and NsNDB1 is delayed in this parameter. The phenotype was strongly correlated with stem-specific changes in NADPH reduction levels. These changes were in turn correlated to the levels of a relatively small set of metabolites and affected the expression of genes associated with floral phase transitions. Specific changes in the wild-type apical NADPH/NADP+ ratio during bolting as well as a high abundance of the StNDB1 protein in potato stem mitochondria provide further support for an important role of mitochondrial NADPH oxidation in stem development.
RESULTS
Overexpression and Suppression of StNDB1 Induce Bolting Phenotypes in N. sylvestris
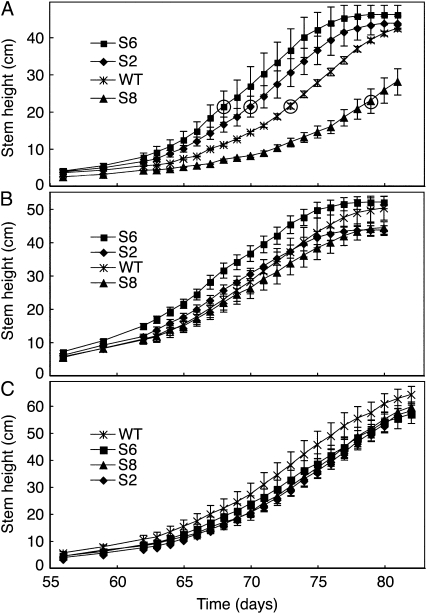
Transformation of N. sylvestris with StNDB1 in the sense orientation has allowed the isolation of homozygous lines S2 and S6 that show a high abundance of StNDB1 mRNA, encoded protein, and associated external NADPH oxidation. In contrast, line S8, which carries the same construct, displays low mRNA levels for StNDB1 and NsNDB1 coupled to hardly detectable protein and activity levels, indicative of sense cosuppression (Michalecka et al., 2004; Liu et al., 2008). When growing these plants at a higher light intensity (500 μmol m−2 s−1), we noted an obvious variation in the timing of bolting, with the overexpressors S2 and S6 repeatedly bolting earlier and the suppressor line S8 bolting later than wild-type plants (Fig. 1). We additionally compared hemizygous and homozygous S8 plants grown at 500 μmol m−2 s−1. They displayed similar times for bolting, with both being significantly later than the wild type, consistent with NDB1 suppression being a dominant trait (Liu et al., 2008). At 200 μmol m−2 s−1 light, all lines developed identically, whereas at intermediate light, a pattern similar to the situation at higher light was discernible, although not statistically significant (Fig. 1). For plants grown at 500 μmol m−2 s−1, dry weight determinations of root and shoot showed that biomass accumulation was unaffected (Supplemental Fig. S1), indicating that the phenotype was not a consequence of a perturbation of metabolic efficiency. The stem extension rate was similar in all lines (Supplemental Fig. S2), and the difference in bolting time between the lines persisted as a similar variation in flowering time (Fig. 1A). These results thus suggest that the principal effect was on the timing of the transition of the stem apex from rosette stage to bolting.
Figure 1.
Time course for shoot growth of N. sylvestris plants. Plants were grown at 23°C under 16 h of light per day at 500 (A), 300 (B), and 200 (C) μmol m−2 s−1. Stem height is presented as means ± se for three plants (A) or six plants (B and C). Median day for appearance of first flower is denoted by circled symbols in A. WT, Wild type.
NADPH/NADP+ Ratios Are Specifically Modified in the Stem
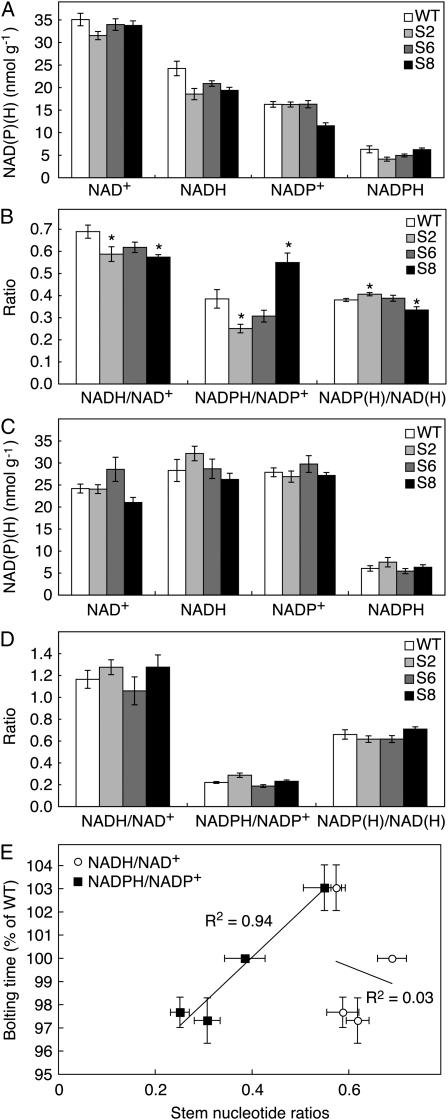
Plants grown at 500 μmol m−2 s−1 light were analyzed for changes in tissue NAD(P)(H) levels (Fig. 2). In leaves, no difference could be observed between the transgenic lines for the NAD(H) and NADP(H) redox couple, regarding neither reduction levels nor total amount of each nucleotide. However, in stems, the NADPH/NADP+ ratio was lowered in the overexpressor lines and increased in the suppressor line S8. The changes in lines S2 and S8 were significant at P < 0.05. The total amounts of NADP(H) and NAD(H) were little changed, whereas both the overexpression lines and the suppression line had somewhat lowered NADH/NAD+ ratios. The results were investigated in relation to the bolting phenotype as seen over four separate growth experiments. Some variation existed in the number of days needed for the wild type to bolt, and the phenotypic variation was most consistent when expressed as relative difference to the wild type. The reduction level of NADP(H) in the different transgenic lines was strongly correlated to bolting time (Fig. 2E), whereas the NAD(H) reduction level varied independently from bolting time.
Figure 2.
Pyridine nucleotide contents and ratios in N. sylvestris plants. Plants were grown at 500 μmol m−2 s−1 and sampled 4 h into the light period. A and B, Stems were taken from 7-week-old plants. C and D, Mature leaf samples were taken from 6-week-old plants. Values are means ± se for six to seven plants. For ratios (B and D), significant differences from the wild type (WT; P < 0.05) are denoted by asterisks. E, Correlation of stem NADPH/NADP+ ratio to bolting time. Bolting time was measured as the time required for plants to grow to 8 cm in height and is expressed as percentage of the time needed for the wild type in each experiment. On the y axis, error bars denote se of four independent experiments, each comprising three to six plants. On the x axis, error bars are as in A to D. r2 values are given by the linear regression lines.
Metabolic Profiles Display Tissue Specificity and Distinct Domains of NADP(H)-Metabolite Correlations
The differences in stem NADPH/NADP+ ratio in the transgenic lines constituted an almost 2-fold variation. In order to analyze for consequences of modified NADPH levels, the same tissue samples that were used for NAD(P)(H) analyses were also subjected to metabolic profiling, rendering levels for 42 and 46 hydrophilic metabolites in stem and leaf, respectively (Supplemental Tables S1 and S2). In stems, significant changes included decreased Glu and trehalose in the overexpression lines S2 and S6 and decreased Pro in S2 (Fig. 3). Regarding these metabolites, the suppression line S8 was similar to the wild type. In contrast, 2-oxoglutarate and tyramine were lower in the S8 line, whereas the overexpression lines were similar to the wild type. Ascorbate was lower in S8, but also in S6 a lower value was observed, although in this instance it was not significantly different from the wild type. For Ala and Ile, no clear pattern could be distinguished in relation to the transgenic modification. In leaves, several metabolites were present in amounts varying significantly from the wild type in at least one transgenic line (Fig. 3). However, with the exception of citramalate and maltose, which were lowered in at least one overexpression line but not in S8, the changes in overexpressors and the suppression line were not consistent with the genetic modification. For example, shikimate was significantly increased in lines S2, S6, and S8.
Figure 3.
Metabolite changes in stem and leaf. The tissue samples used in Figure 2 were analyzed by metabolic profiling. The figure shows levels of the metabolites in stem (A) and leaf (B) for which a significant (P < 0.05) difference from the wild type (WT) was seen in any transgenic line (asterisks). The transgenic lines are ordered according to the NADPH/NADP+ ratio in Figure 2. Error bars denote se for six plants. 2-OG, 2-Oxoglutarate; Asc., ascorbate; Tyra., tyramine; Treh., trehalose; Citramal., citramalate; Mal., malate; Sorb. sorbitol; Fruc., fructose; Malt., maltose; Shik., shikimate.
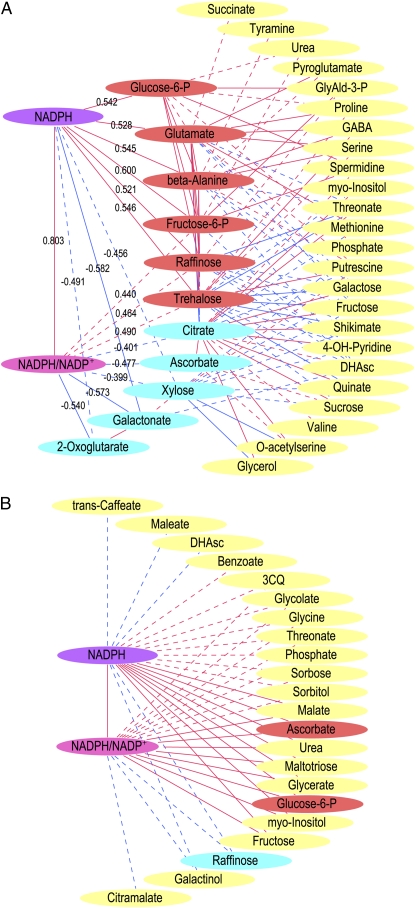
In order to account also for plant-to-plant variation in the analysis, we determined Pearson coefficients and significances for the correlation of both the NADPH/NADP+ ratio and the absolute NADPH level to the quantified metabolites. Figure 4A displays the stem metabolites that correlated significantly to NADP(H) as well as those that correlated to these metabolites in turn. Eleven metabolites correlated to NADPH/NADP+ and/or NADPH, including hexose-Ps and sugars, intermediates of the citric acid cycle and ascorbate metabolism, and amino acids. Consistent with the analysis of Figure 3, positive correlations to Glu and trehalose and negative correlations to 2-oxoglutarate and ascorbate were observed. An obvious stem-specific pattern seen was that 2-oxoglutarate, galactonate, and ascorbate correlated negatively to the NADPH/NADP+ ratio but to very few of the other metabolites analyzed.
Figure 4.
Correlation of NADP(H) parameters to metabolite levels. For stem (A) and leaf (B), correlation was determined by Pearson analyses on a plant-by-plant basis between NADP(H) and metabolite profile data. In A and B, metabolites correlated to the NADPH/NADP+ ratios and/or NADPH levels are shown. Additionally in A, correlations between these metabolites and other metabolites are denoted, but these were for clarity omitted in B. Pearson coefficients are denoted for the direct correlations in A. Connecting lines denote significant correlation and are continuous for P < 0.01 and dashed for P < 0.05. Red and blue lines denote positive and negative correlations, respectively. Nodes for metabolites correlating to NADP(H) in A are colored for positive (red) or negative (blue) correlation. Other metabolite nodes are yellow. 3CQ, Cis-3-caffeolyl quinate; DHAsc, dehydroascorbate; GABA, γ-amino butyrate; GlyAld-3-P, glyceraldehyde-3-P.
Leaves displayed a correlation pattern highly different from stems (Fig. 4B). Hexose-P, especially Glc-6-P, was positively correlated to NADP(H) in both organs. However, raffinose was positively correlated to NADP(H) in stem but negative in leaves, and the opposite pattern was seen for ascorbate. In leaves, 20 additional metabolites correlated to NADPH/NADP+ and/or NADPH, including intermediates from both primary and intermediary metabolism. These results thus show that the integration of NADP(H) into the metabolic systems is completely different in stem and leaf.
The Overexpression and Suppression of StNDB1 Induce Up- and Down-Regulation of Flowering-Associated Genes
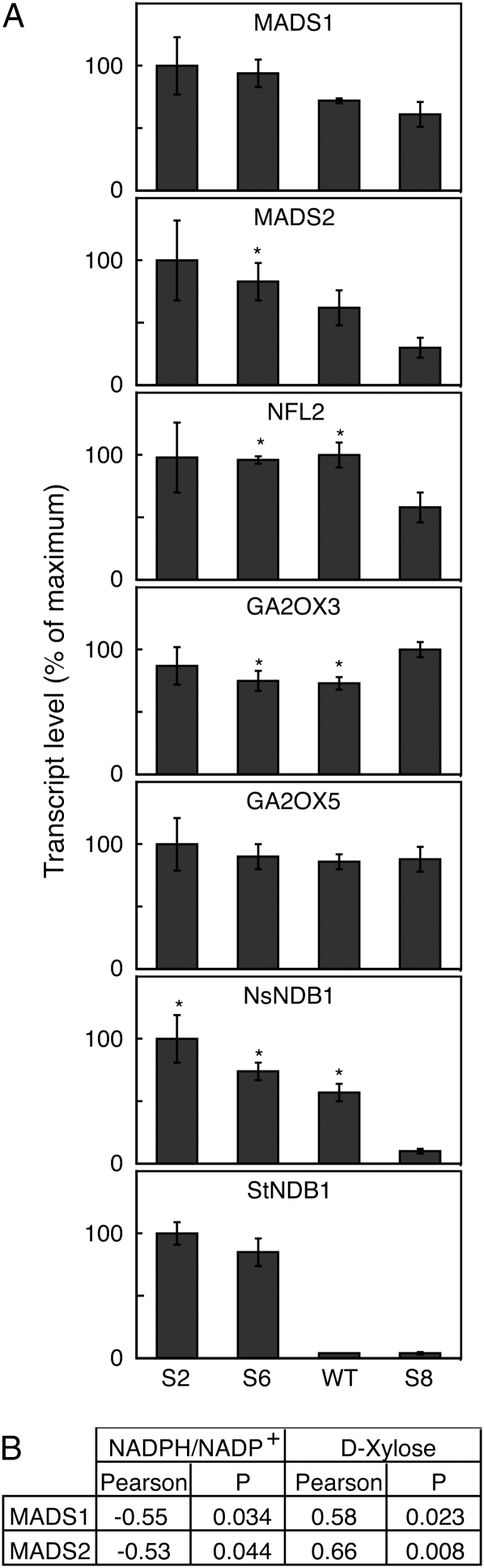
To determine the mode of influence of the observed redox changes on bolting, we analyzed gene expression in stems from the transgenic plants by real-time reverse transcription (RT)-PCR. The transcript levels for StNDB1 and NsNDB1 were consistent with previous results for leaves (Liu et al., 2008), confirming the overexpression of StNDB1 in lines S2 and S6 as well as the suppression of both genes in line S8 in stem tissue (Fig. 5). On searching of databases, we found two floral induction-associated MADS box genes in N. sylvestris (Jang et al., 1999, 2002; Gallego-Giraldo et al., 2007). The NsMADS2 transcript was 3-fold higher in the overexpressor lines (S2 and S6) than in the suppressor line (S8), with the wild type being intermediate (Fig. 5). A similar but weaker trend was observed for NsMADS1. We also used primers against NtNFL2, a homolog of Arabidopsis LEAFY present in Nicotiana tabacum but originating from the N. sylvestris parent (Kelly et al., 1995). Transcript levels for NFL2 were lower in S8 as compared with the other lines. The expression of NsMADS1 and NsMADS2 was significantly correlated to the NADPH/NADP+ ratio (Fig. 5B). This suggests that a lower stem NADPH/NADP+ ratio promotes the expression of floral phase transition genes, inducing the observed phenotype. To investigate if any of the metabolites correlating to NADPH levels (Fig. 4A) were likely to mediate the effect, we additionally performed Pearson analyses for these, which revealed correlation of NsMADS1 and NsMADS2 to d-Xyl but not to the other metabolites (Fig. 5B).
Figure 5.
Gene expression changes in stems of N. sylvestris transgenic lines. A, RNA was analyzed by real-time RT-PCR for expression of floral induction-associated (NFL2, MADS1, and MADS2) and GA-induced (GA2OX3 and GA2OX5) genes. The figure shows average relative mRNA levels ± se for four plants, except for S2 (three plants). Significant differences from the S8 line are denoted by asterisks, except for the transgene StNDB1. B, Significant correlations of MADS1 and MADS2 expression to NADPH/NADP+ ratio and d-Xyl are denoted as Pearson coefficients and P values over 15 individual plants. WT, Wild type.
GAs induce bolting and floral phase transitions in plants, including the rosette plant N. sylvestris (Lee and Zeevaart, 2005). Also, ascorbate and 2-oxoglutarate, which were observed to vary in the stems of the transgenic lines, are cofactors in GA biosynthesis. Therefore, we analyzed gene expression with primers against N. tabacum GA2OX3 and GA2OX5, which are induced by GA in N. tabacum (Gallego-Giraldo et al., 2008). Both genes were unresponsive in the transgenic lines, except for a higher GA2OX3 expression in S8 (Fig. 5). This indicates that GA levels were unchanged in the overexpressors but may be elevated in line S8, which would have partially counteracted the late-bolting phenotype observed in S8. The relative unresponsiveness of GA-induced genes was consistent with the fact that S8 bolted significantly later than S6 also when grown with the GA synthesis inhibitor paclobutrazol. For four plants, the average bolting time with paclobutrazol was 56.7 ± 0.6 and 52.9 ± 1.2 d for S8 and S6, respectively. Thus, the observed phenotypic change was not mediated by GA.
NADP(H) Reduction Varies Temporally and Spatially in the Stem Apex
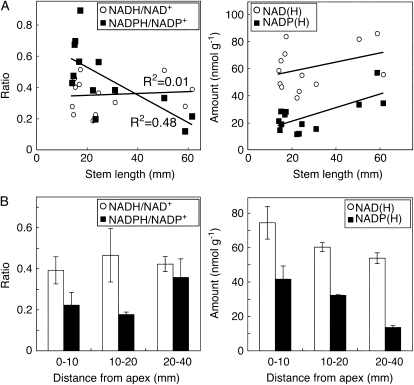
To further investigate the importance of cellular NADPH reduction level during bolting, we compared the apical 10-mm parts of wild-type stems at different stages (Fig. 6A). A 3-fold decrease in the NADPH/NADP+ ratio was observed during the growth of the stem from 15 mm (i.e. just before bolting) to 60 mm length, whereas NADH/NAD+ ratio was unchanged. In the same developmental span, the total amounts of NADP(H) in the apex increased, whereas the NAD(H) amount was little affected.
Figure 6.
Changes in pyridine nucleotides during bolting of wild-type plants. Plants were sown at different times and grown at 500 μmol m−2 s−1. A, The top 10 mm of the stem of plants with different heights was analyzed for ratios and contents of pyridine nucleotides. For each data point, stem apices from four plants were pooled. r2 values were determined by linear regression. B, The distribution of nucleotides in different parts of 5- to 6-cm-long wild-type stems was determined after dissection. Error bars denote se for three pools of four plants.
The spatial distribution of nucleotides and reduction levels was investigated in 50- to 60-mm-high stems that were dissected before extraction. NAD(H) total amount and reduction levels were relatively similar over the investigated length (Fig. 6B). However, the total amounts of NADP(H) were concentrated to the apex, whereas the segment 20 to 40 mm below the apex contained only a small part of the total NADP(H). In the same extracts, the NADPH/NADP+ ratio displayed somewhat higher values in the basal parts.
StNDB1 Is Highly Abundant in Mitochondria Purified from Green Stems
We wanted to investigate if the observed effects of NDB1 in the stem were mirrored in the expression pattern of the encoding gene. The StNDB1 transcript is unresponsive to several treatments in potato leaves (Svensson and Rasmusson, 2001; Svensson et al., 2002; Geisler et al., 2004), and the Arabidopsis ortholog (AtNDB1; At4g28220) is stably expressed upon stress treatments of cell cultures (Clifton et al., 2005). Comparison of the Arabidopsis ecotype Columbia-0 arrays present in Genevestigator (https://www.genevestigator.ethz.ch) revealed expression of AtNDB1 in most tissues, including the apical meristem. For 1,357 arrays, the standard deviation of the signal was 35% of average signal intensity for AtNDB1 but 101% for both the energy-bypass genes AtNDB2 and AtAOX1a. Thus, the AtNDB1 transcript appears relatively unresponsive over a wide range of stresses and conditions.
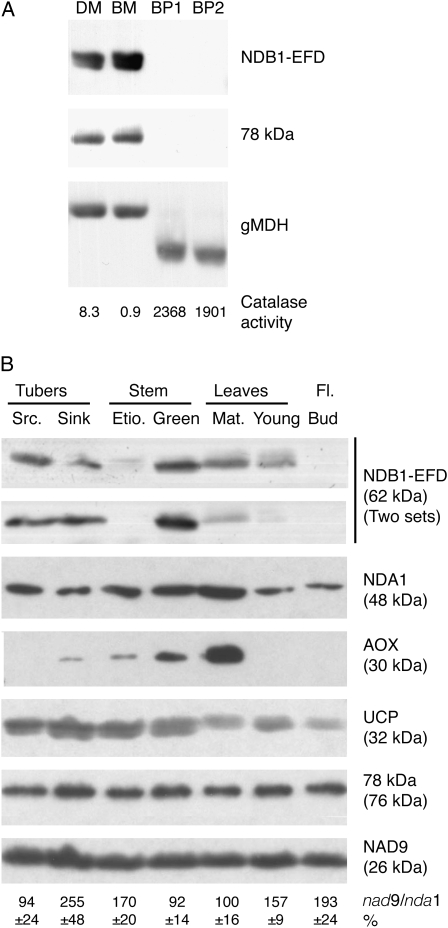
We previously reported the use in western blotting of an antiserum specifically detecting StNDB1 but not N. sylvestris homologs (Michalecka et al., 2004) and which is suitable for analyzing the StNDB1 protein distribution in its species of origin. Since a recent targeting analysis using a C-terminal peptide of the Arabidopsis NDB1 suggested that NDB1 may reside also in peroxisomes (Carrie et al., 2008), we also compared purified potato tuber mitochondria and peroxisomes (Fig. 7A). The peroxisomes had high catalase activities, as observed previously (Struglics et al., 1993), and no mitochondrial contamination. Antibodies against a glyoxysomal malate dehydrogenase detected a 37-kD band in peroxisomes and a larger band in mitochondria, consistent with previous observations (Gietl et al., 1996). However, even at film exposure times where the StNDB1 antibodies gave a strong signal in mitochondria, no NDB1 signal could be detected in peroxisomes (Fig. 7A). Therefore, we conclude that StNDB1 resides in mitochondria and not peroxisomes in potato, consistent with previous results using less specific antibodies against the conserved C terminus of StNDB1 (Rasmusson and Agius, 2001), and that the phenotype observed in this investigation was due to enzymatic changes in the mitochondrial ETC.
Figure 7.
Distribution of StNDB1 in organelles and organs of potato. A, Purified potato tuber mitochondria from cv Desirée (DM) and cv Bintje (BM) and peroxisomes from cv Bintje (BP; two independent preparations) were analyzed for content of NDB1. As controls, the antibody against glyoxysomal malate dehydrogenase (gMDH) detected bands of 37 and 44 kD in peroxisomes and mitochondria, respectively, and catalase activity is denoted for each preparation in μmol min−1 mg−1 protein. B, Mitochondria were purified from seven organs of potato plants (Desirée) in three independent preparation sets. Twenty micrograms of protein per lane was subjected to western blotting using antibodies against different mitochondrial proteins. Antibody designations and apparent molecular masses are denoted to the right. Results from one representative set of preparations are shown except for NDB1-EFD, where two sets are shown to indicate the extent of variation. The ratios of mitochondrial to nuclear genome were determined by real-time PCR against StNAD9 and StNDA1 and are denoted as percentage of the mature leaf value ± se (n = 3) below the blots. AOX, Alternative oxidase; EFD, EF-hand domain; Etio., etiolated; Fl., flower; Mat., mature; Src., source; UCP, uncoupling protein.
To determine organ distribution of StNDB1 in potato, mitochondria were purified in triplicate from seven potato organs: sink and source tubers, etiolated and green stems, young and mature leaves, and flower buds. The mitochondria were analyzed by western blotting, comparing StNDB1 with other ETC proteins (Fig. 7B). Consistent between preparations, StNDB1 was most abundant in green stems, with lower levels being detectable in source and sink tubers and mature leaves. A weak band was repeatedly detected in young leaves but not in etiolated stems and flower buds. Immunoprobing was also carried out with antibodies against the alternative oxidase and the type II NAD(P)H dehydrogenase StNDA1, the latter of which most likely also detects a second potato NDA protein (Svensson and Rasmusson, 2001; Svensson et al., 2002; Geisler et al., 2004). These immunosignals were for both proteins strongest in mature leaves, with the NDA1 signal showing a smaller difference between organs, possibly due to the detection of differently regulated NDA-type proteins in a single band. The uncoupling protein was relatively evenly distributed but somewhat elevated in tubers and stems. The 78-kD and NAD9 subunits of complex I, the proton-pumping NADH dehydrogenase of the ETC, were evenly distributed in mitochondria from different organs, consistent with their role in basic energy production. To estimate whether mitochondria were differently prevalent in the organs, we analyzed the copy number ratio of the mitochondrial NAD9 and the nuclear NDA1 genes by real-time PCR. The results indicate a higher mitochondrial DNA copy number per nuclear DNA in sink tubers and flower buds as compared with other organs. If normalized in this respect, the highest levels of StNDB1 would be present in green stems and sink tubers, but the difference between green stems and leaves is unchanged. The results, therefore, strongly suggest that StNDB1 levels are specifically elevated in green stem mitochondria, consistent with a specific function in stems. Also, the organ distribution of StNDB1 deviates substantially from that of energy-bypass pathways as well as proteins of the energy-conserving part of the potato ETC.
DISCUSSION
Transgenic N. sylvestris Displays Modified NADPH Reduction and a Bolting Phenotype
We present here three consistent lines of evidence suggesting that the NDB1 external NADPH dehydrogenase is important for stem development and specifically accelerates bolting: (1) the NADPH/NADP+ ratio is lowered 3-fold during bolting and onset of stem elongation; (2) the overexpression and suppression of NDB1 accelerate and retard bolting, respectively, coupled to specific changes in stem NADPH/NADP+ ratio and expression of flowering induction-associated genes; and (3) the investigated StNDB1 protein is specifically highly abundant in stem mitochondria of the transgene donor species, potato.
StNDB1 is an external calcium-dependent NADPH dehydrogenase that is theoretically able to modulate the NADPH/NADP+ ratio in the cytosol irrespective of the cellular energy charge (Michalecka et al., 2004). Consistently, the N. sylvestris plants overexpressing StNDB1 displayed decreased leaf NADPH/NADP+ ratios when grown at 200 μmol m−2 s−1 (Liu et al., 2008). The absence of such an effect in leaves at higher light (Fig. 2) could be due to NDB1 being inactive (e.g. calcium availability being insufficient). Alternatively, a larger photon flux may increase the export of chloroplast reductant via a triose-P shuttle (Krömer, 1995), compensating for an elevated mitochondrial NADPH oxidation. In contrast, the stem NADPH/NADP+ ratio varies between the genotypes, suggesting that NDB1 is active under these conditions and affecting the NADPH/NADP+ ratio beyond what other reactions can compensate for. The simplest interpretation of the phenotype, therefore, is that higher levels of external mitochondrial NADPH dehydrogenase will speed up a decrease in NADPH/NADP+ ratio that normally takes place in the wild-type apex during bolting.
Integration of NDB1 and NADP(H) in the Cellular Metabolic Systems
In this investigation, stem NADPH/NADP+ ratios showed specific changes between the transgenic lines. This material gave an opportunity to use metabolic profiling for identifying interaction points between NADP(H) and central carbon metabolism. In both leaves and stems, we found a positive correlation between NADPH and Glc-6-P (in stems also the isomer Fru-6-P). This was likely mediated by NADPH inhibition of Glc-6-P dehydrogenase of the pentose-P pathway, which is believed to be modulated by the NADPH/NADP+ ratio (Kruger and von Schaewen, 2003; Wakao and Benning, 2005), although direct in vivo support has not yet been provided. Previously, a surprising increase in NADPH/NADP+ ratio in an Arabidopsis mutant lacking the NADP+-reducing nonphosphorylating glyceraldehyde-3-P dehydrogenase was attributed to an elevated Glc-6-P dehydrogenase activity (Rius et al., 2006). The Glc-6-P level is a marker for carbon status in Arabidopsis and is located at the connection point between growth, respiration, and carbon storage (Stitt et al., 2007). Thus, the correlation between Glc-6-P and NADPH reduction level appears to be a general property rather than a specific one directly involved in the mediation of bolting.
Apart from the hexose-Ps, the correlation of NADPH to metabolites is highly different in stem and leaf. Differences in individual metabolites may be consequences of the NADPH/NADP+ ratio being outside the response range of a critical enzyme in one organ, variations in isoenzymes expressed, or switches between metabolic pathways using the metabolites in question. Particularly marked differences between leaf and stem were seen for sugars. We further observed that the stem NADPH/NADP+ ratio specifically correlates (positively or negatively) to a small set of metabolites, three of which (2-oxoglutarate, galactonate, and ascorbate) show little correlation to other metabolites in the data set. In contrast, the leaf NADPH/NADP+ couple appears to interact with a large number of metabolites that represent numerous pathways (Fig. 4). This is consistent with NADP(H) having a regulatory role in stems (as suggested by the effect on bolting) whereas a general role in metabolic redox transfer would dominate in the leaves.
In stems, we observed a positive correlation of the NADPH/NADP+ ratio to Glu and a negative one to 2-oxoglutarate and citrate (Figs. 3 and 4), metabolites that are central intermediates connecting the citric acid cycle and nitrogen assimilation. Thus, the Glu/2-oxoglutarate redox pair responds to the NADPH reduction level. A potentially NADPH-mediated functional connection between the oxidative pentose-P pathway and Glu synthase was reported for barley (Hordeum vulgare) and pea (Pisum sativum) roots (Esposito et al., 2003; Bowsher et al., 2007). However, an NADPH effect on the Glu/2-oxoglutarate redox pair being mediated by an NADP(H)-Glu dehydrogenase (Purnell et al., 2005) cannot be excluded at present. The similarity in response between 2-oxoglutarate and citrate may also be derived from an NADP(H) effect on the formation or removal of these metabolites, for example via the control of citric acid cycle proteins by thioredoxin, which is reductively activated by NADPH (Balmer et al., 2004). Overall, the observations here emphasize the metabolic differences between leaves and stems and provide in vivo support for an influence of NADP(H) redox homeostasis on sugar metabolism, the pentose-P pathway, and Glu metabolism.
Redox Modulation of Bolting
The NADP(H) redox effect observed in stems of the transgenic lines must have been relayed to the floral transition signaling pathways in order to affect bolting and flowering time. We observed higher and lower transcript levels in the overexpressor and suppressor lines, respectively, for N. sylvestris MADS1 and MADS2 as well as a decrease of NFL2 in the suppressor line. NsMADS1 and NsMADS2 belong to the SQUAMOSA subfamily of floral meristem and organ identity proteins, including Arabidopsis APETALA1 (AP1; Becker and Theissen, 2003; Smykal et al., 2007). NsMADS1 is an ortholog to the floral transition marker NtMADS11 (Jang et al., 2002; Gallego-Giraldo et al., 2007). NsMADS2 is a transcriptional activator that upon overexpression in N. tabacum causes early flowering, and the ortholog NtMADS5/NAP1-2 is transcriptionally induced in the stem apex during floral transition (Cho et al., 1999; Jang et al., 1999; Smykal et al., 2007). Thus, NsMADS2 has a central function for bolting and flowering induction in N. sylvestris, possibly similar to AP1 in Arabidopsis (He and Amasino, 2005). AP1 cooperates with its activator LEAFY to integrate the converging signals from the photoperiod, GA, autonomous, and vernalization pathways (Bernier and Perilleux, 2005; He and Amasino, 2005; Parcy, 2005). The LEAFY homolog NtNFL2 is expressed in meristems (Kelly et al., 1995), and overexpression of the close homolog NtNFL1 in N. tabacum induces earlier flowering (Ahearn et al., 2001). In summary, the expressional change for NsMADS2 in the transgenic lines (Fig. 5) is sufficient to explain the bolting phenotype observed (Fig. 1), but NsMADS1 and NFL2 are likely to contribute to the phase transition. These results thus show that floral meristem identity genes respond to the stem NADPH/NADP+ ratio and should be promoted by its decrease in the wild-type apex during bolting.
The observed changes in stem NADP(H) may be mediated by metabolite changes to affect the expression of meristem identity genes. Several intermediates of primary metabolism have been implied to affect floral transitions. Examples include ascorbate (Yamamoto et al., 2005; Barth et al., 2006), nitric oxide (He et al., 2004), Pro (Mattioli et al., 2008), and sugars (Francis and Halford, 2006; Aluri and Buttner, 2007), especially metabolites of the trehalose pathway (van Dijken et al., 2004; Veyres et al., 2008). Whereas the trehalose synthase mutation influenced flowering downstream of LEAFY (van Dijken et al., 2004), ascorbate and nitric oxide affected LEAFY expression (He et al., 2004; Barth et al., 2006). Also, Suc exported from leaves has been suggested to affect meristem identity genes in the stem apex (Bernier and Perilleux, 2005). For other metabolites, there is little insight into how the effects are mediated. Our investigation identifies a pattern of relatively few compounds correlated to NADPH levels in the stems of the transgenic lines. These are candidates for mediating, individually or in combination, the NADP(H)-imposed redox regulation of floral integrator genes and consequentially bolting. Especially, the correlation of MADS gene expression to Xyl suggests an involvement of sugar metabolism, complementing the previously known sugar effects on shoot meristems (Francis and Halford, 2006). The changes in ascorbate content are also particularly interesting given that the terminal step of ascorbate biosynthesis appears to be physically (Millar et al., 2003) and functionally (Nunes-Nesi et al., 2005) linked to the ETC. Therefore, ascorbate synthesis may be influenced downstream of NDB1 in the electron transport path, whereas the majority of the other metabolic changes described here are most likely a direct consequence of the modified NADPH/NADP+ ratio.
An option worth considering would be that NADPH and/or NADP+ are directly sensed in the cell. This scenario is not without precedence, since sensors for NADH/NAD+ such as Streptomyces coelicolor Rex (Brekasis and Paget, 2003) and the human CtBP protein (Zhang et al., 2002) have been long identified. Furthermore, human HSCARG was recently reported to be a sensor for NADPH/NADP+ and via protein binding to decrease nitric oxide formation in response to lowered NADPH/NADP+ ratios (Zhao et al., 2008). When the results of these and our current study are considered together, it seems reasonable to postulate that an NADPH-sensing system also exists in plants. Identifying the exact mechanism linking the NADPH/NADP+ redox couple will probably require considerably more research effort. However, irrespective of which of these models is ultimately correct, the results we present here clearly demonstrate that manipulation of the mitochondrial external NADPH dehydrogenase StNDB1 has profound effects on stem NADPH/NADP+ ratio and, as a consequence, floral integrator genes and bolting time. Thus, NADPH constitutes a connection between redox signaling and floral phase transitions, both of which integrate multiple influences from the plant environment (Bernier and Perilleux, 2005; Foyer and Noctor, 2005).
NDB1 Deviates from Other Mitochondrial Enzymes Regarding Phenotype and Expression
It is relevant to compare the StNDB1 transgenic investigation here with known phenotypes of plants modified for mitochondrial proteins. Mitochondrial effects on plant development have so far mainly been concerned with flower morphology. Cytoplasmic male sterility traits involve chimeric genes in the mitochondrial genome that modify expression of flower organ identity genes, leading to conversions of floral organs without affecting the vegetative plant body (Linke and Börner, 2005; Carlsson et al., 2008). In a second class of genetic alterations, N. sylvestris and Zea mays genotypes lacking subunits of energy-conserving ETC complexes display retardations regarding growth, leaf shape, photosynthesis, and chloroplast greening (Marienfeld and Newton, 1994; Gutierres et al., 1997; Karpova et al., 2002; Dutilleul et al., 2003; Noctor et al., 2004). Third, repression of citric acid cycle proteins has resulted in variable effects. Examples in tomato (Solanum lycopersicum) include a decrease in stomatal function, photosynthesis, and growth rate in fumarase antisense plants and an increase of photosynthesis and growth rate in plants suppressed for malate dehydrogenase (Nunes-Nesi et al., 2005, 2007). Arabidopsis suppressed for pyruvate dehydrogenase kinase displayed decreased vegetative growth and early flowering (Zou et al., 1999). In a fourth category, genetic modifications of energy-bypass pathways in Arabidopsis have uncovered the importance of the uncoupling protein AtUCP1 for optimal photosynthesis and growth rate (Sweetlove et al., 2006), while the alternative oxidase gene AtAOX1a is essential for optimal growth at lower temperatures (Fiorani et al., 2005) and combined light and drought stress (Giraud et al., 2008). In summary, plants modified for enzymes of the mitochondrial respiratory pathways have previously displayed phenotypes of modified photosynthesis and growth but not specific developmental changes without showing a modified biomass accumulation.
In combination, the specific bolting phenotype, the lack of an effect on the biomass accumulation seen here and at lower light (Liu et al., 2008), and the protein distribution in potato organs suggest that StNDB1 deviates in physiological role from ETC proteins of both the energy-conserving and the energy-bypass pathways. A specialized function of NDB1 is also consistent with the distribution of external NADPH dehydrogenases, being present in plants and Neurospora but not in mammals and yeasts (Rasmusson et al., 2008).
CONCLUSION
We demonstrate that the mitochondrial external NADPH dehydrogenase StNDB1 has capacity to impose changes in the cellular NADPH/NADP+ ratio that in turn can modulate a developmental process. Specifically, we observed an apical NADPH-induced redox modulation of bolting via changes in expression of floral meristem integrator genes. This was consistent with wild-type temporal changes in NADPH/NADP+ ratio during bolting. Also, major differences between stem and leaf were identified regarding points of interaction between the NADP(H) redox couple and primary metabolism. However, further investigations are needed to define how NADP(H) is integrated into the metabolic and regulatory systems of plant cells. Regarding the enzymatic properties, gene expression, and the phenotype induced by expressional modification, StNDB1 strongly deviates from other mitochondrial respiratory proteins. This supports specific, including regulatory, physiological roles of the external mitochondrial NADPH dehydrogenase.
MATERIALS AND METHODS
Plant Growth
Seeds from homozygous transgenic Nicotiana sylvestris overexpressing or cosuppressing potato (Solanum tuberosum) NDB1 (Michalecka et al., 2004; Liu et al., 2008) were sown on soil and grown in a greenhouse with 16 h of supplementary light per day at 200 μmol m−2 s−1 at 23°C. Two-week-old seedlings with similar sizes were transferred to individual pots and grown at 23°C at 200 μmol m−2 s−1 or 500 μmol m−2 s−1 in a greenhouse with supplementary light or at 300 μmol m−2 s−1 in a climate chamber. Shoot height was measured as the length from the stem bottom, just below the lowest leaf, to the top of the shoot apex. For dry weight determination, shoots and roots were dried at 90°C for 21 h. Paclobutrazol was applied to soil by adding 100 mL of a 10 μm solution to each plant two times weekly, starting at day 17.
Determination of Pyridine Nucleotide Levels
Transgenic lines were compared by sampling stems that were approximately 4 cm long, whereas leaf samples were taken from the first and second uppermost fully developed leaves. Pyridine nucleotide levels were measured as described previously (Liu et al., 2008). Additional powder aliquots were stored at −80°C for other measurements.
Metabolic Profiling
Metabolites were extracted principally as described previously (Roessner-Tunali et al., 2003). One hundred to 150 mg of sample powder was mixed with 1.4 mL of methanol, and 60 μL of ribitol (0.2 mg mL−1) was added as an internal standard. The mixture was extracted for 15 min at 70°C and then centrifuged for 10 min at 14,000 rpm. The supernatant was transferred to a glass vial, and 750 μL of chloroform and 1,500 μL of water were added. The mixture was centrifuged for 15 min at 4,000 rpm. The upper polar phase was dried in vacuo and stored at −80°C. Metabolite analysis was performed by gas chromatography-time of flight-mass spectrometry as described previously (Lisec et al., 2006), and the relative metabolite contents were evaluated by comparison with libraries housed in the Golm Metabolome Database (Kopka et al., 2005; Schauer et al., 2005).
Transcript Quantification
Total RNA isolation, DNase treatment, cDNA synthesis, and real-time RT-PCR were carried out using the RNeasy Plant mini kit (Qiagen), DNase I (New England Biolabs), RevertAid H minus first-strand cDNA synthesis kit (Fermentas), and GoTaq DNA polymerase (Promega), respectively, but otherwise as described previously (Svensson and Rasmusson, 2001; Svensson et al., 2002; Geisler et al., 2004). Primer pairs used were as reported previously (Liu et al., 2008) or derived from N. sylvestris or Nicotiana tabacum cDNA or EST sequences as follows (with GenBank accession nos.): NsMADS1 (AF068725), 5′-GCATTGCAAGAGCAAAACAA-3′ and 5′-CCATTAAGATGGCGAAGCA-3′; NsMADS2 (AF068726), 5′-GTTGAATGGCATCAGCAAAA-3′ and 5′-TGAACTCCAGCAAAAGGACA-3′; NtNFL2 (U16173, U16174), 5′-GGGAGCATCCTTTTATCGTG-3′ and 5′-GTCGCATTTTTGGCTTGTTT-3′; NtGA2OX3 (EF471117), 5′-GGTGCACATACTGACCCACA-3′ and 5′-TGGTGGTCCTCCAAAATAAATC-3′; and NtGA2OX5 (EF471118), 5′-GGAGAGCATACAGACCCACAA-3′ and 5′-AGGTGGCCCTCCAAAATAAA-3′. The results were normalized to the expression of a control gene. For this, we used N. sylvestris UBI11 (Genschik et al., 1992; M74101, 5′-GGGAAGACCATCACTTTGGA-3′ and 5′-GCCACTAAAGGAAAGCACAGA-3′) that is homologous to Arabidopsis UBQ10 (Czechowski et al., 2005).
Organelle Analyses
Potato mitochondria from growing sink tubers and source tubers (sprouting in darkness at 25°C) were purified as described (Struglics et al., 1993). Young leaf (1–3 cm), mature leaf (13–15 cm), green stem (the part carrying the uppermost eight leaves), etiolated stem (growing in darkness at 25°C), and flower bud (sampled approximately 1 d before flower opening) mitochondria were purified principally as reported (Boutry et al., 1984). Peroxisomes were purified as described (Struglics et al., 1993). Protein analysis was performed by the bicinchoninic acid method (Sigma). Mitochondria were analyzed by SDS-PAGE and western blotting (Michalecka et al., 2004). Antibodies used were against potato NDB1 EF-hand domain (Michalecka et al., 2004), potato NDA1 (Rasmusson and Agius, 2001), Sauromatum guttatum alternative oxidase (Elthon et al., 1989), Arabidopsis (Arabidopsis thaliana) uncoupling protein (Borecky et al., 2001), glyoxysomal malate dehydrogenase (Gietl et al., 1996), and the mitochondrial complex I subunits NAD9 from wheat (Triticum aestivum; Lamattina et al., 1993) and 78-kD protein from Neurospora crassa (Friedrich et al., 1989). The latter detects the 76-kD subunit in potato (Rasmusson et al., 1998). Catalase activity was detected in the presence of Triton X-100 as reported previously (Struglics et al., 1993). Mitochondrial and nuclear genome copy number ratio was determined by isolating total cellular DNA with the DNeasy kit (Qiagen), followed by real-time PCR using primers against the potato mitochondrial gene NAD9 (5′-CGATCGATATTTGAGGAGTT-3′ and 5′-TCCAATAGTTCAGCAACAGA-3′; accession no. X79774) and the nuclear gene NDA1 (5′-CACAATGCCATGGTTCAA-3′ and 5′-TCCAATAGGTTCAGCAACAGA-3′; AJ245861) at 60°C annealing, as described previously (Michalecka et al., 2003). The analysis was made using three biological replicates and two to four machine replicates.
Data Treatment
Comparisons of values for significant differences were made using Student's t test in Excel (Microsoft) at P < 0.05, unless otherwise denoted. Pearson correlation analyses were carried out using SPSS 16.0 (SPSS). Correlation networks were built using Cytoscape 2.6 (Cline et al., 2007).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biomass accumulation of N. sylvestris lines.
Supplemental Figure S2. Stem extension rates in N. sylvestris transgenic lines.
Supplemental Table S1. Metabolite changes in leaves.
Supplemental Table S2. Metabolite changes in stems.
Supplementary Material
Acknowledgments
Drs. T.E. Elthon (University of Nebraska, Lincoln, NE), T. Friedrich (Albert-Ludwigs Universität, Freiburg, Germany), C. Gietl (Technical University, Munich), J.-M. Grienenberger (Université Louis-Pasteur, Strasbourg, France), and A.E. Vercesi (Universidade Estadual de Campinas, Campinas, Brazil) are acknowledged for generous donations of antibodies. Dr. T. Moritz (Umeå Plant Science Center, Umeå, Sweden) is acknowledged for fruitful suggestions and Ms. A. Goldyn (Lund University, Lund, Sweden; present affiliation, Max Planck Institute for Metals Research, Stuttgart, Germany) for technical assistance.
This work was supported by the Swedish Research Council (grant no. 621–2006–4597).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Allan G. Rasmusson (allan.rasmusson@cob.lu.se).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahearn KP, Johnson HA, Weigel D, Wagner DR (2001) NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure. Plant Cell Physiol 42 1130–1139 [DOI] [PubMed] [Google Scholar]
- Aluri S, Buttner M (2007) Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc Natl Acad Sci USA 104 2537–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schurmann P, Droux M, et al (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, De Tullio M, Conklin PL (2006) The role of ascorbic acid in the control of flowering time and the onset of senescence. J Exp Bot 57 1657–1665 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29 464–489 [DOI] [PubMed] [Google Scholar]
- Bernier G, Perilleux C (2005) A physiological overview of the genetics of flowering time control. Plant Biotechnol J 3 3–16 [DOI] [PubMed] [Google Scholar]
- Borecky J, Maia IG, Costa AD, Jezek P, Chaimovich H, de Andrade PB, Vercesi AE, Arruda P (2001) Functional reconstitution of Arabidopsis thaliana plant uncoupling mitochondrial protein (AtPUMP1) expressed in Escherichia coli. FEBS Lett 505 240–244 [DOI] [PubMed] [Google Scholar]
- Boutry M, Faber AM, Charbonnier M, Briquet M (1984) Microanalysis of plant mitochondrial protein synthesis products: detection of variant polypeptides associated with cytoplasmic male sterility. Plant Mol Biol 3 445–452 [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Lacey AE, Hanke GT, Clarkson DT, Saker LR, Stulen I, Emes MJ (2007) The effect of Glc6P uptake and its subsequent oxidation within pea root plastids on nitrite reduction and glutamate synthesis. J Exp Bot 58 1109–1118 [DOI] [PubMed] [Google Scholar]
- Brekasis D, Paget MS (2003) A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J 22 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, Leino M, Sohlberg J, Sundström JF, Glimelius K (2008) Mitochondrial regulation of flower development. Mitochondrion 8 74–86 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582 3073–3079 [DOI] [PubMed] [Google Scholar]
- Cho S, Jang S, Chae S, Chung KM, Moon YH, An G, Jang SK (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol Biol 40 419–429 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58 193–212 [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protocols 2 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhafez D, Murcha MW, Clifton R, Soole KL, Day DA, Whelan J (2006) Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell Physiol 47 43–54 [DOI] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, Mcintosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S, Massaro G, Vona V, Di Martino Rigano V, Carfagna S (2003) Glutamate synthesis in barley roots: the role of the plastidic glucose-6-phosphate dehydrogenase. Planta 216 639–647 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Halford NG (2006) Nutrient sensing in plant meristems. Plant Mol Biol 60 981–993 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Hofhaus G, Ise W, Nehls U, Schmitz B, Weiss H (1989) A small isoform of NADH:ubiquinone oxidoreductase (complex I) without mitochondrially encoded subunits is made in chloramphenicol-treated Neurospora crassa. Eur J Biochem 180 173–180 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Garcia-Martinez JL, Moritz T, Lopez-Diaz I (2007) Flowering in tobacco needs gibberellins but is not promoted by the levels of active GA1 and GA4 in the apical shoot. Plant Cell Physiol 48 615–625 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Ubeda-Tomas S, Gisbert C, Garcia-Martinez JL, Moritz T, Lopez-Diaz I (2008) Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol 49 679–690 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Broselid C, Hederstedt L, Rasmusson AG (2007) Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J Biol Chem 282 28455–28464 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Johansson FI, Svensson AS, Rasmusson AG (2004) Antimycin A treatment decreases respiratory internal rotenone-insensitive NADH oxidation capacity in potato leaves. BMC Plant Biol 4 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Parmentier Y, Durr A, Marbach J, Criqui MC, Jamet E, Fleck J (1992) Ubiquitin genes are differentially regulated in protoplast-derived cultures of Nicotiana sylvestris and in response to various stresses. Plant Mol Biol 20 897–910 [DOI] [PubMed] [Google Scholar]
- Gietl C, Seidel C, Svendsen I (1996) Plant glyoxysomal but not mitochondrial malate dehydrogenase can fold without chaperone assistance. Biochim Biophys Acta 1274 48–58 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Amasino RM (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10 30–35 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305 1968–1971 [DOI] [PubMed] [Google Scholar]
- Jang S, An K, Lee S, An G (2002) Characterization of tobacco MADS-box genes involved in floral initiation. Plant Cell Physiol 43 230–238 [DOI] [PubMed] [Google Scholar]
- Jang S, Hong MY, Chung YY, An G (1999) Ectopic expression of tobacco MADS genes modulates flowering time and plant architecture. Mol Cells 9 576–586 [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner DR (1995) NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell 7 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21 1635–1638 [DOI] [PubMed] [Google Scholar]
- Krömer S (1995) Respiration during photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 46 45–70 [Google Scholar]
- Kruger NJ, von Schaewen A (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6 236–246 [DOI] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur J Biochem 217 831–838 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Zeevaart JA (2005) Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol 138 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke B, Börner T (2005) Mitochondrial effects on flower and pollen development. Mitochondrion 5 389–402 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protocols 1 387–396 [DOI] [PubMed] [Google Scholar]
- Liu YJ, Norberg FE, Szilagyi A, De Paepe R, Åkerlund HE, Rasmusson AG (2008) The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol 49 251–263 [DOI] [PubMed] [Google Scholar]
- Marienfeld JR, Newton KJ (1994) The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli R, Marchese D, D'Angeli S, Altamura MM, Costantino P, Trovato M (2008) Modulation of intracellular proline levels affects flowering time and inflorescence architecture in Arabidopsis. Plant Mol Biol 66 277–288 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Duarte M, Møller IM, Prokisch H, Dolan PL, Pinto L, Nelson MA, Videira A (2001) The external calcium-dependent NADPH dehydrogenase from Neurospora crassa mitochondria. J Biol Chem 276 3947–3951 [DOI] [PubMed] [Google Scholar]
- Michalecka AM, Agius SC, Møller IM, Rasmusson AG (2004) Identification of a mitochondrial external NADPH dehydrogenase by overexpression in transgenic Nicotiana sylvestris. Plant J 37 415–425 [DOI] [PubMed] [Google Scholar]
- Michalecka AM, Svensson ÅS, Johansson FI, Agius SC, Johanson U, Brennicke A, Binder S, Rasmusson AG (2003) Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol 133 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, Foyer CH (2003) Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Dutilleul C, De Paepe R, Foyer CH (2004) Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J Exp Bot 55 49–57 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F (2005) Flowering: a time for integration. Int J Dev Biol 49 585–593 [DOI] [PubMed] [Google Scholar]
- Purnell MP, Skopelitis DS, Roubelakis-Angelakis KA, Botella JR (2005) Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222 167–180 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Agius SC (2001) Rotenone-insensitive NAD(P)H dehydrogenases in plants: immunodetection and distribution of native proteins in mitochondria. Plant Physiol Biochem 39 1057–1066 [Google Scholar]
- Rasmusson AG, Geisler DA, Møller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8 47–60 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Heiser V, Irrgang KD, Brennicke A, Grohmann L (1998) Molecular characterisation of the 76 kDa iron-sulphur protein subunit of potato mitochondrial complex I. Plant Cell Physiol 39 373–381 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55 23–39 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Svensson AS, Knoop V, Grohmann L, Brennicke A (1999) Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J 20 79–87 [DOI] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61 945–957 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579 1332–1337 [DOI] [PubMed] [Google Scholar]
- Smykal P, Gennen J, De Bodt S, Ranganath V, Melzer S (2007) Flowering of strict photoperiodic Nicotiana varieties in non-inductive conditions by transgenic approaches. Plant Mol Biol 65 233–242 [DOI] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn JE, Piques M (2007) Multilevel genomics analysis of carbon signalling during low carbon availability: coordinating the supply and utilisation of carbon in a fluctuating environment. Funct Plant Biol 34 526–549 [DOI] [PubMed] [Google Scholar]
- Struglics A, Fredlund KM, Rasmusson AG, Møller IM (1993) The presence of a short redox chain in the membrane of intact potato-tuber peroxisomes and the association of malate dehydrogenase with the peroxisomal membrane. Physiol Plant 88 19–28 [Google Scholar]
- Svensson ÅS, Johansson FI, Møller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517 79–82 [DOI] [PubMed] [Google Scholar]
- Svensson ÅS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28 73–82 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR (2006) Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci USA 103 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Benning C (2005) Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J 41 243–256 [DOI] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC (2004) Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48 703–734 [DOI] [PubMed] [Google Scholar]
- Vercesi AE, Borecky J, de Godoy Maia I, Arruda P, Cuccovia IM, Chaimovich H (2006) Plant uncoupling mitochondrial proteins. Annu Rev Plant Biol 57 383–404 [DOI] [PubMed] [Google Scholar]
- Veyres N, Danon A, Aono M, Galliot S, Karibasappa YB, Diet A, Grandmottet F, Tamaoki M, Lesur D, Pilard S, et al (2008) The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J 55 665–686 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Bhuiyan NH, Waditee R, Tanaka Y, Esaka M, Oba K, Jagendorf AT, Takabe T (2005) Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J Exp Bot 56 1785–1796 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH (2002) Regulation of corepressor function by nuclear NADH. Science 295 1895–1897 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang J, Li H, Li Y, Ren J, Luo M, Zheng X (2008) An NADPH sensor protein (HSCARG) down-regulates nitric oxide synthesis by association with argininosuccinate synthetase and is essential for epithelial cell viability. J Biol Chem 283 11004–11013 [DOI] [PubMed] [Google Scholar]
- Zou JT, Qi QG, Katavic V, Marillia EF, Taylor DC (1999) Effects of antisense repression of an Arabidopsis thaliana pyruvate dehydrogenase kinase cDNA on plant development. Plant Mol Biol 41 837–849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.