Abstract
To gain insight into the molecular mechanism of RNA editing, we have characterized the low psii accumulation66 (lpa66) Arabidopsis (Arabidopsis thaliana) mutant, which displays a high chlorophyll fluorescence phenotype. Its perturbed chlorophyll fluorescence is reflected in reduced levels of photosystem II (PSII) proteins. In vivo protein labeling showed that synthesis rates of the PSII reaction center protein D1/D2 were lower, and turnover rates of PSII core proteins higher, than in wild-type counterparts. The assembly of newly synthesized proteins into PSII occurs in the lpa66 mutant but with reduced efficiency compared with the wild type. LPA66 encodes a chloroplast protein of the pentatricopeptide repeat family. In lpa66 mutants, editing of psbF that converts serine to phenylalanine is specifically impaired. Thus, LPA66 is specifically required for editing the psbF transcripts in Arabidopsis, and the amino acid alternation due to lack of editing strongly affects the efficiency of the assembly of PSII complexes.
PSII is a large pigment-protein complex found in the membranes of chloroplasts, containing more than 20 subunits, which catalyzes light-driven water oxidation and the reduction of plastoquinone concomitant with oxygen evolution. Some PSII proteins are encoded by the nuclear genome and others by the chloroplast genome in higher plants (Wollman et al., 1999; Nelson and Yocum, 2006). Thus, coordinated regulation of nuclear and chloroplast gene expression is essential for the biogenesis and assembly of photosynthetically competent protein complexes (Goldschmidt-Clermont, 1998). Chloroplast gene expression is regulated mainly at posttranscriptional levels by various mechanisms (Deng and Gruissem, 1987), and a number of nucleus-encoded factors have been shown to be involved in RNA splicing, processing, editing, degradation, and translation in plants (Barkan and Goldschmidt-Clermont, 2000; Zerges, 2000; Choquet and Wollman, 2002; Manuell et al., 2004; Marín-Navarro et al., 2007). Among them, a PPR superfamily has received much attention recently, because of its members' involvement in posttranscriptional regulation of gene expression in plastids (Shikanai, 2006; Andrés et al., 2007; Delannoy et al., 2007). In Chlamydomonas reinhardtii, MCA1, the only PPR protein from this organism characterized to date, is involved in the regulation of petA gene expression by interacting with the first 21 nucleotides of the 5′ untranslated region of its transcripts, thereby protecting them from 5′→3′ degradation (Loiselay et al., 2008). In Arabidopsis (Arabidopsis thaliana), numerous PPR proteins have been shown to play various roles, including the following. CRR2 and HCF152 have been shown to be involved in cleavage and splicing of their specific mRNA targets, respectively (Hashimoto et al., 2003; Meierhoff et al., 2003; Nakamura et al., 2003). PGR3 is involved in the stabilization of petL RNA operons and the translation of petL (Yamazaki et al., 2004). CRR4, CRR21, CRR22, CRR28, CLB19, YS1, and RARE1 have been shown to be specifically required for editing their corresponding RNA targets (Kotera et al., 2005; Okuda et al., 2006, 2007, 2009; Chateigner-Boutin et al., 2008; Robbins et al., 2009; Zhou et al., 2009). In addition, DG1 and pTAC2 have been shown to be involved in the regulation of plastid-encoded RNA polymerase-dependent transcript accumulation (Pfalz et al., 2005; Chi et al., 2008). PPRs have also been found to regulate plastid gene expression in other organisms, such as ZmCRP1, ZmPPR2, ZmPPR4, and ZmPPR5 in maize (Zea mays; Fisk et al., 1999; Williams and Barkan, 2003; Schmitz-Linneweber et al., 2006; Beick et al., 2008), OsPPR1 in rice (Oryza sativa; Gothandam et al., 2005), and PPR531-11 in the moss Physcomitrella patens (Hattori et al., 2007).
PPR proteins, which constitute one of the largest families of proteins in plants, are defined by the tandem array of PPR motifs with a highly degenerate unit consisting of 35 amino acids (Lurin et al., 2004). There are about 100 members of the family in P. patens and over 450 members in both Arabidopsis and rice (Lurin et al., 2004; Merchant et al., 2007; O'Toole et al., 2008). The plant PPR protein family can be divided into two subfamilies on the basis of their motif content and organization, the P subfamily and combinatorial and modular proteins (PCMP) subfamilies (Lurin et al., 2004; Rivals et al., 2006). In proteins of the P subfamily, such as HCF152 and PGR3 in Arabidopsis, the 35 amino acid repeats are organized as tandem repeats. Members of the PCMP subfamily (also referred to as the PLS subfamily) contain PPR-like motifs with either short (PPR-like S) or longer (PPR-like L) repeats. This subfamily is specific to land plants and is subdivided into three subgroups according to their C-terminal contents: PLS, E, and DYW proteins (Lurin et al., 2004; Rivals et al., 2006). In Arabidopsis, the PCMP subfamily contains about 200 members, including the recently characterized CRR4, CRR21, CRR22, CRR28, CLB19, YS1, MEF1, and RARE1 (Kotera et al., 2005; Okuda et al., 2007, 2009; Chateigner-Boutin et al., 2008; Robbins et al., 2009; Zehrmann et al., 2009; Zhou et al., 2009). CRR4, CRR21, and CLB19 belong to the E subgroup and share a common E domain required for editing the target site, but not for target specificity, which suggests that physical recruitment of the editing machinery may involve non-PPR domains (Okuda et al., 2007). The other five members belong to the DYW subgroup, which additionally contains the DYW domain besides the E domain.
RNA editing is a posttranscriptional process that, in plants, alters specific C nucleotides to U (Maier et al., 1996; Brennicke et al., 1999; Bock, 2000; Shikanai, 2006) or occasionally U to C in bryophytes and ferns (Kugita et al., 2003; Wolf et al., 2004). Such processes have been observed in both plastids and mitochondria of the land plants examined except the Marchantiidea, but not in any of the investigated algae and cyanobacteria (Schmitz-Linneweber et al., 2004). Usually, RNA editing results in the modification of the encoded amino acid sequences (Bock et al., 1994; Sasaki et al., 2001; Okuda et al., 2007; Chateigner-Boutin et al., 2008) or the generation of either a translational start codon (Hoch et al., 1991; Kotera et al., 2005) or stop codon (Wintz and Hanson, 1991). There are about 20 to 50 known editing sites in plastid transcripts and more than 400 known sites in mitochondrial transcripts in the angiosperms that have been studied (Giege and Brennicke, 1999; Handa, 2003; Sugiura, 2008). In some ferns, there may be hundreds of editing sites in plastid transcripts (Wolf et al., 2004), while in the moss Physcomitrella, there are probably less than 10 sites in mitochondrial transcripts (Terasawa et al., 2007). In total, 34 editing sites have been identified to date in Arabidopsis chloroplast transcripts (Chateigner-Boutin and Small, 2007), almost half of which are in subunits of NDH transcripts ndhB, ndhF, ndhD, and ndhG, while there are only three known sites in the transcripts encoding the PSII proteins psbE, psbF, and psbZ (Chateigner-Boutin and Small, 2007). Several mutants deficient in specific RNA editing sites in plastid transcripts have been identified, and the genes responsible for them have been characterized (Bock et al., 1994; Kotera et al., 2005; Okuda et al., 2007, 2009; Chateigner-Boutin et al., 2008; Zhou et al., 2009). Here, we report the identification of a high chlorophyll fluorescence Arabidopsis mutant, low psii accumulation66 (lpa66), with reduced levels of PSII. We present evidence that LPA66 is specifically involved in editing psbF and that the amino acid alteration due to lack of editing is responsible for the perturbation of efficient assembly of PSII complexes in the mutant.
RESULTS
PSII Activity Was Impaired in the lpa66-1 Mutant
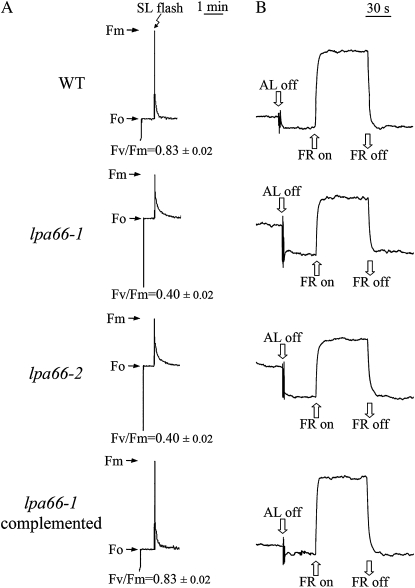
The lpa66-1 mutant was isolated by screening for mutants from the Scheible and Somerville T-DNA Arabidopsis lines (Weigel et al., 2000) with a high chlorophyll fluorescence phenotype (Meurer et al., 1996; Peng et al., 2006; Ma et al., 2007). The mutant plants showed reduced growth, and the leaves appeared pale green under optimal growth conditions (Supplemental Fig. S1). The ratio of variable fluorescence to maximum fluorescence (Fv/Fm), reflecting the maximum potential capacity of the photochemical reactions of PSII (Krause and Weis, 1991), was significantly lower in the mutant (0.40 ± 0.02) than in wild-type plants (0.83 ± 0.02; Fig. 1A), indicating that PSII functions were perturbed in the mutant. However, P700 could be oxidized in the mutant plants (Fig. 1B), suggesting that PSI was functional, as reportedly observed in both lpa1 and lpa2 mutants (Peng et al., 2006; Ma et al., 2007).
Figure 1.
Spectroscopic analysis of wild-type (WT), lpa66-1 and lpa66-2 mutant, and lpa66-1 complemented transgenic plants. A, Chlorophyll fluorescence induction. Fm, Maximum fluorescence yield; Fo, minimum fluorescence yield when PSII centers are open; SL, saturating light. The ratios of variable to maximum fluorescence, reflecting the maximum potential of PSII photochemical reactions, were calculated from Fv/Fm = (Fm − Fo)/Fm. B, Redox kinetics of P700. The oxidation state of P700 was investigated by measuring absorbance changes of P700 at 820 nm induced by far-red light (FR; 720 nm).
Molecular Cloning of the LPA66 Gene
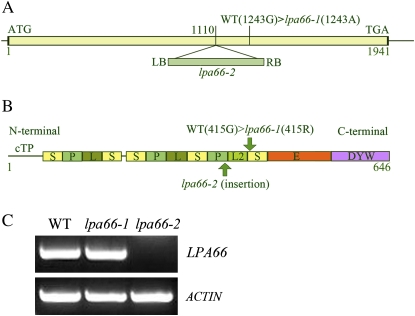
Genetic analysis showed that the lpa66-1 mutation was recessive and that the lpa66-1 mutant phenotype did not cosegregate with the phosphinothricin resistance marker, indicating that the mutated LPA66 gene is not tagged by the T-DNA (data not shown). Map-based cloning of the lpa66-1 mutant based on simple sequence length polymorphism molecular markers revealed a nucleotide substitution in the gene At5g48910 (Fig. 2A), which led to an amino acid change of Gly to Arg (Fig. 2B). Reverse transcription (RT)-PCR analysis showed that the abundance of At5g48910 transcripts in the lpa66-1 mutant was comparable to that in wild-type plants (Fig. 2C). An independent T-DNA insertion line carrying a T-DNA insertion at nucleotide position 1,110 bp of the At5g48910 gene relative to the ATG codon from the Arabidopsis Biological Resource Center (ABRC) was designated lpa66-2. The phenotype of lpa66-2 mutants was pale green and indistinguishable from that of lpa66-1 (Supplemental Fig. S1). No expression of At5g48910 in the lpa66-2 mutant was detected by RT-PCR analysis (Fig. 2C), indicating that it is a null mutant. Therefore, attention was mainly focused in the analyses reported here on the lpa66-1 mutant, and the discussed results apply solely to this mutant, unless otherwise specified.
Figure 2.
Identification of the lpa66 mutation. A, Schematic diagram of the LPA66 gene. The box (1–1,941bp) represents the coding region of LPA66 from ATG to TGA without introns. The lpa66-1 and lpa66-2 mutations are indicated. LB and RB represent the left and right borders, respectively, of the inserted T-DNA in the lpa66-2 line. The diagram is not drawn to scale. B, Predicted motif structure of LPA66. The designations of the P, L, L2, S, C-terminal E motif, and DYW domain correspond to those proposed by Lurin et al. (2004). cTP indicates the plastid transit peptide. The start and end positions of LPA66 are indicated as 1 and 646, respectively. C, RT-PCR analysis of mutant plants. RT-PCR was performed with specific primers for LPA66 and actin genes. WT, Wild type.
To confirm that the phenotype of the lpa66 mutant was due to the mutation in At5g48910, a complementation experiment was carried out using wild-type genomic At5g48910 DNA. Eight successfully complemented transgenic plants had similar growth rates and chlorophyll fluorescence induction kinetics (Fig. 1; Supplemental Fig. S1). Thus, the inactivation of the At5g48910 gene is responsible for the lpa66 mutant phenotype.
LPA66 Encodes a Chloroplast PPR Protein with an E Motif and a DYW Domain
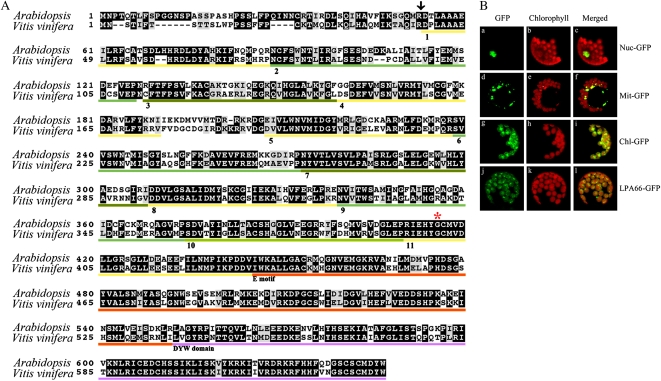
The LPA66 (At5g48910) gene is not disrupted by any introns and encodes a protein of 646 amino acids. The N-terminal 53 amino acids are predicted to be a chloroplast transit peptide by the programs TargetP 1.1 and ChloroP 1.1 (Fig. 3A). To determine the subcellular localization of the LPA66 protein, a fragment of the 257 N-terminal amino acids of LPA66 was fused to the N terminus of the synthetic GFP (sGFP) with a S65T mutation. The LPA66-GFP fusion proteins were transiently expressed in protoplasts under the control of the cauliflower mosaic virus 35S promoter, and GFP fluorescence was found to be exclusively colocalized with the chloroplastic chlorophyll, in accordance with results obtained when the GFP was fused to the transit peptide of the small subunit of ribulose bisphosphate carboxylase (Lee et al., 2002b). When GFP was fused to the targeting signals of the fibrillarin and FRO1 proteins from Arabidopsis (Pih et al., 2000; Lee et al., 2002a), GFP signals were found to be located specifically in the nucleus and mitochondria, respectively (Fig. 3B). Thus, these results indicate that LPA66 is localized to the chloroplast.
Figure 3.
LPA66 sequence alignment and subcellular localization. A, Amino acid alignment of LPA66 (At5g48910). The amino acid sequence of At5g48910 was compared with the homologous sequence from grapevine (LOC100261359). The predicted cleavage site for the plastid transit peptide is indicated by an arrow. PPR motifs are numbered. The orange line beneath the sequences indicates the E motif. The DYW domain is purple underlined, and the mutated amino acid in lpa66 is marked by the red asterisk. B, Subcellular localization of the LPA66 protein according to fluorescence signals visualized using confocal laser scanning microscopy. The indicated fusion proteins with sGFP were transiently expressed in protoplasts under the control of the cauliflower mosaic virus 35S promoter, and the green GFP signals were obtained by confocal microscopy (A, D, G, and J). The chloroplasts were visualized by chlorophyll autofluorescence (B, E, H, and K). The colocalization of GFP and chloroplasts is indicated in merged images (C, F, I, and L). The constructs used for transformation are indicated to the right: Nuc-GFP, control with nuclear localization signal of fibrillarin; Mit-GFP, control with mitochondrial localization signal of FRO1; Chl-GFP, control with the transit peptide of the ribulose bisphosphate carboxylase small subunit; LPA66-GFP, signals from the LPA66-GFP fusion protein.
BLAST searches revealed that LPA66 is a member of the PPR protein family. It contains 11 PPR and PPR-like motifs followed by an E motif and a DYW domain in its C terminus (Figs. 2B and 3A). Thus, according to the classification of PPR proteins (Lurin et al., 2004; Rivals et al., 2006), LPA66 is a DYW protein of the PLS (PCMP) subfamily. Genomic database searches and protein sequence alignments revealed that it shares significant sequence identity with an unknown protein in grapevine (Vitis vinifera; 66% identity, 81% similarity; Fig. 3A). However, our efforts to identify potential LPA66 orthologs in P. patens, spinach (Spinacia oleracea), tobacco (Nicotiana tabacum), rice, wheat (Triticum aestivum), and maize, based on BLAST search of genome or related ESTs, were unsuccessful (data not shown).
Editing of psbF mRNA Transcripts Is Impaired in lpa66
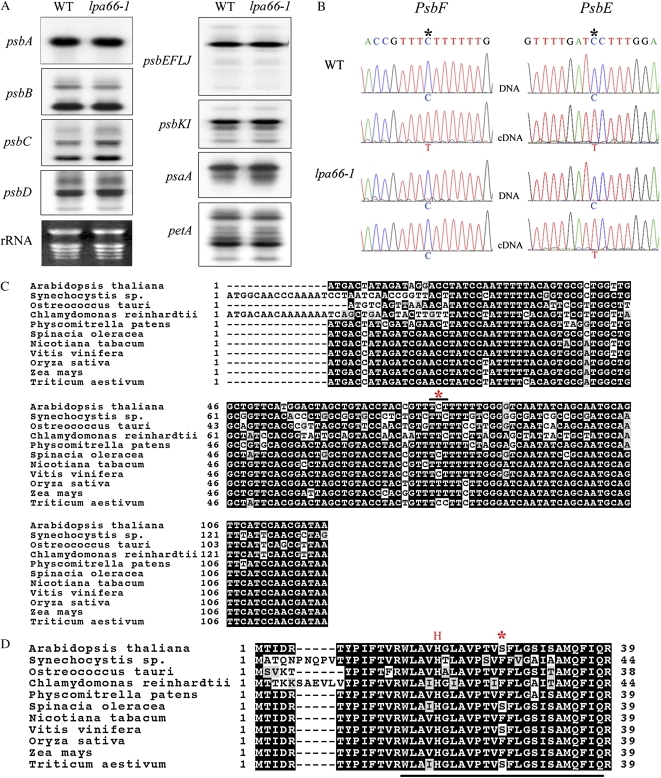
To assess the possibility that the defective PSII function in lpa66 mutants is due to a defect at the RNA transcript level, we compared the abundance and patterns of chloroplast mRNA transcripts in the mutants and wild-type counterparts by RNA gel-blot hybridization analysis. Our results showed that there were similar amounts of psbA and psbC transcripts (encoding D1 and CP43 proteins of PSII, respectively) in the mutant and wild-type plants (Fig. 4A). In addition, similar abundance and patterns of PSII transcripts, including psbD/C (encoding D2 and CP43 proteins, respectively), psbEFLJ (encoding the α- and β-subunits of cytochrome b559, PsbL, and PsbJ proteins, respectively), and psbKI (encoding the PsbK and PsbI proteins, respectively), were observed in the lpa66 mutant and wild-type plants (Fig. 4A). There were also no significant differences in levels of psaA and petA transcripts (encoding PSI subunit PsaA and cytochrome b6f subunit cytochrome f, respectively) between the mutant and wild-type plants (Fig. 4A). Several PPR proteins of the PLS family have been shown to be involved in RNA editing (Kotera et al., 2005; Okuda et al., 2007, 2009; Chateigner-Boutin et al., 2008; Zhou et al., 2009). Therefore, we examined the possibility that LPA66 has such a role by directly sequencing RT-PCR products encompassing the 34 editing sites identified to date in Arabidopsis chloroplast transcripts, in both mutant and wild-type plants, using the primers listed in Supplemental Table S1. In the wild-type plants, nucleotide 77C was edited to 77U in psbF transcripts, which introduces a conserved Phe (TTT) instead of Ser (TCT) in the β-subunit of cytochrome b559 (Fig. 4B). However, the editing of this target site in psbF transcripts was blocked in lpa66 mutant plants, while in the same operon, the other editing site in psbE transcripts was processed normally when compared with the wild type (Fig. 4B). The other 32 editing sites, including the remaining PSII site in psbZ transcripts, were “correctly” edited in the mutant (i.e. in an identical manner to the editing in wild-type plants; data not shown).
Figure 4.
RNA transcripts and editing analysis in lpa66 mutants. A, RNA gel blot hybridization with total RNA from leaves of wild-type (WT) and lpa66-1 mutant plants. Ten micrograms of total leaf RNA per well from 5-week-old wild-type and lpa66-1 plants was loaded per well. The probes for the genes psbA, psbB, psbC, psbD, psbEFLJ, psbKI, psaA, and petA are indicated to the left. rRNA was visualized by staining with ethidium bromide as an equal loading control. B, Analysis of RNA editing of psbF and psbE transcripts. RT-PCR products containing the psbF and psbE editing sites (asterisks) were directly sequenced. C and D, Alignments of psbF gene sequences (C) and predicted proteins directly translated from genomic sequences (D) from various organisms. The chloroplast genomic accession numbers are NC_000932 (Arabidopsis thaliana), NC_000911 (Synechocystis PCC 6803), NC_008289 (Ostreococcus tauri), NC_005353 (Chlamydomonas reinhardtii), NC_005087 (Physcomitrella patens), NC_002202 (Spinacia oleracea), NC_001879 (Nicotiana tabacum), DQ424856 (Vitis vinifera), NC_001320 (Oryza sativa), NC_001666 (Zea mays), and NC_002762 (Triticum aestivum). The editing site and its corresponding amino acid residue are indicated by asterisks (C and D, respectively); the His residue for heme binding is indicated by the red H, and the transmembrane region is underlined in D.
Reduced Levels of PSII Proteins in lpa66
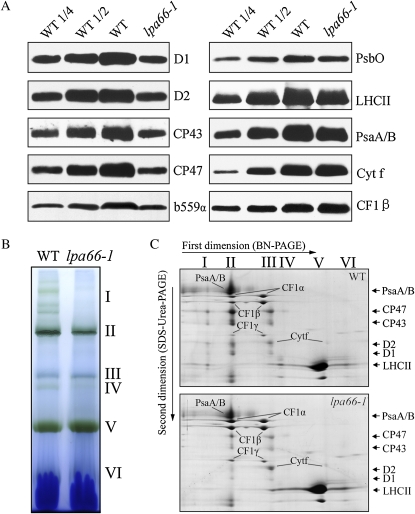
To assess the possibility that impairment of the PSII function might be reflected at the level of chloroplast proteins, immunoblot analysis was performed with specific antibodies against the subunits of photosynthetic protein complexes using total protein extraction prepared from the leaves of mutant and wild-type plants. The protein contents of the chloroplast-encoded PSII subunits D1, D2, cytochrome b559, CP47, and CP43 were found to be reduced to approximately 25% of wild-type levels. The levels of nucleus-encoded PSII proteins, the 33-kD protein of the oxygen-evolving complex, and LHCII were slightly reduced in the mutants (Fig. 5A). The contents of PSI reaction center PsaA/B proteins were also slightly reduced compared with wild-type levels (Fig. 5A), but the levels of cytochrome f of the cytochrome b6f complex and the CF1β-subunit of the ATP synthase were slightly increased (Fig. 5A).
Figure 5.
Levels of chloroplast proteins. A, Immunodetection of chloroplast proteins. The total protein extracts (10 μg of proteins) from 5-week-old wild-type (WT) and lpa66-1 leaves were separated by SDS-urea-PAGE and blotted, and the blots were probed with specific antibodies: anti-D1, anti-D2, anti-CP43, anti-CP47, anti-cytochrome b559α, anti-PsbO, anti-LHCII, anti-PsaA/B, anti-cytochrome f, and anti-CF1β. B, BN gel analysis of thylakoid membrane protein complexes. Thylakoid membranes (10 μg of chlorophyll) from 5-week-old wild-type and lpa66-1 leaves were solubilized with 1% dodecyl-β-d-maltoside and separated by 6% to 12% gradient BN gel electrophoresis. The positions of protein complexes representing PSII supercomplexes (I), monomeric PSI and dimeric PSII (II), monomeric PSII (III), CP43 minus PSII (IV), trimeric LHCII/PSII reaction center (V), and monomeric LHCII (VI) are identified as described previously (Guo et al., 2005; Peng et al., 2006). C, Two-dimensional separation of protein complexes in the thylakoid membranes. After separated on the BN gel, the complex proteins in a single lane were further separated by 15% SDS-urea-PAGE and stained with Coomassie Brilliant Blue. Names of the proteins resolved by the second-dimension SDS-PAGE, previously identified, are indicated by arrows (Peng et al., 2006).
In further analyses, the composition of photosynthetic protein complexes in the thylakoid membranes of mutant and wild-type plants was analyzed by blue-native (BN)-PAGE electrophoresis (Schagger et al., 1994; Guo et al., 2005), which resolved six major bands corresponding to PSII supercomplexes (band I), monomeric PSI and dimeric PSII (band II), monomeric PSII (band III), CP43-free PSII (band IV), trimeric LHCII/PSII reaction center (band V), and monomeric LHCII (band VI; Guo et al., 2005). As shown in Figure 5B, the amounts of chlorophyll-containing protein complexes (labeled I and II) were reduced in the lpa66 mutant compared with that in wild-type plants when thylakoid membranes containing equal amounts of chlorophyll were compared. Further analyses of the two-dimensional SDS-urea-PAGE gels after Coomassie Brilliant Blue staining showed that the PSII core proteins (D1/D2/CP47/CP43) in PSII supercomplexes, dimers, and monomers were significantly reduced in the mutant (Fig. 5C). In addition, there was more LHCII accumulated in band V in the mutant than in wild-type plants.
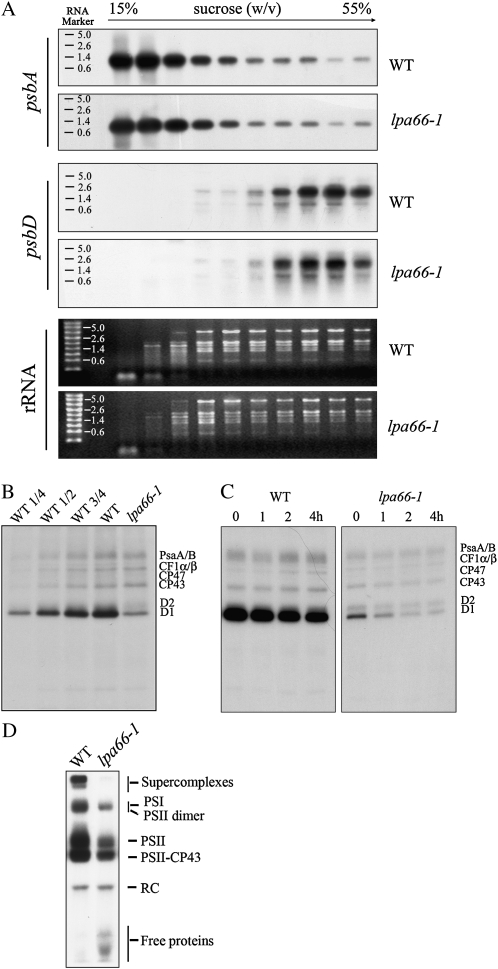
Protein Synthesis Rates of D1 and D2 Were Reduced in lpa66
Reduced levels of PSII complexes may be due to either reduced rates of protein synthesis or increased degradation of PSII subunits. The possible effect of the mutation on the protein synthesis capacity of chloroplasts, therefore, was investigated by analyzing the polysome association patterns of psbA and psbD transcripts after Suc gradient fractionation. The results showed no obvious alterations of polysome association between the mutant and wild-type plants (Fig. 6A). The synthesis and degradation of plastid-encoded thylakoid membrane proteins were further studied in wild-type and mutant leaves by in vivo pulse-chase labeling experiments in the presence of cycloheximide, which inhibits the translation of nucleus-encoded proteins. After a 20-min pulse labeling, the incorporation of [35S]Met into PSII proteins D1 and D2 was dramatically reduced, to about 25% of wild-type levels, while the synthesis rates of other PSII subunits CP43/CP47, PSI reaction center PsaA/B proteins, and the α- and β-subunits of the chloroplast ATP synthase (CF1α/β) were comparable to those in their wild-type counterparts (Fig. 6B). As shown in Figure 6C, the turnover rates of core PSII proteins CP47, CP43, D1, and D2 were increased in the lpa66 mutant during the chase with unlabeled Met after pulse labeling for 20 min.
Figure 6.
Polysome association and in vivo labeling of wild-type (WT) and lpa66-1 plants. A, Association of psbA and psbD mRNAs with polysomes. Ten fractions of equal volume were collected from the top to bottom of 15% to 55% Suc gradients, and equal proportions of the RNA purified from each fraction were analyzed by gel-blot hybridization. rRNAs were detected by ethidium bromide staining. The RNA size markers are indicated to the left. B, Pulse labeling of thylakoid membrane proteins. After pulse labeling young Arabidopsis seedlings in the presence of cycloheximide for 20 min, thylakoid membranes were isolated, and the proteins were separated by SDS-urea-PAGE and visualized autoradiographically. C, Pulse and chase labeling of thylakoid membrane proteins. After pulse labeling for 20 min followed by 1-, 2-, or 4-h chases with cold Met, thylakoid membranes were isolated, and the proteins were separated by SDS-urea-PAGE and visualized autoradiographically. D, BN gel analysis of labeled thylakoid membrane protein complexes after pulse labeling. After a 20-min pulse in Arabidopsis young seedlings in the presence of cycloheximide, the thylakoid membranes were isolated and solubilized with dodecyl-β-d-maltoside, then the protein complexes were separated by BN-PAGE and visualized autoradiographically. Bands corresponding to various PSII assembly complexes are indicated to the right.
Assembly of PSII Complexes Was Impaired in lpa66
In the wild-type plants, PSII assembly is very efficient (Fig. 6D). After a 20-min pulse labeling, most of the radioactivity detected in PSII components was incorporated in PSII protein complexes, and no visible radioactivity was detected in unassembled proteins. In lpa66 the PSII protein complexes were clearly labeled; however, there was a considerable amount of radioactivity in unassembled proteins after pulse labeling for 20 min. These results indicate that the assembly of PSII proteins into PSII complexes was less efficient in lpa66 mutants (Fig. 6D).
DISCUSSION
psbF editing changes a genomically encoded Ser codon into a Phe codon, which is evolutionarily conserved, in the psbF-encoded protein (β-subunit of cytochrome b559) of many photosynthetically active organisms but not in wild-type tobacco plants, in which the correct Phe codon is already specified at the DNA level (Fig. 4, C and D). Previous studies have shown that replacement of part of the tobacco psbF gene with the homologous region from spinach results in the production of unedited psbF transcripts, suggesting that there is a trans-factor for the psbF site in spinach and presumably many other plants (Bock et al., 1994). Further analysis showed that psbF editing could be restored by the presence of the spinach nucleus through an interspecific protoplast fusion approach (Bock and Koop, 1997). Thus, it is possible that this trans-specific factor has been lost in tobacco due to the lack of selective pressure following loss of the psbF editing site and that this factor is specific for psbF editing. In this study, we report the identification of a PPR protein, LPA66, which is specifically required for editing psbF transcripts in Arabidopsis.
LPA66 is a DYW-class PPR protein containing single E and DYW domains. Similar PPR proteins have been shown to be involved in RNA editing or RNA cleavage. CRR4 and CRR21 are specifically involved in editing sites ndhD-1 and ndhD-2 (Kotera et al., 2005; Okuda et al., 2007), YS1 is required for editing of rpoB-1 (Zhou et al., 2009), while CLB19 has been demonstrated to be required for the editing of two independent sites in rpoA and clpP transcripts (Chateigner-Boutin et al., 2008). CRR22 is involved in editing of the sites ndhB-7, ndhD-5, and rpoB-3, and CRR28 is required for editing of ndhB-2 and ndhD-3 (Okuda et al., 2009). RARE1 is required for editing of accD1 transcripts (Robbins et al., 2009). MEF1, the first plant mitochondrial editing factor, is involved in editing of three specific sites, rps4-956, nad7-963, and nad2-1160, of mRNAs (Zehrmann et al., 2009). The E motif has been suggested to interact with an editing enzyme catalyzing an alteration of C to U, rather than to be the motif that catalyzes the RNA editing reaction (Okuda et al., 2007). Comparison of the C-terminal regions of LPA66 and these factors revealed that LPA66, like YS1, CRR22, CRR 28, MEF1, and RARE1, also contains a DYW domain (Supplemental Fig. S2). The C-terminal DYW domain has been proposed to be an essential (“missing”) catalytic domain for RNA editing activity (Salone et al., 2007), but there is no direct evidence that proteins containing DYW domains have such activity; for example, CRR2 (a PPR protein containing both E and DYW domains) has been reported to be specifically involved in RNA cleavage but not RNA editing (Hashimoto et al., 2003, Okuda and Shikanai, 2008). It appears that, despite their sequence similarity, the functions of different DYW family members have diverged (Okuda et al., 2009).
psbE and psbF belong to the same operon, psbEFLJ, and there is only a 123-bp distance between the editing sites of the transcripts they encode in Arabidopsis. BLAST searches and sequence analysis detected no obvious contiguous conserved cis-elements surrounding the editing sites in psbE and psbF transcripts (data not shown), implying that these two sites may be independently edited. In both lpa66-1 and lpa66-2 mutants, only the psbF site is unmodified by RNA editing, but it was successfully edited in wild-type plants (Fig. 4B). Accordingly, the 26th amino acid of the β-subunit of cytochrome b559 is a hydrophilic Ser in the lpa66 mutants, while it is a hydrophobic Phe in wild-type plants. In cyanobacteria and alga, in which no evidence of RNA editing has been published, the psbF gene encodes a Phe at the corresponding site (Fig. 4, C and D). However, alignment of the predicted sequences of proteins directly translated from the genomes of various higher plant species has shown that this site encodes a Phe in some species (both dicotyledonous and monocotyledonous) and a Ser in others (Fig. 4D). Such distribution of the Phe and Ser codons implies that this editing site existed before the divergence of monocots and dicots. Coupled with the data from tobacco, this suggests a relatively recent loss of the site and the associated editing factor (LPA66) from certain lineages (e.g. tobacco). A protein putatively orthologous to LPA66 is encoded by the genome of grapevine, where a Ser codon is present at the position corresponding to the psbF editing site. This would suggest that in grapevine, this codon is also edited, and that the grapevine protein LOC100261359 is likely to be involved. In contrast, a Phe codon is already present at the position corresponding to the psbF editing site in P. patens, rice, and maize (Fig. 4). Although P. patens, rice, and maize contain many DYW-class PPR proteins with homology to LPA66 (O'Toole et al., 2008), BLAST searches revealed that there are no putative LPA66 orthologs in these species (data not shown). Thus, LPA66 is likely to be a site-specific factor for RNA editing of psbF in the higher plants that require this site to be edited and probably has no other function, as it appears to have been lost in those plants that do not require editing of psbF.
It has been previously demonstrated that cytochrome b559 is essential for the stable accumulation of PSII protein complexes. Deletion of the genes encoding the α- and β-subunit, or both, has been shown to abolish PSII activity completely (Pakrasi et al., 1990; Morais et al., 1998; Swiatek et al., 2003), and mutants examined by Pakrasi et al. (1991), in which one of the heme-ligating His residues was mutated, displayed inefficient PSII assembly and impaired PSII activity. Our results show that defective psbF editing results in reduced growth rates and pale green coloration (Supplemental Fig. S1), phenotypic traits that are associated with reduced levels of PSII proteins (Fig. 5). These results are consistent with previous studies on a tobacco mutant in which the conserved Phe was changed to Ser (Bock et al., 1994; Bondarava et al., 2003), and suggest that the amino acid substitution caused by the lack of editing leads to the reduced levels of PSII proteins observed in the mutants.
The detection of similar amounts and patterns of PSII transcripts (Fig. 4A) in the wild-type and mutant plants indicates that the reduced levels of PSII proteins in the latter may be due to posttranscriptional regulation. The decreased synthesis of D1 and D2 proteins in the mutant may be due to the decreased efficiency of PSII assembly. In Chlamydomonas, cytochrome f synthesis has been shown to be regulated by the level of unassembled cytochrome f in the thylakoid membranes, via interactions with the 5′ untranslated region of petA transcripts (Choquet et al., 1998). A similar mechanism, which has been termed “control by epistasy of synthesis” (CES), has been found to influence the biogenesis of photosynthetic protein complexes in Chlamydomonas (Wostrikoff et al., 2004; Minai et al., 2005). Recently, evidence has been presented indicating that the CES mechanism also operates in higher plant chloroplasts in the regulation of Rubisco large subunit translation in response to its assembly state (Wostrikoff and Stern, 2007). The CES mechanism is crucial for the stoichiometric synthesis and assembly of photosynthetic protein complexes (Choquet and Wollman, 2002; Pogson et al., 2008). Similarly, in Arabidopsis, the presence of unassembled PSII proteins due to the impairment of assembly efficiency in the lpa66 mutant could down-regulate the translation of their corresponding transcripts. It is also interesting that the accumulation of unassembled D1 also leads to the increased turnover rates of D1 in the mutant. Since the accumulation of PSII core protein appears to occur in a coordinated manner, a lesion in one of the main integral subunits of PSII will result in a concomitant decrease of other PSII core proteins (Jensen et al., 1986; de Vitry et al., 1989; Yu and Vermaas, 1990, 1993). In the mutant, the stability of other PSII core proteins was reduced, which may account for the reduced PSII protein levels.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The lpa66-1 mutant was screened from a collection of pSKI015 T-DNA-mutagenized Arabidopsis (Arabidopsis thaliana ecotype Columbia) lines from the ABRC based on the high chlorophyll fluorescence phenotypes using a chlorophyll fluorescence imaging system (Ma et al., 2007). The lpa66-2 mutant (T-DNA insertion line CS813518) was obtained from the ABRC, and the homozygous mutant was verified by PCR using primers LP (5′-GTGAAAACGTTTCCGGTCTCGTACC-3′) and RP (5′-GATGATATCGGGTTATTCGCTGAACGG-3′). The T-DNA insertion was confirmed by PCR and sequencing with primers LB (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′) and RP. Wild-type and homozygous mutant plants were grown in soil under short-day conditions (10 h of light/14 h of dark) with a photon flux density of 120 μmol m−2 s−1 in a growth chamber at 22°C.
Chlorophyll Fluorescence
Chlorophyll fluorescence was measured with a PAM-2000 portable chlorophyll fluorometer (Walz) connected to leaves, which were dark adapted for 30 min before measurements, by a leaf-clip holder (model 2030-B; Walz). The variables Fo, Fm, Fv, and the Fv/Fm ratio were measured and calculated basically according to Meurer et al. (1996). For measurement of light-induced P700 absorbance changes at 820 nm, the PAM chlorophyll fluorometer was equipped with an ED 800T emitter-detector unit (Walz), and the measurements were performed according to Meurer et al. (1996).
Map-Based Cloning and Complementation
The lpa66-1 mutation was mapped with a series of simple sequence length polymorphism markers based on the polymorphisms between two Arabidopsis ecotypes, Columbia and Landsberg erecta (Lukowitz et al., 2000). The mutant plants (Columbia) were crossed with wild-type Landsberg erecta to generate F1 plants. The heterozygotes were allowed to self-fertilize to generate a segregation population. Homozygous F2 mutant plants (lpa66/lpa66) were screened based on the high chlorophyll fluorescence phenotype described above. Genomic DNA was extracted from about 1,024 mutant plants and subjected to PCR with specific molecular markers. The mutation was mapped to a 216-kb region between two bacterial artificial chromosomes (MJE7 and K19E20) on chromosome 5. Candidate genes with a predicted chloroplast transit peptide were sequenced and analyzed with genomic DNA from lpa66 and wild-type plants (Columbia). To confirm the mutation, a complementation experiment was performed as follows. A fragment containing the full-length lpa66 coding sequence was amplified with primers 5′-GTTGAGGATCCATGAACCCAACACAGAC-3′ and 5′-ATGTCTCGAGTCACCAATAATCCATACACG-3′ and subcloned into the pBI121 vector under the control of the cauliflower mosaic virus 35S promoter. The constructs were then transformed into Agrobacterium tumefaciens strain C58 and introduced into the lpa66 mutant plants by a floral dip method (Clough and Bent, 1998). Transgenic plants were planted on Murashige and Skoog medium containing 40 μg mL−1 kanamycin monosulfate. Resistant plants were transferred to soil and grown in a growth chamber to produce seeds. The success of the complementation was confirmed by chlorophyll fluorescence analysis.
Analysis of RNA Editing
Total extracted RNA from leaves of lpa66 mutant and wild-type plants was treated with DNase I and then reverse transcribed with random hexamers (Takara). A series of specific primers (Supplemental Table S1) for the genes encompassing the editing sites in Arabidopsis (Tillich et al., 2005; Chateigner-Boutin and Small, 2007; Okuda et al., 2007) were used to amplify these gene sequences from the cDNA by RT-PCR, and the products were sequenced directly.
Thylakoid Membrane Preparation
Thylakoid membranes were prepared according to Zhang et al. (1999). Briefly, the leaves were homogenized on ice in isolation buffer (400 mm Suc, 50 mm HEPES-KOH, pH 7.8, 10 mm NaCl, and 2 mm MgCl2), and filtered through two layers of cheesecloth. The resulting filtrates were centrifuged at 5,000g for 10 min at 4°C, and their chlorophyll contents were measured according to Porra et al. (1989).
BN-PAGE, SDS-PAGE, and Western-Blot Analysis
BN-PAGE was performed using acrylamide gels with linear 6% to 12% gradients basically according to Peng et al. (2006). For western-blot analysis, proteins in the thylakoid membranes were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with specific primary antibodies, and signals from secondary conjugated antibodies were detected by the enhanced chemiluminescence method.
Northern-Blot and Polysome Association Analyses
Total RNA was extracted from fresh leaf tissues using Trizol reagent, and polysomes were isolated from leaf tissues according to Barkan (1988). RNA in each fraction was isolated, separated, and transferred onto nylon membranes (Amersham Pharmacia Biotech), which were probed with 32P-labeled probes prepared according to Peng et al. (2006) and then exposed to x-ray films.
In Vivo Labeling Assays
In vivo protein labeling was performed essentially as described previously (Meurer et al., 1998). For pulse labeling, primary leaves from 12-d-old plants were incubated in 1 μCi μL−1 [35S]Met in the presence of 20 μg mL−1 cycloheximide for 20 min at 25°C after preincubation with cycloheximide for 30 min. Pulse labeling was followed by a chase in the presence of 1 mm unlabeled Met. After labeling, thylakoid membranes were isolated according to Peng et al. (2006), and the proteins were subjected to SDS-PAGE or BN gel analysis.
GFP Fusion Constructs for Transient Expression in Protoplasts
A fragment encoding the N-terminal 1 to 257 amino acids of LPA66 was amplified by RT-PCR with primers 5′-ACGTCGACATCTTTGTTGATTCTCAATG-3′ and 5′-CCATGGAATCTTTGAAAAACCCGTTCAG-3′ and subcloned into the pUC18-35s-sGFP vector to generate a fusion protein with the GFP as a reporter in the C terminus. In addition, the N-terminal part (1–282 amino acids) of AtFbr1, the transit peptide (1–81 amino acids) of the small subunit of ribulose bisphosphate carboxylase, and the entire coding region of FRO1 except the termination codon were amplified (using primer pairs 5′-ACAACTCGAGATGAGACCCCCAGTTACAGG-3′ and 5′-TCCATGGTCACCTGTTCTGCTGGCTTAAAC-3′, 5′-CACGTCGACAAACCTCAGTCACACAAAGAG-3′ and 5′-TCCATGGATTCGGAATCGGTAAGGTCAGG-3′, and 5′-AATGTCGACGATTTCTCTAATTGACGATGG-3′ and 5′-GTCCATGGAGTTTTCTGGTTGAGGATTGC-3′, respectively) and subcloned into the same vector as nuclear, chloroplast, and mitochondria controls, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LPA66 (At5g48910; NP_199702) and Vitis protein LOC100261359 (XP_002268530).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Photograph showing the phenotypes of 5-week-old wild-type, lpa66-1, lpa66-2, and lpa66-1 mutant complemented with LPA66 cDNA plants.
Supplemental Figure S2. Comparison of the predicted E motif of LPA66 with the corresponding regions of CLB19, CRR2, CRR4, CRR21, CRR22, CRR28, YS1, MEF1, and RARE1.
Supplemental Table S1. Primers used for analyzing RNA editing in Arabidopsis chloroplasts.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the Arabidopsis seeds.
This work was supported by the National Natural Science Foundation of China (grant no. 30725003) and National Basic Research Program of China (grant no. 2009CB118500).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lixin Zhang (zhanglixin@ibcas.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andrés C, Lurin C, Small ID (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol Plant 129 14–22 [Google Scholar]
- Barkan A (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J 7 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572 [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82 549–557 [DOI] [PubMed] [Google Scholar]
- Bock R, Koop HU (1997) Extraplastidic site-specific factors mediate RNA editing in chloroplasts. EMBO J 16 3282–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Kössel H, Maliga P (1994) Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J 13 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarava N, De Pascalis L, Al-Babili S, Goussias C, Golecki JR, Beyer P, Bock R, Krieger-Liszkay A (2003) Evidence that cytochrome b559 mediates the oxidation of reduced plastoquinone in the dark. J Biol Chem 278 13554–13560 [DOI] [PubMed] [Google Scholar]
- Brennicke A, Marchfelder A, Binder S (1999) RNA editing. FEMS Microbiol Rev 23 297–316 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56 590–602 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I (2007) A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res 35 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L (2008) The pentatricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA (1998) Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA 95 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wollman FA (2002) Translational regulations as specific traits of chloroplast gene expression. FEBS Lett 529 39–42 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID (2007) Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans 35 1643–1647 [DOI] [PubMed] [Google Scholar]
- Deng XW, Gruissem W (1987) Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell 49 379–387 [DOI] [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman FA (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol 109 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Walker MB, Barkan A (1999) Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J 18 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege P, Brennicke A (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA 96 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177 115–180 [DOI] [PubMed] [Google Scholar]
- Gothandam KM, Kim ES, Cho H, Chung YY (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58 421–433 [DOI] [PubMed] [Google Scholar]
- Guo J, Zhang Z, Bi Y, Yang W, Xu Y, Zhang L (2005) Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett 579 3619–3624 [DOI] [PubMed] [Google Scholar]
- Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36 541–549 [DOI] [PubMed] [Google Scholar]
- Hattori M, Miyake H, Sugita M (2007) A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J Biol Chem 28 10773–10782 [DOI] [PubMed] [Google Scholar]
- Hoch B, Maier RM, Appel K, Igloi GL, Kossel H (1991) Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353 178–180 [DOI] [PubMed] [Google Scholar]
- Jensen KH, Herrin DL, Plumley FG, Schmidt GW (1986) Biogenesis of photosystem II complexes: transcriptional, translational and posttranslational regulation. J Cell Biol 103 1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330 [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42 313–349 [Google Scholar]
- Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K (2003) RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res 31 2417–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK (2002. a) A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Kim DH, Lee SW, Kim ZH, Hwang I (2002. b) In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol Cells 14 388–397 [PubMed] [Google Scholar]
- Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y (2008) Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol 28 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maier RM, Zeltz P, Kössel H, Bonnard G, Gualberto JM, Grienenberger JM (1996) RNA editing in plant mitochondria and chloroplasts. Plant Mol Biol 32 343–365 [DOI] [PubMed] [Google Scholar]
- Manuell A, Beligni MV, Yamaguchi K, Mayfield SP (2004) Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem Soc Trans 32 601–605 [DOI] [PubMed] [Google Scholar]
- Marín-Navarro J, Manuell AL, Wu J, Mayfield SP (2007) Chloroplast translation regulation. Photosynth Res 94 359–374 [DOI] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Wosterikoff K, Wollman FA, Choquet Y (2005) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais F, Barber J, Nixon PJ (1998) The chloroplast-encoded alpha subunit of cytochrome b-559 is required for assembly of the photosystem two complex in both the light and the dark in Chlamydomonas reinhardtii. J Biol Chem 273 29315–29320 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Meierhoff K, Westhoff P, Schuster G (2003) RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem 270 4070–4081 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystem I and II. Annu Rev Plant Biol 57 521–565 [DOI] [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T (2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem 281 37661–37667 [DOI] [PubMed] [Google Scholar]
- Okuda K, Shikanai T (2008) PPR proteins function as a trans-factor in chloroplast RNA editing. In JF Allen, E Gantt, JH Golbeck, B Osmond, eds, Photosynthesis. Energy from the Sun: 14th International Congress on Photosynthesis. Springer, Dordrecht, The Netherlands, pp 1211–1214
- O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25 1120–1128 [DOI] [PubMed] [Google Scholar]
- Pakrasi HB, De Ciechi P, Whitmarsh J (1991) Site directed mutagenesis of the heme axial ligands of cytochrome b559 affects the stability of the photosystem II complex. EMBO J 10 1619–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakrasi HB, Nyhus KJ, Granok H (1990) Targeted deletion mutagenesis of the beta subunit of cytochrome b559 protein destabilizes the reaction center of photosystem II. Z Naturforsch C 45 423–429 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2005) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih KT, Yi MJ, Liang YS, Shin BJ, Cho MJ, Hwang I, Son D (2000) Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol 123 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13 602–609 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Rivals E, Bruyere C, Toffano-Nioche C, Lecharny A (2006) Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol 141 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA (in press) [DOI] [PMC free article] [PubMed]
- Salone V, Rudinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett 581 4132–4138 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y (2001) Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J Biol Chem 276 3937–3940 [DOI] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, Vonjagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217 220–230 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Herrmann RG, Maier RM (2004) Evolutionary fluctuation of plastid RNA editing. Endocytobiosis Cell Res 15 246–255 [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol Life Sci 63 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M (2008) RNA editing in chloroplasts. Nucleic Acids Mol Biol 20: 123–142
- Swiatek M, Regel RE, Meurer J, Wanner G, Pakrasi HB, Ohad I, Herrmann RG (2003) Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: the psbEFLJ operon in Nicotiana tabacum. Mol Gen Genet 268 699–710 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24 699–709 [DOI] [PubMed] [Google Scholar]
- Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM (2005) Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J 43 708–715 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Barkan A (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36 675–686 [DOI] [PubMed] [Google Scholar]
- Wintz H, Hanson MR (1991) A termination codon is created by RNA editing in the petunia atp9 transcript. Curr Genet 19 61–64 [DOI] [PubMed] [Google Scholar]
- Wolf PG, Rowe CA, Hasebe M (2004) High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene 339 89–97 [DOI] [PubMed] [Google Scholar]
- Wollman FA, Minai L, Nechushtai R (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim Biophys Acta 1411 21–85 [DOI] [PubMed] [Google Scholar]
- Wostrikoff K, Girad-Bascou J, Wollman FA, Choquet Y (2004) Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J 23 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplast. Proc Natl Acad Sci USA 104 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tasaka M, Shikanai T (2004) PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J 38 152–163 [DOI] [PubMed] [Google Scholar]
- Yu J, Vermaas W (1990) Transcript levels and synthesis of photosystem II components in cyanobacterial mutants with inactivated photosystem II genes. Plant Cell 2 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vermaas W (1993) Synthesis and turnover of photosystem II reaction centre polypeptides in cyanobacterial D2 mutants. J Biol Chem 268 7407–7413 [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M (2009) A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W (2000) Translation in chloroplasts. Biochimie 82 583–601 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58 82–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.