Abstract
Searches of sequenced genomes of diverse organisms revealed that the moss Physcomitrella patens is the most primitive organism possessing oleosin genes. Microscopy examination of Physcomitrella revealed that oil bodies (OBs) were abundant in the photosynthetic vegetative gametophyte and the reproductive spore. Chromatography illustrated the neutral lipids in OBs isolated from the gametophyte to be largely steryl esters and triacylglycerols, and SDS-PAGE showed the major proteins to be oleosins. Reverse transcription-PCR revealed the expression of all three oleosin genes to be tissue specific. This tissue specificity was greatly altered via alternative splicing, a control mechanism of oleosin gene expression unknown in higher plants. During the production of sex organs at the tips of gametophyte branches, the number of OBs in the top gametophyte tissue decreased concomitant with increases in the number of peroxisomes and level of transcripts encoding the glyoxylate cycle enzymes; thus, the OBs are food reserves for gluconeogenesis. In spores during germination, peroxisomes adjacent to OBs, along with transcripts encoding the glyoxylate cycle enzymes, appeared; thus, the spore OBs are food reserves for gluconeogenesis and equivalent to seed OBs. The one-cell-layer gametophyte could be observed easily with confocal microscopy for the subcellular OBs and other structures. Transient expression of various gene constructs transformed into gametophyte cells revealed that all OBs were linked to the endoplasmic reticulum (ER), that oleosins were synthesized in extended regions of the ER, and that two different oleosins were colocated in all OBs.
Eukaryotes and prokaryotes contain neutral lipids in subcellular droplets as food reserves and/or for other purposes (Hsieh and Huang, 2004; Martin and Parton, 2006; Goodman, 2008; Rajakumari et al., 2008). These lipid droplets are present in seeds, pollens, fruits, and flowers of higher plants; the vegetative and reproductive organs of lower plants, algae, fungi, and nematodes; mammalian organs/tissues, such as mammalian glands and adipose tissues; and bacteria. Among all these lipid droplets, oil bodies (OBs) in seeds are the most prominent and have been extensively studied.
Seeds of diverse plant species store oils (triacylglycerols [TAGs]) as food reserves for germination and postgermination growth (Napier et al., 1996; Frandsen et al., 2001; Murphy, 2001; Hsieh and Huang, 2004). The TAGs are present in small subcellular, spherical OBs of approximately 0.5 to 2 μm in diameter. Each OB has a matrix of TAGs surrounded by a layer of phospholipids (PLs) and the structural protein oleosins. The massive oleosins completely cover the surface of the OBs and prevent them from coalescence; so, a large surface area per unit TAG is available for lipase binding and catalysis during germination. Each oleosin molecule has a characteristic long central hydrophobic stretch, which forms a hairpin penetrating into the matrix TAGs for stable anchorage.
Other than being present in the seeds of plants, oleosin-coated OBs are also present in pollen (probably for storage of acyl moieties for tube elongation [Kim et al., 2002]) and the tapeta of Brassica (Brassica napus and Brassica rapa) and Arabidopsis (Arabidopsis thaliana; Hsieh and Huang, 2005, 2007). Inside each tapetum cell, many oleosin-coated OBs associate with numerous flavonoid-containing vesicles to form large subcellular particles termed tapetosomes, each 2 to 3 μm in diameter. Tapetosomes temporarily store lipids and flavonoids, which are deposited onto the maturing pollen as a pollen coat for water-proofing and UV protection, respectively. In fruits of some species, such as olive (Olea europaea), avocado (Persea americana), and oil palm (Syagrus cocoides), the fleshy mesocarp possesses much larger (10–50-μm diameter) subcellular lipid particles of TAGs, which are devoid of surface oleosins and apparently are for attracting animals for seed dispersal (Murphy, 2001; Hsieh and Huang, 2004). OBs are also present, although generally in low abundance, in leaves of diverse plant species, and their structures and functions are unknown (Lersten et al., 2006).
Oleosins of all plant species contain a conserved central hydrophobic hairpin of approximately 72 residues flanked by less-conserved amphipathic N and C termini of highly variable length (Hsieh and Huang, 2004). Within the hairpin, the turn consists of 12 most-conserved residues (PX5SPX3P), of which the three Pro and one Ser residues (termed the Pro knot) are completely conserved without a single exception among hundreds of examined oleosins of various species. Paralogs of oleosin genes are present within each species, and individual paralogs are expressed in a tissue-specific manner. For example, Arabidopsis has 17 oleosin genes, which are selectively expressed in seed, pollen, and the tapetum (Kim et al., 2002).
Oleosins and TAGs are generally believed to be synthesized on the endoplasmic reticulum (ER) inside a seed cell. Whereas the nascent oleosins are attached to the ER surface via the long hydrophobic hairpin stretch, TAGs are sequestered between the two PL layers of the ER membrane. These oleosins and TAGs migrate to and are eventually concentrated in confined ER regions, which are detached to form mature OBs (Napier et al., 1996; Murphy, 2001; Abell et al., 2004; Hsieh and Huang, 2004). What is uncertain is the location of the ER on which oleosins and TAGs are synthesized. These major OB components could be synthesized in specific ER subdomains, as interpreted from results of immunodetection of oleosins with transmission electron microscopy (TEM; Herman, 1987), biochemical isolation, and enzymatic analysis of ER subfractions (Lacey et al., 1999) and fluorescence detection of TAG-synthesizing enzymes with confocal laser scanning microscopy (CLSM; Shockey et al., 2006). Existence of OB-synthesizing ER subdomains would raise the possibility that each subdomain produces an OB with only one of the several oleosin isoforms in the cell. Alternatively, oleosin and TAG synthesis could occur in nonspecific, extended regions of the ER (Hsieh and Huang, 2004).
Lipid droplets in cells of nonplant organisms, such as mammals and yeast, also possess surface proteins with structural and/or metabolic functions (Martin and Parton, 2006; Goodman, 2008; Rajakumari et al., 2008). These surface proteins are not related to oleosins (Ting et al., 1997) and do not possess a long hydrophobic stretch. Their polypeptides wrap around rather than penetrate into the lipid droplets. Whereas seed OBs possess only TAGs as the matrix lipids, the lipid droplets in mammals and yeast contain both TAGs and steryl esters (SEs). The evolutionary relationship of plant OBs and nonplant lipid droplets is unknown.
To explore the evolutionary trends of OBs and oleosins in primitive plants, algae, and fungi, we searched for genes encoding oleosins in these organisms having completely sequenced genomes. Only the moss Physcomitrella patens (three paralogs) and the primitive fern (fern ally; Selaginella moellendorffii; eight paralogs) possess genes encoding oleosins. Primitive plants, including bryophytes (mosses) and ferns, contain neutral lipids and OBs (Swanson et al., 1976; Pihakaski et al., 1987; Dembitsky, 1993). We chose the more primitive Physcomitrella for intensive study. The three oleosin genes are expressed in a tissue-specific manner, which is further regulated via alternative splicing, a process unknown with oleosin genes in higher plants. OBs in the dehydrated spore resemble those in seeds in being food reserves for germination. OBs in the nondehydrated, photosynthetic gametophyte, although harboring surface oleosins, possess both TAGs and SEs esters and thus resemble more the lipid droplets in mammals and yeasts. The gametophyte OBs are for gluconeogenesis when food reserves are needed, such as during sex organ production. The one-cell-layer gametophyte can be used for transient expression of oleosin genes for convenient microscopy exploration. The approach shows that all OBs are linked to extended regions of the ER, on which different oleosins are synthesized concurrently. Here, we report our findings.
RESULTS
Abundant OBs Are Present in the Photosynthetic Gametophyte and Dehydrated Spore
We used light microscopy and TEM to observe OBs in Physcomitrella cells throughout the life cycle (Fig. 1). Cells of the protonema, which were young tissues grown from spore after germination for 10 d, contained no or few OBs. The cells had conspicuous plastids with large starch grains. Cells of the mature gametophyte, which was the conspicuous photosynthetic branches, contained numerous OBs. These OBs could be observed after Sudan Black staining and were as numerous as the larger chloroplasts (approximately 150 per 100 μm × 100 μm). The spherical OBs had heterogeneous sizes, of <1 μm to several micrometers in diameter. Cells of the antheridium, the male reproductive structure, had one to two OBs per cell; they were strongly electron dense after osmium fixation, presumably possessing highly unsaturated lipids. Early cells of the archegonium, the female sex structure, contained no or few OBs. Some internal cells of the archegonium differentiated into spore mother cells and began to accumulate OBs, even before meiosis. After meiosis, the spore continued to accumulate OBs, and the mature, dehydrated spores were packed with OBs of various sizes, from 0.2 to 3 μm in diameter. Upon germination, the spore became less hydrated, and peroxisomes (glyoxysomes) appeared.
Figure 1.
Microscopy images of different tissues of Physcomitrella during its life cycle, showing the presence or absence of subcellular OBs. Samples were photographed with light microscopy (all in color) or TEM (black and white). A, Haploid cells (from top to bottom) of protonema (immature tissue grown from spore after germination for 10 d), mature gametophytes (dark OBs [stained with Sudan Black B] among green chloroplasts), antheridium (dark OBs), and archegonia (no OBs). B, Sporophytes during maturation (from top to bottom). Left column shows the whole sporophyte, and right column reveals diploid cells enclosing the locule in which spore mother cells (smc) became spore. C, Mature spore packed with OBs but no peroxisomes (left two images) and germinating spore with numerous peroxisomes (P; right two images).
OBs Isolated from the Photosynthetic Gametophyte Contain Oleosins, SEs, and TAGs
We could not collect enough spores, which were of minute sizes (20 μm in diameter), and then crack their hard shell gently for isolation of the internal OBs. Thus, we isolated the OBs from the photosynthetic gametophyte after gentle homogenization of the cells and floatation centrifugation. TEM of the floated OB fraction revealed OBs of heterogeneous sizes, ranging from 0.5 to 5 μm (Fig. 2A). SDS-PAGE showed that the OB fraction was highly enriched with protein(s) of approximately 17 kD (Fig. 2B). The protein was extracted from the gel and subjected to trypsin digestion. The resulting fragments were analyzed with matrix-assisted laser-desorption/ionization time of flight (MALDI-TOF) mass spectrometry and identified with use of the predicted protein database derived from the Physcomitrella genome (http://moss.nibb.ac.jp/). They corresponded to the N-terminal fragments of OLE1 and OLE2 (whole proteins predicted to be 13–21 kD; to be described). Thus, the approximately 17-kD proteins highly enriched in the OB fraction were OLE1 and OLE2.
Figure 2.
Analyses of an OB fraction isolated from mature, green gametophyte branches. A, TEM of the OB fraction, showing spherical OBs of heterogeneous sizes. B, SDS-PAGE gel of proteins in total extract and OB fraction. Molecular marker positions are indicated. C, TLC plate of neutral lipids in total extract and OB fraction. Approximate locations of lipid groups, SEs (cholesteryl palmitate as marker), TAGs (triolein), free fatty acids (FFA; oleic acid), sterols (cholesterol), DAGs (1,3- and 1,2-diolein), and monoacylglycerols (MAGs; 1-monoolein) are indicated.
The neutral lipids of the OB fraction were analyzed with thin-layer chromatography (TLC) and visualized after sulfuric acid spraying (Fig. 2C). The major lipids were SEs and TAGs, and diacylglycerols (DAGs) were in lesser amounts. The SEs were identified with HPLC-mass spectrometry (HPLC-MS) to be lanosterol esters (42.7% of all SEs), stigmasterol esters (30.32%), β-sitosterol esters (19.97%), and campesterol esters (8.89%). The TAGs contained the common acyl moieties of oleic (O), linoleic (L), linolenic (Ln), and palmitic (P) acids and were O/L/Ln (28.68% of all TAGs), L/L/O and O/O/L (not separated, 20.73%), P/L/L (20.73%), L/L/L (16.90%), L/L/Ln (7.76%), and O/O/O (5.21%).
Physcomitrella Is the Most Primitive Organism Possessing Oleosins
We used (1) the conserved Pro knot sequence (PX5SPX3P) and (2) the complete hairpin sequence of oleosins from different plant species as queries to search for oleosins in the Joint Genome Initiative database. Attention was paid to organisms whose genomes have been completely sequenced. The searched primitive species included lycophytes (S. moellendorffii), bryophytes (P. patens), algae and related organisms (Aureococcus anophagefferens, Chlamydomonas reinhardtii, Chlorella sp., Emiliania huxleyi, Micromonas pusilla, Ostreococcus lucimarinus, Phaeodactylum tricornutum, and Volvox carteri), fungi and related organisms (Aspergillus niger, Batrachochytrium dendrobatidis, Cochliobolus heterostrophus, Laccaria bicolor, Mycosphaerella fijiensis, Nectria haematococca, Phanerochaete chrysosporium, Phycomyces blakesleeanus, Saccharomyces cerevisiae, Pichia stipitis, Postia placenta, and Sporobolomyces roseus), and oomycetes (Phytophthora ramorum). Among these species, only the primitive fern (fern ally; S. moellendorffii; eight genes) and the moss P. patens (three genes) had oleosin genes. Clearly, no oleosin gene was present in algae, fungi, and oomycetes. Thus, the moss Physcomitrella was the most primitive organism found to possess oleosin genes.
An unrooted phylogenetic tree of oleosins in species with completely sequenced genomes was constructed on the basis of their conserved hairpin sequences plus the moderately conserved sequences immediately flanking the hairpin (Fig. 3). The tree includes 17 oleosins from Arabidopsis, six from rice (Oryza sativa), eight from Populus, eight from Selaginella, and three from Physcomitrella. The oleosin genes in the higher plants Arabidopsis, rice, and Populus have had more variations. Physcomitrella has the least variations and the fewest oleosin genes. A pileup of the amino acid sequences of these oleosins revealing the conserved and nonconserved residues is in Supplemental Table S1.
Figure 3.
An unrooted phylogenetic tree of oleosins from Physcomitrella and several other representative species constructed on the basis of their predicted amino acid sequences. Arabidopsis (At; a nonwoody dicot), Populus trichocarpa (Pt; a woody dicot), O. sativa (Os; a monocot), Selaginella mutica (Sm; a primitive fern), and P. patens (Pp) genomes have been completely sequenced, and all their oleosins are included. One pine (Pinus ponderosa) oleosin is used to represent gymnosperm proteins. Nomenclature of the Arabidopsis oleosins follows that reported (Kim et al., 2002); S, T, and SM denote oleosins present specifically in seed, tapetum, and seed and microspore, respectively. The phylogenetic tree was constructed from aligned sequences of oleosins (the conserved hairpin sequence plus its immediately flanking semiconserved sequences) by a distance method (neighbor-joining) using PHYLIP and 1,000 bootstrap replicates. Bootstrap values higher than 50 are indicated.
The Three Oleosin Genes Are Expressed in a Tissue-Specific Manner, Which Can Be Altered via Alternative Splicing
Reverse transcription (RT)-PCR with use of gene-specific primers was performed to examine the levels of transcripts encoding oleosins and related proteins in various tissues throughout the life cycle of Physcomitrella (Fig. 4). For each of the three oleosin transcripts, the RT-PCR primers detected the sequence encoding the oleosin hairpin region and thus would detect both oleosin isoforms generated via alternative splicing (see next paragraph). OLE1 and OLE2 transcripts were present in all tissues, and their levels in zygotes and spore increased during spore maturation. The OLE3 transcript was present only in the spore samples. Transcripts of genes encoding malate synthase and isocitrate lyase (one gene each per haploid genome) were also present in all tissues but at higher levels in mature and germinated spore. Physcomitrella has four genes encoding putative DAG acyltransferase with use of acyl-CoA as the acyl donor (DAGAT1a, DAGAT1b, DAGAT1c, and DAGAT1d), two genes encoding putative DAG acyltransferase with use of PLs as the acyl donor (DAGAT2a and DAGAT2b), and one gene encoding a putative steryl acyltransferase (SEAT). These genes were annotated on the basis of their sequence similarities with the annotated genes in yeast (Rajakumari et al., 2008). The transcripts of these acyltransferases were present at different levels in diverse tissues. Only the expression of DAGAT1d and SEAT had a clear pattern, similar to that of OLE1 and OLE2, of increasing in level from zygotes to maturing spore. Thus, the data strongly suggest that DAGAT1d and SEAT encode the acyltransferases for synthesis of the storage TAGs and SEs, respectively, in maturing spore.
Figure 4.
RT-PCR of transcripts encoding oleosins and related proteins in various tissues. Tissues shown from left to right are protonema (P); top, middle, and bottom leafy tissues of gametophyte branches (T, M, and B, respectively); antheridia (A); antheridia and archegonia (AA); zygotes (Z), sporophytes of early, middle, and late developmental stages (S1, S2, and S3, respectively); and mature spore (MS) and germinated spore (GS). Transcripts are those encoding oleosins (OLE1, OLE2, and OLE3), malate synthase (MS), isocitrate lyase (ICL), DAG acyltransferases (DAGAT1 [four paralogs] and DAGAT2 [two paralogs] using acyl-CoA and PLs as the acyl donors, respectively), and steryl acyltransferase (SEAT). Approximately equal amounts of the transcript encoding actin (ACT2) were present in the samples.
OLE1 and OLE2 could each produce two different transcripts via alternative splicing, which would result in two oleosin isoforms of different sizes (Fig. 5). The alternative splicing sites occurred downstream of the sequence encoding the hydrophobic hairpin stretch; thus, the resulting two oleosin isoforms still possessed the hairpin stretch and the structural characteristics of an oleosin. Alternative splicing of OLE1 gave OLE1a of 15.3 kD (predicted) and OLE1b of 16.1 kD, and that of OLE2 gave OLE2a of 12.5 kD and OLE2b of 21.7 kD. Importantly, transcripts encoding OLE1a and OLE2a were restricted to spore, whereas those encoding OLE1b and OLE2b were ubiquitous. Thus, the tissue-specific presence of the machineries of alternative splicing (Barbazuk et al., 2008) allows for substantial changes in expression and thus, presumably, differentiation of the oleosin genes and oleosin functions. Preliminary testing of OLE3 for alternative splicing generated negative results, and the predicted OLE3 had 13.8 kD.
Figure 5.
Gene structures of OLE1 and OLE2 and alternative splicing of each gene resulting in two tissue-specific transcripts. A, Arrangement of OLE1 and OLE2 in two scaffolds. Occurrence of two open reading frames (shaded boxes) in each of the two genes via alternative splicing is indicated. The dotted lines represent the sequences encoding the hairpin region. Primers for RT-PCR are shown. B, A pileup of OLE1a, OLE1b, OLE2a, OLE2b, and OLE3. The hairpin sequences in the second row are dotted, and the 3P and 1S in the central hairpin turn, PX5SPX3P, are highlighted with asterisks. C, RT-PCR of transcripts of OLE1a and OLE1b, OLE2a and OLE2b, as well as ACT2 (a loading control) in various tissues (see Fig. 4 legend for labels). Primers were 1HF and 1aR for OLE1a and 1HF and 1bR for OLE1b (see A); and 2HF and 2aR for OLE2a and 2HF and 2bR for OLE2b.
OBs in Spore Are Equivalent Metabolically to Those in Seeds
During the life cycle of Physcomitrella, haploid spores were produced via meiosis. Each spore became dehydrated and packed with OBs (Fig. 1C) and could stay dormant or germinate in favorable conditions. Many of these physiological aspects are similar to those of seeds. In spore that had just germinated, peroxisomes appeared adjacent to the OBs (Fig. 1). These peroxisomes were most likely the glyoxysomes, in reference to those in germinated seeds (Pracharoenwattana and Smith, 2008). Attempts to use antibodies against cotton malate synthase to detect the enzyme in Physcomitrella spore peroxisomes (glyoxysomes) via immunofluorescence microscopy were unsuccessful, presumably because the antibody-antigen reaction was not strong enough. Nevertheless, the levels of transcripts encoding malate synthase and isocitrate lyase, two marker enzymes of the glyoxysomes, in spore increased substantially during germination (Fig. 4). Thus, the spore OBs are present as food reserves for future gluconeogenesis via the glyoxysomes and other metabolic machineries.
OBs in the Photosynthetic Gametophyte Serve as Food Reserves and Are Mobilized via the Glyoxysomes, as Exemplified in the Production of Sex Organs
The vegetative gametophyte was induced to produce sex organs by switching the culture temperature from 25°C to 15°C. Within a 7-d period, brown antheridia (to produce sperms) and greenish archegonia (not easily visible) were produced (Fig. 6). OBs and peroxisomes in the uppermost leafy tissue were observed during this period of induction with BODIPY dye (for OBs) and antibodies against cotton catalase (for peroxisomes). During the 7-d period, the OB number rapidly deceased, concomitant with increased number of peroxisomes (glyoxysomes; Fig. 6).
Figure 6.
Images of the gametophyte after induction of sporophyte development. The gametophyte was examined after induction of sporophyte development on cold treatment for 0, 5, and 7 d. A, The uppermost row shows images of the tip of gametophyte, which was producing antheridia (brown color) and archegonia (not visible). The subsequent rows are fluorescence CLSM images of several cells in an uppermost leafy tissue of a branch. The cells were examined for OBs using the lipid probe BODIPY 493/503 (green), peroxisomes with rabbit anticatalase antibodies and then anti-rabbit IgG antibodies conjugated to Cy3 (red), and chloroplasts with autofluorescence (blue). Each column shows identical cells after the indicated days of cold treatment. Photos were taken to reveal OBs or peroxisomes alone or in combination (merge-1 for OBs and peroxisomes, and merge-2 for chloroplasts also and with dotted lines to outline the cell circumference). B, TEM pictures of portions of cells in an uppermost leafy tissue of a gametophyte branch showing the presence of OBs (OB) at day 0 and a peroxisome (P) at day 7.
The above changes in OBs and peroxisomes occurred only in the uppermost leafy tissues but not in the middle leafy tissues of a standup branch (Fig. 7A). During the 7-d period, the OB number in the uppermost leafy tissue decreased by 80%, concomitant with a marked increase in peroxisome number. No such changes of the two organelles occurred in the mid leafy tissues of a standup branch. In both the uppermost and mid leafy tissues, the number of chloroplasts remained unchanged. Thus, mobilization of lipid reserves to initiate production of sex organs in a standup branch required only OBs in the uppermost leafy tissue. Presumably, a longer sustained sexual reproduction process would require mobilization of the lipid reserves in the lower leafy tissues of a branch. The plant was cultivated in a sugar-rich medium; thus, there was a lesser need for mobilizing all lipid reserves in a branch for sexual reproduction.
Figure 7.
Changes in the number of organelles and levels of gene transcripts in the uppermost and mid leafy tissues of gametophyte branches upon induction of sporophyte development for 0, 3, 5, and 7 d. A, Number of OBs, peroxisomes (PEX), and chloroplasts (CLP) per cell area. B, RT-PCR results of transcripts encoding malate synthase (MS), isocitrate lyase (ICL), OLE1 (OLE1), OLE2 (OLE2), and actin (ACT2, as a loading control).
During the 7-d period, transcripts encoding malate synthase and isocitrate lyase, markers of glyoxysomes, in the uppermost leafy tissue rapidly increased in level, concomitant with a decrease in levels of transcripts encoding OLE1 and OLE2 (Fig. 7B). These changes in transcript levels did not occur in the mid leafy tissue.
All of the OBs in a Gametophyte Cell Are Linked to Extended Regions of the ER, on Which Different Oleosins Are Synthesized Concurrently
The leafy tissue of the gametophyte consists of only one cell layer. We tried to establish the leafy gametophyte as a transient expression system for cells that contain abundant OBs and can be transformed easily with bombardment and observed clearly with CLSM. Such a plant system has not been previously established (Miao and Jiang, 2007).
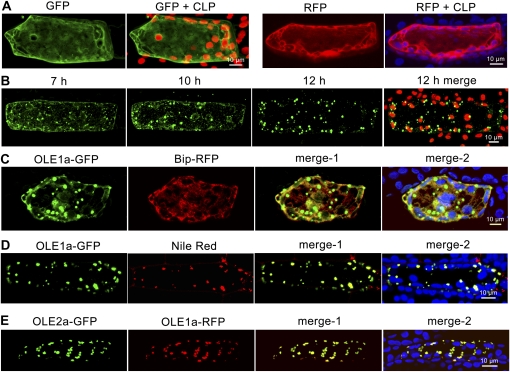
When cells were transformed with GFP or red fluorescent protein (RFP) driven by a 35S promoter, GFP or RFP was observed in the cytosol and was not associated with specific subcellular structures (Fig. 8A). When GFP was attached to the 3′ terminus of a complete OLE1a open reading frame, OLE1a-GFP initially appeared in a network and the associated droplets (Fig. 8B). Gradually, from 7, 10, to 12 h, proportionally less OLE1a-GFP was present in the network and more in the associated droplets. The network and the associated droplets were the ER and OBs, respectively, because after cotransformation with OLE1a-GFP and BiP-RFP (chaperone binding protein [BiP], an ER marker, from Arabidopsis [Kim et al., 2001]), OLE1a-GFP overlapped with BiP-RFP in the network and was highly enriched in the droplets (Fig. 8C). When OLE1a-GFP was used, OLE1a-GFP and the lipid dye Nile Red superimposed in all the droplets (Fig. 8D). When OLE1a-RFP and OLE2a-GFP were cotransformed, their encoded proteins appeared in all the droplets (Fig. 8E). The overall findings indicate that different oleosins are synthesized in extended regions of the ER and move to the associated OBs.
Figure 8.
Transient expression of various GFP and RFP constructs in individual gametophyte cells. Chloroplast autofluorescence is shown in red or blue in merged pictures. The speed of transient expression varied among experiments, and CLSM images were obtained at 6 to 8 (early), 8 to 10 (mid), and 10 to 12 h (late time point) after bombardment. A, Expression of control GFP or RFP not attached to OLE1a at a late time point. B, Expression of OLE1a-GFP at time intervals. GFP (green) was present largely in a cellular network at an early time point but was associated more with subcellular droplets at a late time point. C, Coexpression of OLE1a-GFP and BiP-RFP. GFP (green) and RFP (red) at a mid time point are shown. D, Expression of OLE1a-GFP. GFP (green) and OBs (red, stained with Nile Red) at a late time point are shown. E, Coexpression of OLE1a-GFP and OLE2a-RFP. GFP (green) and RFP (red) at a late time point are shown.
DISCUSSION
The OBs in both Physcomitrella and seeds apparently are similar in having a matrix of oils enclosed by a layer of oleosins and presumably also PLs. However, Physcomitrella OBs have the following early evolutionary trends. (1) The sizes of OBs in both the photosynthetic gametophyte and mature spore vary substantially, from 0.2 to 5 μm in diameter. Seed OBs have a narrow size range within a species (Tzen et al., 1993). Physcomitrella might not have evolved a mechanism to control the sizes of OBs. OBs in the photosynthetic gametophyte may resemble the lipid droplets in yeast and mammal cells in that the droplet sizes are related to the metabolic conditions of the cells. (2) Among all examined plant species, Physcomitrella has the fewest number of oleosin genes with minimal diversification. Alternative splicing that alters the tissue-specific expression of the oleosin genes in Physcomitrella may represent a mechanism for gene diversification. (3) Whereas seed OBs contain mostly TAGs, Physcomitrella OBs possess largely SEs and TAGs and some DAGs. This lipid composition of Physcomitrella OBs is similar to that of the intracellular and/or extracellular lipid droplets in yeast and mammals (with SEs and TAGs) and the extracellular lipid droplets in insects (largely DAGs; Ryan, 1994). (4) All the OBs within a Physcomitrella gametophyte cell apparently are physically linked to the ER, a trend suspected to occur in yeast, mammals, and other nonplant organisms (Martin and Parton, 2006; Goodman, 2008; Rajakumari et al., 2008). Actually, this characteristic may be normal for lipid droplets in all nondehydrated vegetative cells, in which metabolic dynamics is expected. No such information is available for the OBs in not-yet-dehydrated, maturing seed cells, and the OBs in mature seed cells are apparently not linked to the ER, which disappears during dehydration. (5) Unlike mature seeds, the Physcomitrella gametophyte possesses oleosin-coated OBs in highly hydrated vegetative cells; this situation is similar to the lipid droplets in yeast and mammalian cells. Oleosins on seed OBs may protect the OBs from dehydration (Napier et al., 1996; Murphy, 2001; Hsieh and Huang, 2004). This idea could be applied to the oleosins on OBs in the vegetative nondehydrated Physcomitrella gametophyte because many moss tissues can undergo extreme and prolonged dehydration and still resuscitate upon water uptake. The evolutionary acquisition of oleosins, which are absent in algae, would represent one of the desiccation and stress tolerance features adapted by Physcomitrella (Rensing et al., 2008). Whether during early evolution, OBs coated with oleosins appeared first in vegetative cells (photosynthetic gametophyte and then leaves) or in desiccated sexual organs (spore and then seed) is unknown.
Lipid droplets are present in green leaves of diverse species, although they are less obvious and abundant (Lersten et al., 2006). Whether these lipid droplets possess oleosins and other characteristics and play a similar physiological role of food storage as do Physcomitrella gametophyte OBs remain to be elucidated. No or few (authentic or simply background) oleosin transcripts are present in high-quality massively parallel signature sequencing or sequencing-by-synthesis leaf transcriptomes of Arabidopsis (Meyers et al., 2004) and rice (Nobuta et al., 2007). Regardless, oleosin encoded by an Arabidopsis gene transformed into tobacco (Nicotiana tabacum) leaf cells targets to lipid droplets via the ER (Wahlroos et al., 2003). In some species, the leaf lipid droplets may contain hydrophobic secondary metabolites (e.g. rubber droplets in guayule) instead of TAGs. In some other species, the leaf lipid droplets may be remnants of OBs in primitive plants and may be induced to proliferate under special situations, such as starvation and senescence. Under the latter situations, glyoxysomes and other machineries appear and convert degraded lipids into sugar for internal use or for export to nonsenescing tissues (Pracharoenwattana and Smith, 2008). Overall, leaf lipid droplets in diverse species, unlike those in seeds, are heterogeneous in structures and lipid contents and have diverse functions under different developmental, physiological, and environmental conditions.
The haploid Physcomitrella spore is genetically and physiologically similar to the haploid pollen in higher plants. Both the spore and pollen also contain storage OBs coated with oleosins. However, the function of Physcomitrella spore OBs is for gluconeogenesis via the glyoxysomes. Pollen OBs are not metabolized via the glyoxysomes and likely act as reserves of acyl moieties for synthesis of new plasma membrane during pollen tube elongation.
Physcomitrella can be easily transformed and examined with CLSM for transient expression of genes that are involved in storage TAG and SE metabolism and contain abundant OBs. Such a plant system was not previously available (Miao and Jiang, 2007), and transient expression of genes in Physcomitrella has been performed only with juvenile protonema (e.g. Marella et al., 2006), which contain few or no OBs (Fig. 1A). Earlier, maturing embryos of flax and microspore cultures of Brassica were used for transient expression of modified oleosin genes, and the transformed plant materials were examined with in vitro biochemical analyses (Abell et al., 2004). The OB-containing internal cells in an embryo may not be transformed easily with bombardment or observed clearly with CLSM, and the microspore culture is highly artificial, and the microspore (pollen) OBs are not for gluconeogenesis. Another major advantage of the Physcomitrella transient expression system is that the growth condition of the plant can be altered easily for study of storage lipid metabolism and packaging.
The sizes of OBs in the Physcomitrella gametophyte and spore are highly variable, whereas those of OBs in seeds of individual species are more confined. Presumably, Physcomitrella has not evolved a mechanism to control the coordinate synthesis of TAGs and oleosins, and thus the sizes of OBs, within the same cell. In seeds, OB sizes are directly related to the ratio of TAGs to oleosins, as in kernels of maize lines that were bred for high or low oils (Ting et al., 1997) and seeds of Arabidopsis mutants whose oleosin genes were knocked out or down (Siloto et al., 2006; Shimada et al., 2008). In maize lines bred for low oils (resulting in a low ratio of oils to oleosins), the OBs are not only smaller but also have irregularly shaped surface that could accommodate more surface oleosins per unit of matrix TAGs. In high-oil maize lines, the OBs are spherical and substantially larger. Similarly, in mutant Arabidopsis seeds with lesser amounts of oleosins, the OBs are considerably bigger. Occasional dumbbell-shaped OBs are present and looked upon as fusing detached OBs that do not have sufficient surface oleosins. An alternative explanation is that the apparent fusion occurred among budding OBs that were still attached to the ER.
In maturing seeds, whether oleosins and TAGs are synthesized in extended regions or restricted subdomains of the ER is uncertain. Earlier, OB synthesis in restricted ER subdomains has been suggested on the basis that immuno-TEM reveals more oleosins in the ER near ER-OB structures (Herman, 1987) and that a low-density, isolated subfraction (ER-OB structures) could synthesize more TAGs in vitro (Lacey et al., 1999). However, these results could also be interpreted as OBs being synthesized in extended ER regions, such that there is a concentration gradient in the ER, with more oleosins near the budding OBs; the abundant oleosins in these ER-OB structures, upon isolation, would facilitate in vitro TAG synthesis. More recently, fluorescence microscopy revealed specific DAGAT for TAG synthesis in highly defined ER subdomains in transformed BY2 cells (Shockey et al., 2006); uncertainty exists because BY2 cells do not contain OBs. On the contrary, in cells of both Physcomitrella gametophyte (this report) and Brassica tapetum (Hsieh and Huang, 2005), oleosins are synthesized in extended regions of the ER. Nevertheless, neither of these systems is directly related to maturing seeds, which is specialized to produce massive OBs. It is also possible that oleosins are synthesized in extended ER regions and diffuse to restricted TAG-synthesizing ER subdomains from which nascent OBs detach to become solitary entities.
MATERIALS AND METHODS
Plant Materials
Spore of Physcomitrella patens subsp. patens was kindly provided by Dr. Eugene Nothnagel of the University of California, Riverside, CA. Gametophytes were grown axenically on a solid Knop's medium containing 125 mg L−1 KNO3, 125 mg L−1 KH2PO4, 125 mg L−1 MgSO47H2O, 500 mg L−1 Ca(NO3)24H2O, and 10 g L−1 Glc supplemented with 1 mL L−1 1,000× Hunter's metal 49 micronutrients [76 mg L−1 5-sulfosalicylic acid dihydrate, 7 g L−1 Fe(NH4)2(SO4)26H2O, 3.04 g L−1 MnSO4H2O, 2.2 g L−1 ZnSO47H2O, 0.025 mg L−1 (NH4)6Mo7O244H2O, 616 mg L−1 CuSO45H2O, 238 mg L−1 CoSO47H2O, 57.2 mg L−1 H3BO3, and 18 mg L−1 Na3VO4; Basile, 1978] and 1.2% (w/v) agar, pH 4.6. Plants were cultured at 25°C ± 1°C under a 16-h light (60 to approximately 100 μE m−2 s−1)/8-h dark cycle. Sexual development was carried out with cold stimulation. Cultures of 45 d were half-submerged in water and cultured at 15°C. After the cold treatment for 60 d, mature sporophytes were harvested from the apex. From these sporophytes, spore was collected.
Tissues for RT-PCR analysis were protonema (tissue grown from spore after germination for 10 d); mature gametophytes (60-d-old culture) subdivided into top, middle, and bottom leafy tissues; antheridia; antheridia and archegonia; zygote; young, maturing, and old sporophytes (S1, S2, and S3 obtained after 32, 45, and 56 d of cold stimulation, respectively); and mature spore and germinating spore (on a solid Knop's medium covered with a layer of cellophane at 25°C and with continuous light for 2 d).
Staining of OBs in Situ
OBs in situ were stained with Sudan Black B, Nile Red (Greenspan et al., 1985), or 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503, D-3922 from Invitrogen). For Sudan Black staining, fresh tissues were placed in 70% (v/v) propylene glycol for 5 min, transferred to a saturated Sudan Black B solution (in 70% propylene glycol) for 10 min, washed with 50% propylene glycol twice, and observed with light microscopy. For Nile Red or BODIPY staining, fresh tissue or fixed tissue (after immunofluorescence treatment, to be described) were placed in a solution consisting of Nile Red stock (100 μg/mL acetone) or BODIPY 493/503 stock (10 mg/mL DMSO) diluted 100× with 1× phosphate buffered saline (PBS; 10 mm potassium phosphate, pH 7.4, 138 mm NaCl, and 2.7 mm KCl) for 10 min, washed with PBS twice, and observed with a Zeiss LSM 510 META NLO confocal microscope. Nile Red and BODIPY 493/503 were excited with the 543- and 488-nm lines, and its emission was detected with filter band-pass 565 to 615 and band-pass 500 to 530, respectively.
Isolation of OBs from the Gametophyte
All procedures were performed at 4°C. Fresh, 60-d-old gametophytes were soaked in a grinding medium (0.6 m Suc, 0.1 m HEPES-NaOH, and 4 mm dithiothretol, pH 7.5) for 20 min and chopped with a razor blade and then ground with a mortar and pestle. The ground sample was filtered through a layer of Nitex cloth (50 × 50 μm) to yield a total extract. The total extract was placed at the bottom of a centrifuge tube, and a lighter solution (0.4 m Suc and 0.1 m HEPES-NaOH, pH 7.5) was loaded above the extract. The tube was centrifuged at 18,000 rpm for 45 min in a Beckman SW28 rotor. Floated OBs at the top of the gradient were collected with a spatula.
Analysis of Lipids
Lipids in the total gametophyte extract and isolated OB fraction were extracted with 1.2× volume of lipid extraction buffer (chloroform/heptanes/methanol, 4/3/2, v/v/v) three times. The extract was evaporated to dryness with a stream of nitrogen gas and redissolved in ether or acetone for TLC or HPLC, respectively.
Lipid samples were applied to TLC plates (silica gel 60A; Whatman), which were developed in hexane:diethyl ether:acetic acid (80:20:2, v/v/v). Lipids on the plates were visualized after sulfuric acid spray. HPLC-atmospheric pressure chemical ionization/MS was performed with Agilent 1100 series liquid chromatography coupled with ThermoFinnigan LCQ Advantage ion trap mass spectrometer with an atmospheric pressure chemical ionization interface. HPLC was carried out with a 5-μL sample (20 mg/mL) and an RP-18 column (Phenomenex Luna 3μ C18, 150 × 2.0 mm) at 30°C. Elution was performed with isocratic acetone-acetonitrile (1:1, v/v) at a flow rate of 0.2 mL/min. The peaks were analyzed with diode-array detection at 205 nm and then MS detection. Ionization was performed in the positive ion mode for all analyses.
Electron Microscopy
Tissues (cut into approximately 2- × 2-mm pieces) and the isolated OB fraction (in 0.4 m Suc and 0.01 m potassium phosphate buffer, pH 7.4) were fixed with 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.1 m potassium phosphate, pH 7.0, at 4°C for 24 h. The samples were washed with 0.1 m potassium phosphate buffer for 10 min two times and then treated with 1% OsO4 and 0.1 m potassium phosphate, pH 7.0, at room temperature for 4 h. The fixed samples were rinsed with 0.1 m potassium phosphate buffer and dehydrated through an acetone series and embedded in Spurr's medium. Ultrathin sections (70–90 nm) were obtained with a Leica Reichert Ultracut S or Leica EM UC6 ultramicrotome. Sections were stained with uranyl acetate and lead citrate and examined with a Philips CM 100 transmission electron microscope at 80 kV.
Immunofluorescence CLSM
All antibody treatments were performed with 1:50 dilution of the IgG fraction (isolated and resuspended into the same original anticatalase serum volume), 1% (w/v) milk powder, and 1× PBS. Each wash was performed with PBST (1× PBS and 0.05% [w/w] Tween 20) for 10 min. Tissues were fixed in 4% paraformaldehyde, 1× PBS, and 0.15 m Suc at 4°C for 16 h. After two washes, the tissues were treated with 1% cellulase R10 (Yakult) in 1× PBS for 20 min at 25°C. After two washes, the tissues were incubated with 1× PBS with 0.1% Tween 20 for 20 min at 25°C. After two washes, the tissues were treated with a blocking solution (3% milk and 1× PBS) at 25°C for 1 h and then rabbit antibodies against cottonseed catalase at 4°C for 16 h. After three washes, the tissues were treated with cyanine 3-conjugated donkey antibodies against rabbit IgG (Jackson Immuno Research Laboratories) for 1 h at 25°C. After three washes, the tissues were stained with BODIPY 493/503 for OB staining. The tissues were placed on a slide and observed with a LSM 510 META confocal microscope (Carl Zeiss). BODIPY 493/503, cyanine 3, and chloroplasts were excited with the Argon 488-, HeNe 543-, and Argon 488-nm lines, respectively, and the emissions were detected with emission filters of band-passes 500 to 530, 565 to 615, and 650 to 710 nm, respectively.
SDS-PAGE and Identification of Oleosins with MALDI-TOF
Proteins in the total cellular extract and the isolated OB fraction were separated with 12% (w/v) SDS-PAGE (Wu et al., 1997). The gel was stained with Coomassie Brilliant Blue. The gel containing the visible approximately 17-kD proteins of the OB fraction was cut. The proteins were extracted and subjected to trypsin digestion and MS (MALDI-TOF) analysis with the Voyager DE-STR (PerSeptive Biosystems).
Searches for Oleosin Genes of Physcomitrella and Other Organisms
Sequences of the conserved hairpin domain of oleosins and several complete oleosins of Arabidopsis (Arabidopsis thaliana; Kim et al., 2002) were used as query sequences for the BLAST program (tBLASTn) against genome and transcriptome databases of P. patens (http://www.cosmoss.org/). Three oleosin genes, PpOLE1, PpOLE2, and PpOLE3, were found on scaffold 84, scaffold 21, and scaffold 180, respectively. Similar searches yielded oleosin genes of Oryza sativa (from http://rice.plantbiology.msu.edu/), Arabidopsis, Selaginella mutica, and Populus trichocarpa (from the Joint Genome Initiative Eukaryotic Genomics database, http://www.jgi.doe.gov/). An oleosin of Pinus ponderosa, Pinus-OLE, was obtained from an earlier study (Lee et al., 1994). A phylogenetic tree of the above oleosins was constructed on the basis of protein sequence similarities (of the conserved hairpin sequence plus the moderately conserved sequences immediate flanking the hairpin) and constrictions with the Clustal method, a distance method (neighbor-joining), and PHYLIP with 1,000 bootstrap replicates.
RT-PCR Analyses
RNA was extracted from tissues with use of an RNeasy Mini Kit (Qiagen). Total RNA (2 μg) was first treated with DNaseI (Invitrogen) for 35 min at 37°C. The proteins were removed with phenol/chloroform/isoamyl alcohol (25:24:1, pH 4.5) and the phenol with chloroform-isoamyl alcohol (24/1, v/v). The RNA was precipitated with 0.1× volume of 3 m NaOAc (pH 5.2) and 2.5× volume of absolute ethanol at −20°C for 16 h. The RNA (1 μg) was used to make cDNA with the SuperScript III RT-PCR system (Invitrogen). The RNA was preincubated with oligo(dT)12,18 and deoxynucleotide triphosphate at 65°C for 5 min and then placed on ice. cDNA Synthesis Mix was added to the RNA, and the mixture was incubated at 50°C for 1 h. The reaction was terminated by heating at 75°C for 5 min. PCR was carried out with use of 0.2 μL of the cDNA as template and DyNazyme DNA polymerase with deoxynucleotide triphosphate and primers. From the sequence information of the genes and their transcripts, primers were selected and synthesized. Primer pairs for amplifying full-length cDNA of PpOLE1a, PpOLE2a, and PpOLE3 are shown in Supplemental Table S1. Amplified DNA fragments were subcloned into pGEM-T Easy (Promega) and subjected to DNA sequencing with use of M13 forward and reverse primers. Primer pairs for amplification of specific gene fragments in the study of gene expression patterns are shown in Supplemental Table S1. Amplified DNA fragments of approximately 200 bp were analyzed on a 1.8% agarose gel.
Transient Expression Assays
DNA sequences encoding the complete coding region of PpOLE1a and PpOLE2a were amplified by PCR with use of primers shown in Supplemental Table S1. The resulting coding fragments were digested with BamHI and cloned into the expression site of a GFP expression vector (Chiu et al., 1996) or an RFP expression vector (Lee et al., 2001) to be driven by a 35S promoter of Cauliflower mosaic virus. A BiP-RFP expression vector of a similar construct (Kim et al., 2001) was obtained from Dr. David Ho, Institute of Plant and Microbial Biology, Taipei. Transformation of the gametophyte was carried out with particle bombardment. Sixty-day-old gametophyte tissues were placed on solid Knop's medium. Plasmid DNA (5 μg) was coated onto the surface of 1.25 mg of 1.6-nm gold particles, which would be used for six different shootings. The gold particles were bombarded with 900 psi under 28-inch Hg vacuum onto the gametophyte from a distance of 6 cm in PDS-1000 (Bio-Rad). After bombardment, the tissues were left on the culture medium and observed with CLSM at time intervals. GFP and RFP were excited with the Argon 488- and HeNe 543-nm lines, and their emissions were detected by emission filters of band-passes 500 to 530 and 565 to 615, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A pileup of 45 oleosins from Physcomitrella, Arabidopsis, Populus, Oryza, Selaginella, and pine.
Supplemental Table S1. Primers for PCR and RT-PCR.
Supplementary Material
Acknowledgments
We greatly appreciate the assistance from Dr. Eugene Nothnagel for introducing Physcomitrella to us, Drs. Wann-Neng Jan and Tuan-Nan Wen (Institute of Plant and Microbial Biology, Academia Sinica) for electron microscopy (Plant Cell Biology Core Lab) and proteomics (Proteomics Core Lab), respectively, Dr. Chia-Chin Hou (Metabolomics Core Lab, Agricultural Biotechnology Research Center, Academia Sinica) and Dr. Bruce Whitaker (U.S. Department of Agriculture, Beltsville) for HPLC/MS, Lin-yun Kuang (Transgenic Plant Lab, Institute of Plant and Microbial Biology, Academia Sinica) for growth of Physcomitrella, Shung-Yee Kung (University of California, Riverside) for TLC, Dr. Richard Trelease (Arizona State University) for antibodies against cotton catalase, and Dr. Tuan-hua David Ho (Institute of Plant and Microbial Biology) for the BiP-RFP expression vector.
This work was supported by U.S. Department of Agriculture-National Research Initiative Grant 2005–02429 and an Academia Sinica Pilot grant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anthony H.C. Huang (anthony.huang@ucr.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Abell BM, Hahn M, Holbrook LA, Moloney MM (2004) Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J 37 461–470 [DOI] [PubMed] [Google Scholar]
- Barbazuk WB, Fu Y, McGinnis KM (2008) Opportunities and challenges: genome-wide analyses of alternative splicing in plants. Genome Res 18 1381–1392 [DOI] [PubMed] [Google Scholar]
- Basile DV (1978) Culture media for bryophytes. In M Rechcigl, ed, CRC Handbook Series in Nutrition and Food, Section G, Vol III. CRC Press, Cleveland, pp 557–568
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter for plants. Curr Biol 6 325–330 [DOI] [PubMed] [Google Scholar]
- Dembitsky VM (1993) Lipids in bryophytes. Prog Lipid Res 32 281–356 [DOI] [PubMed] [Google Scholar]
- Frandsen GI, Mundy J, Tzen JT (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112 301–307 [DOI] [PubMed] [Google Scholar]
- Goodman JM (2008) The gregarious lipid droplets. J Biol Chem 42 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile Red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM (1987) Immunogold-localization and synthesis of an oil-body membrane protein in developing soybean seeds. Planta 172 336–345 [DOI] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2004) Endoplasmic reticulum, oleosins, and oils in seeds and tapetum. Plant Physiol 136 3427–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2005) Lipid-rich tapetosomes in Brassica tapetum are composed of oleosin-coated oil droplets and vesicles, both assembled in and then detached from the endoplasmic reticulum. Plant J 43 889–899 [DOI] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC (2002) Expression of Arabidopsis oleosin genes and characterization of their encoded oleosins. J Biol Chem 277 22677–22684 [DOI] [PubMed] [Google Scholar]
- Lacey DJ, Beaudoin F, Dempsey CE, Shewry PR, Napier JA (1999) The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J 17 397–405 [Google Scholar]
- Lee K, Bih FY, Learn GH, Ting JTL, Sellers C, Huang AHC (1994) Oleosins in the gametophytes of Pinus and Brassica and their phylogenetic relationship with those in the sporophytes of various species. Planta 193 461–469 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lersten NR, Czlapinski AR, Curtis JD, Freckmann R, Horner HT (2006) Oil bodies in leaf mesophyll cells of angiosperms: overview and a selected survey. Am J Bot 93 1731–1739 [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46 1032–1044 [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Natl Rev 7 373–378 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YS, Jiang L (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protocols 2 2348–2353 [DOI] [PubMed] [Google Scholar]
- Murphy DJ (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Plant J 13 1–16 [DOI] [PubMed] [Google Scholar]
- Napier JA, Stobart AK, Shewry PR (1996) The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol Biol 31 945–956 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, Belo A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang GL, et al (2007) An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol 25 473–477 [DOI] [PubMed] [Google Scholar]
- Pihakaski K, Pihakaski S, Karunen P (1987) Seasonal changes in leaf lipids of Diphensia lapponica with special reference to storage lipid bodies. Nord J Bot 7 281–292 [Google Scholar]
- Pracharoenwattana I, Smith SM (2008) When is a peroxisome not a peroxisome? Trends Plant Sci 13 522–525 [DOI] [PubMed] [Google Scholar]
- Rajakumari S, Grillitsch K, Daum G (2008) Synthesis and turnover of non-polar lipids in yeast. Prog Lipid Res 47 157–171 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69 [DOI] [PubMed] [Google Scholar]
- Ryan RO (1994) The structures of insect lipoproteins. Curr Opin Struct Biol 4 499–506 [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55 798–809 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerols biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson ES, Anderson WH, Gellerman JL, Schlenk H (1976) Ultrastructure and lipid composition of mosses. Bryologist 79 339–349 [Google Scholar]
- Ting JTL, Balsamo RA, Ratnayake C, Huang AHC (1997) Oleosin of plant seed oil bodies is correctly targeted to the lipid bodies in transformed yeast. J Biol Chem 272 3699–3706 [DOI] [PubMed] [Google Scholar]
- Tzen JTC, Cao YZ, Laurent P, Ratnayake C, Huang AHC (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol 101 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlroos T, Soukka J, Denesyuk A, Wahlroos R, Korpela T, Kilby NJ (2003) Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis 35 125–132 [DOI] [PubMed] [Google Scholar]
- Wu SSH, Platt KA, Ratnayake C, Wang TW, Ting JTL, Huang AHC (1997) Isolation and characterization of novel neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc Natl Acad Sci USA 94 12711–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.