Abstract
In plants, very-long-chain fatty acids (VLCFAs; >18 carbon) are precursors of sphingolipids, triacylglycerols, cuticular waxes, and suberin. VLCFAs are synthesized by a multiprotein membrane-bound fatty acid elongation system that catalyzes four successive enzymatic reactions: condensation, reduction, dehydration, and a second reduction. A bioinformatics survey of the Arabidopsis (Arabidopsis thaliana) genome has revealed two sequences homologous to YBR159w encoding a Saccharomyces cerevisiae β-ketoacyl reductase (KCR), which catalyzes the first reduction during VLCFA elongation. Expression analyses showed that both AtKCR1 and AtKCR2 genes were transcribed in siliques, flowers, inflorescence stems, leaves, as well as developing embryos, but only AtKCR1 transcript was detected in roots. Fluorescent protein-tagged AtKCR1 and AtKCR2 were localized to the endoplasmic reticulum, the site of fatty acid elongation. Complementation of the yeast ybr159Δ mutant demonstrated that the two KCR proteins are divergent and that only AtKCR1 can restore heterologous elongase activity similar to the native yeast KCR gene. Analyses of insertional mutants in AtKCR1 and AtKCR2 revealed that loss of AtKCR1 function results in embryo lethality, which cannot be rescued by AtKCR2 expression using the AtKCR1 promoter. In contrast, a disruption of the AtKCR2 gene had no obvious phenotypic effect. Taken together, these results indicate that only AtKCR1 is a functional KCR isoform involved in microsomal fatty acid elongation. To investigate the roles of AtKCR1 in postembryonic development, transgenic lines expressing RNA interference and overexpression constructs targeted against AtKCR1 were generated. Morphological and biochemical characterization of these lines confirmed that suppressed KCR activity results in a reduction of cuticular wax load and affects VLCFA composition of sphingolipids, seed triacylglycerols, and root glycerolipids, demonstrating in planta that KCR is involved in elongation reactions supplying VLCFA for all these diverse classes of lipids.
Very-long-chain fatty acids (VLCFAs) with chain lengths between C20 and C34 are essential and ubiquitous constituents of eukaryotic cells. They are most commonly found as building blocks of sphingolipids, but they are also important components of glycerophospholipids, triacylglycerols, sterol esters, and wax esters. Depending on their cellular and tissue localization and chain length, VLCFAs perform a wide range of physiological and structural roles. For example, they are involved in the stabilization of highly curved nuclear pore membranes (Schneiter et al., 1996, 2004) and the creation of membrane domains involved in lipid and protein trafficking (Dickson et al., 2006; Toulmay and Schneiter, 2007) and cell signaling (Leonard et al., 2002). In mammals, C20 VLCFAs also serve as precursors of biologically active eicosanoids implicated in inflammation responses (Karimi et al., 2007) and are critical components required for myelin production in the central nervous system (Poulos et al., 1992) as well as for normal functioning of photoreceptor cells in the retina (Zhang et al., 2001; McMahon et al., 2007). Furthermore, VLCFAs play an important structural role in skin barrier function (Wertz and Downing, 1983; Westerberg et al., 2004; Vasireddy et al., 2007). Similarly, in higher plants, VLCFAs give rise to cuticular waxes, lipid molecules deposited on the primary plant surfaces that serve as a barrier preventing excessive water loss and pathogen attack (Jenks et al., 1994), and mediate plant-insect interactions (Eigenbrode and Espelie, 1995) and pollen-stigma signaling required for fertilization (Preuss et al., 1993). Modified VLCFAs are also present in suberin, an extracellular plant polyester that controls water and solute fluxes in plant tissues (Bernards, 2002; Franke and Schreiber, 2007).
VLCFAs are formed by elongation of C16 and C18 fatty acids by endoplasmic reticulum (ER) membrane-bound enzymes (Cinti et al., 1992; Kunst and Samuels, 2003; Tehlivets et al., 2007), thought to be physically associated in a complex referred to as the fatty acid elongase (FAE; von Wettstein-Knowles, 1982). Each elongation cycle involves four successive enzymatic reactions: condensation of malonyl-CoA with an acyl-CoA by β-ketoacyl-CoA synthase (KCS), resulting in β-ketoacyl-CoA, reduction of β-ketoacyl-CoA to β-Zhydroxyacyl-CoA by β-ketoacyl-CoA reductase (KCR), dehydration of β-hydroxyacyl-CoA to an enoyl-CoA by β-hydroxyacyl-CoA dehydratase (HCD), and reduction of enoyl-CoA by enoyl reductase (ECR), thereby generating an acyl chain extended by two carbons (Nugteren, 1965).
Biochemical studies of the VLCFA biosynthesis and analyses of mutants with defects in fatty acid elongation revealed that multiple FAEs with unique substrate chain length specificities are involved in generating the complete array of C20 to C34 acyl chains required by eukaryotic cells (Sprecher, 1974; von Wettstein-Knowles, 1993). The KCS of the elongase determines the substrate specificity of each elongation reaction (Millar and Kunst, 1997). Consistent with the requirement for fatty acyl chains of diverse lengths, a large family of 21 FAE1-like KCS sequences has been annotated in the Arabidopsis (Arabidopsis thaliana) genome, together with an unrelated ELONGATION DEFECTIVE (ELO)-like family of four putative condensing enzymes (Dunn et al., 2004). To date, only a few of these condensing enzymes have been studied in any depth, and with the exception of FAE1, a seed-specific condensing enzyme involved in C20 and C22 fatty acid biosynthesis for seed storage lipids (Kunst et al., 1992), and CER6, a condensing enzyme that catalyzes the elongation of fatty acyl-CoAs longer than C22 in the epidermal cells of Arabidopsis (Millar et al., 1999; Fiebig et al., 2000), the functions the of other condensing enzymes in VLCFA elongation have not been established in planta, although several additional FAE1-like KCSs have been functionally characterized via heterologous expression in yeast (Trenkamp et al., 2004; Paul et al., 2006).
In contrast to the KCSs that have strict substrate and tissue specificities, the other three enzymes of the fatty acyl elongase, the KCR, the HCD, and the ECR, are believed to be common to all VLCFA biosynthetic reactions, have (presumed) broad substrate specificities, and are expressed in all cells (Millar and Kunst, 1997). Although these elongase component enzymes have been identified in Saccharomyces cerevisiae (Kohlwein et al., 2001; Beaudoin et al., 2002; Han et al., 2002; Denic and Weissman, 2007), this hypothesis could not be tested until recently because the corresponding enzymes and genes were not available from a multicellular organism. A single ECR gene has been confirmed and characterized in Arabidopsis and shown to encode a functional ECR that physically interacts with the Elo2p and Elo3p condensing enzymes when expressed in yeast (Gable et al., 2004). The Arabidopsis ECR was shown to be identical to CER10 (Zheng et al., 2005), the protein defective in the cer10 mutant isolated by Koornneef et al. (1989). Biochemical analysis of the cer10 mutant demonstrated that the ECR gene product is involved in VLCFA elongation required for the synthesis of many VLCFA-containing lipids, including cuticular waxes, seed triacylglycerols, and sphingolipids (Zheng et al., 2005). Similarly, the newly identified HCD PASTICCINO2 (Bach et al., 2008) is encoded by a single gene in Arabidopsis. Partial loss of PAS2 function in the pas2-1 mutant is associated with a general reduction of VLCFA content in seed storage triacylglycerols and complex sphingolipids as well as reduced levels of cuticular waxes that are generated from VLCFA precursors. A complete loss of PAS2 activity is embryo lethal (Bach et al., 2008). Two KCR genes, named GL8A and GL8B, are present in the maize (Zea mays) genome (Xu et al., 1997; Dietrich et al., 2005) and are required for cuticular wax accumulation on seedling leaves. However, they are not expressed only in the epidermis, but throughout the plant. Attempts to generate double mutants by crossing gl8a × gl8b single mutant lines failed because embryos carrying both mutations were not viable. Thus, in addition to its wax biosynthetic role, KCR has an essential function in maize, perhaps in the production of sphingolipids (Dietrich et al., 2005), but this idea has not been experimentally verified.
Two KCR orthologs have also been annotated in the Arabidopsis genome (Dietrich et al., 2005). One of these genes, At1g67730, functionally complements the KCR-deficient ybr159wΔ mutant of S. cerevisiae (Beaudoin et al., 2002), but the specific role of the protein that it encodes has not been determined. The other KCR candidate gene, At1g24470, has not been analyzed at all. In this study, we employ Arabidopsis insertional mutants disrupted in the KCR genes and transgenic lines in which KCR genes have been silenced by introduction of RNA interference (RNAi) or overexpression constructs to investigate the in planta functions of the two KCR proteins. We demonstrate that these two proteins are divergent and that At1g67730-encoded enzyme is the functional KCR of the Arabidopsis FAE essential for embryo development and plant morphogenesis.
RESULTS
Arabidopsis Has Two KCR Genes
The isolation of the yeast KCR gene YBR159w resulted in the identification of a single Arabidopsis ortholog, At1g67730 (Beaudoin et al., 2002), which we refer to as AtKCR1. Further exploration of the Arabidopsis genome revealed another KCR-like sequence, At1g24470 (Dietrich et al., 2005), which we designate AtKCR2. AtKCR1 has a 957-bp open reading frame (ORF) that encodes a protein of 318 amino acids with a predicted molecular mass of 35.7 kD. AtKCR2 has a 939-bp ORF that encodes a protein of 312 amino acids with a predicted molecular mass of 35.0 kD. Both these proteins have a putative NADH binding site and contain highly conserved Ser-Tyr-Lys residues essential for catalysis (Beaudoin et al., 2002), though a dilysine motif required for ER retention is obvious only in AtKCR1 (Supplemental Fig. S1). Unlike the maize orthologs GL8A and GL8B that are 97% identical and have partially redundant biochemical functions (Dietrich et al., 2005), AtKCR1 and AtKCR2 exhibit only 45% amino acid identity (Table I), suggesting that they could play diverse roles in VLCFA production in Arabidopsis.
Table I.
Percentage of amino acid sequence identity (similarity) between different KCR proteins
| AtKCR1 | AtKCR2 | YBR159 | GL8a | GL8b | |
|---|---|---|---|---|---|
| AtKCR1 | – | ||||
| AtKCR2 | 45 (80) | – | |||
| YBR159 | 30 (65) | 29 (64) | – | ||
| GL8a | 55 (84) | 48 (76) | 30 (63) | – | |
| GL8b | 56 (84) | 49 (76) | 29 (63) | 97 (99) | – |
Isolation of Mutants Disrupted in the AtKCR Genes
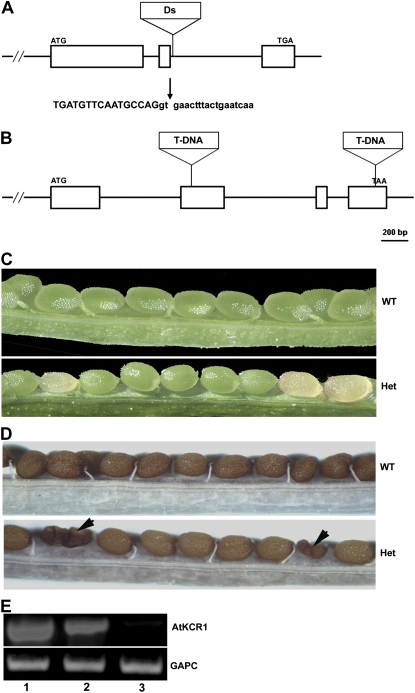
To investigate the in planta function of the two predicted AtKCR enzymes, we initiated a reverse genetic approach by identifying insertional mutants disrupted in AtKCR1 and AtKCR2 genes. A single line, RATH12-5282-1-G, obtained from Plant Functional Genomics Research Group of RIKEN Genomic Sciences Center (Kuromori et al., 2004) was found to carry a Ds transposon in the second intron of the AtKCR1 gene. The exact site of insertion was determined by sequencing the PCR product generated using a combination of gene- and Ds transposon-specific primers (Fig. 1A). Surprisingly, no homozygous atkcr1/atkcr1 plants were identified by PCR genotyping of the progeny of the self-fertilized insertion line, and the segregation of wild-type to heterozygous plants was approximately 1:2 (47:103), suggesting that homozygous kcr1 loss-of-function mutation may be embryo-lethal. To test this hypothesis, we dissected the siliques developing on heterozygous AtKCR1/atkcr1 plants and compared them with the siliques from the wild-type plants. Siliques from the wild type contained uniformly green developing seeds, whereas siliques from the self-fertilized AtKCR1/atkcr1 heterozygotes contained approximately 25% (28/120) of aborted seeds. Initially, aborted seeds appeared white or transparent in color at a stage when wild-type appearing siblings were green (Fig. 1C). Later on, the aborted seeds were dark brown, shrunken, and deformed (Fig. 1D). Attempts to confirm by PCR amplification that these aborted seeds were indeed homozygous kcr1 mutants failed. However, reverse transcription (RT)-PCR analysis using AtKCR1-specific primers showed reduced AtKCR1 transcript levels in AtKCR1/atkcr1heterozygotes and no detectable transcript in the presumed homozygous white seeds (Fig. 1E).
Figure 1.
Characterization of the AtKCR1 and AtKCR2 insertion lines. A, The AtKCR1 gene structure and mutation site. White boxes represent exons. Vertical arrow indicates the exact Ds transposon insertion site. The genomic DNA sequence flanking the Ds insertion is shown with uppercase letters indicating exon sequences and lowercase letters indicating intron sequences. B, The AtKCR2 gene structure and mutation sites. White boxes represent exons. C, Immature siliques of wild-type (WT) and heterozygous (Het) AtKCR1/atkcr1 plants. D, Mature siliques of wild-type and heterozygous AtKCR1/atkcr1 plants. E, RT-PCR analysis of AtKCR1 mRNA levels in green seeds from immature wild-type siliques (lane 1) and green seeds and white seeds from immature heterozygous siliques (lanes 2 and 3, respectively). The bottom panel shows expression of the cytosolic glyceraldehye-3-P dehydrogenase (GAPC) loading control for the corresponding lanes in the top panel.
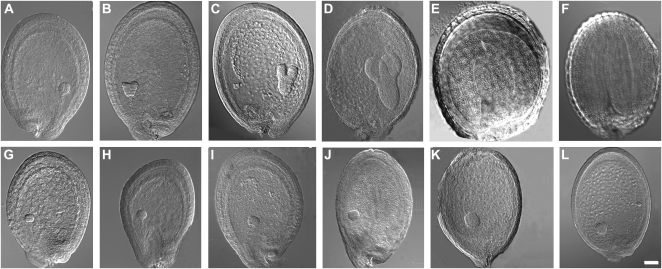
Seeds from siliques developing on heterozygous plants were further analyzed by differential interference contrast (DIC) microscopy (Fig. 2) to compare the progression of embryogenesis in these two seed types. In young siliques, all the seeds were white and contained embryos at the globular stage. However, in older siliques, only wild-type-appearing green seeds continued to develop normally with embryos proceeding from globular stage (Fig. 2A) to maturation (Fig. 2F) over the course of 8 d after flowering. In contrast, embryos from white seeds arrested at the globular stage (Fig. 2, G–L).
Figure 2.
Arrest of atkcr1/atkcr1 embryo development. Six siliques of different developmental ages growing on heterozygous AtKCR1/atkcr1 plants were dissected. A to F, DIC images of wild-type-looking green seeds at globular (A), heart (B), early torpedo (C), late torpedo (D), and bent cotyledon (E) stages of development and at maturity (F). G to L, DIC images of atkcr1/atkcr1 mutant embryos from white seeds developing in the same silique arrested at globular stage. Bar = 50 μm.
We also obtained two T-DNA insertion lines in the AtKCR2 gene, SALK_096487 (Alonso et al., 2003) and SAIL _536_H04 (Sessions et al., 2002), with T-DNA insertions in the second and fourth exons, respectively (Fig. 1B). No visible developmental phenotypes were observed in homozygous atkcr2 progenies from either T-DNA mutant line.
AtKCR1 and AtKCR2 Expression Domains Overlap and They Both Reside in the ER
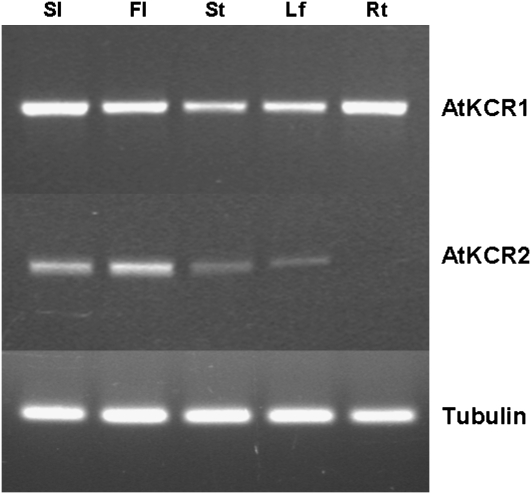
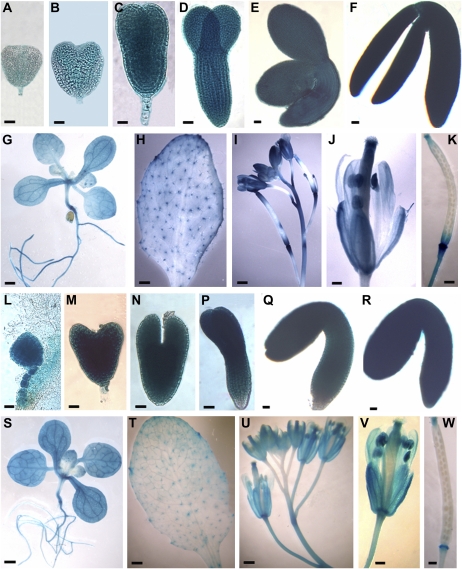
To determine if the two apparent Arabidopsis homologs are both constitutively expressed and to delineate their expression domains, we performed RT-PCR analyses and β-glucuronidase (GUS) assays in transgenic plants transformed with AtKCR1promoter-GUS and AtKCR2promoter-GUS constructs. As shown in Figure 3, both genes are expressed in green siliques, flowers, inflorescence stems, and leaves, with the highest expression levels detected in siliques and flowers. High AtKCR1 transcript levels were also present in roots, whereas AtKCR2 transcript was almost undetectable in root tissues. These expression results were confirmed by GUS activity assays (Fig. 4). In addition, AtKCR1promoter- and AtKCR2promoter-directed GUS activity was also detected in embryos of different stages (Fig. 4, A–F and L–R) and young developing seedlings (Fig. 4, G and S) but was absent from mature seeds (Fig. 4, K and W) and stem bases (data not shown).
Figure 3.
RT-PCR analysis of AtKCR1 and AtKCR2 expression in Arabidopsis. Sl, Green siliques; Fl, flowers; St, stems; Lf, leaves; Rt, roots. The bottom panel shows the expression of tubulin as a loading control for the corresponding lanes in the top panel.
Figure 4.
Tissue-specific expression patterns of AtKCR1 (A–K) and AtKCR2 (L–W) detected in transgenic AtKCR1promoter:GUS and AtKCR2promoter:GUS lines. Tissues were assayed for GUS activity by histochemical staining with the 5-bromo-4-chloro-3-indolyl-β-d-glucuronide substrate. Shown are different stages of embryo development (A–F, AtKCS1; L–R, AtKCS2); seedling with roots (G and S), cauline leaf with trichomes (H and T); inflorescence stem with buds and siliques (I and U); flower (J and V); and siliques (K and W). Bars = 50 μm in A to F and L to R; 5 mm in I and U; 1 mm in G, H, J, K, S, T, V, and W.
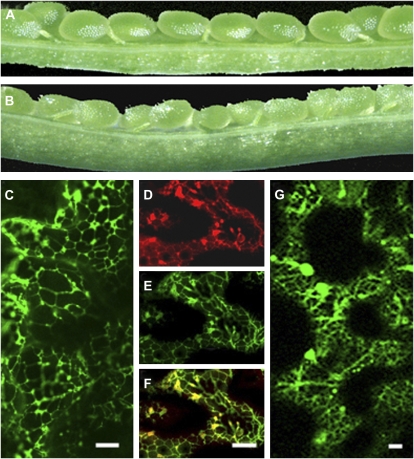
To compare the subcellular localization of the AtKCR1 and AtKCR2, yellow fluorescent protein (YFP)-AtKCR1 and cyan fluorescent protein (CFP)-AtKCR2 fusion constructs were initially transiently expressed in tobacco (Nicotiana tabacum) under the control of the 35S promoter. Visualization of tobacco leaves revealed that both proteins were localized in the ER (data not shown). We also introduced both fluorescent protein fusion constructs into Arabidopsis. To ensure that the YFP-AtKCR1 is fully functional, we expressed it in AtKCR1/atkcr1 heterozygous plants. Sixteen out of 20 recovered independent transgenic lines had siliques with only green wild-type-looking seeds and no white seeds, indicating complementation (Fig. 5B) and demonstrating that the mutation in the AtKCR1 gene is responsible for the embryo lethality. In all the complemented YFP-AtKCR1 transgenic lines examined by confocal microscopy, YFP fluorescence labeled a reticulate network typical of the ER (Fig. 5C). Because the disruption of AtKCR2 did not result in a detectable phenotype, we could not verify if the CFP-AtKCR2 fusion protein is active by complementation of a loss-of-function mutant, but we decided to proceed with its localization. For this purpose, we transformed the 35Spromoter-CFP-AtKCR2 fusion construct into wild-type Arabidopsis. This experiment revealed that the AtKCR2 protein also resides in the ER (Fig. 5G).
Figure 5.
Complementation of the atkcr1 mutant and subcellular localization of the AtKCR1 and AtKCR2. A and B, Immature siliques of wild-type (A) and heterozygous (B) plants carrying 35S:YFP-AtKCR1. C and E, YFP-AtKCR1 in Arabidopsis leaf is localized in the ER (green). D, Hexyl rhodamine B staining of the ER (red). F, Overlay of D and E. G, CFP-AtKCR2 in Arabidopsis leaf is localized in the ER. Bars = 10 μm.
Only AtKCR1 Complements the Yeast ybr159Δ Mutant and Embryo Lethality of the Arabidopsis atkcr1 Mutant
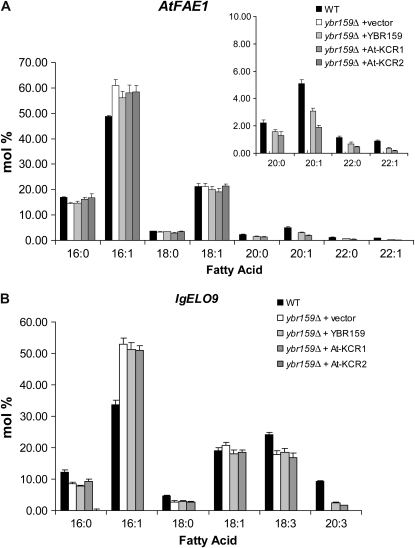
AtKCR1 has been previously shown to restore heterologous elongase activity of the yeast ybr159Δ mutant similar to the expression of native yeast KCR gene YBR159 (Beaudoin et al., 2002), indicating that the Arabidopsis and yeast enzymes are functionally equivalent and that AtKCR1 also likely catalyzes the reduction of the β-ketoacyl-CoA. This result was confirmed in this study (Fig. 6). By contrast, the expression of AtKCR2 failed to restore heterologous fatty acid elongation mediated by either FAE1 or the Isochrysis galbana Δ9-ELO-like elongating activity (Qi et al., 2002) in the ybr159Δ yeast mutant (Fig. 6) and also failed to rescue the slow growth phenotype of ybr159Δ yeast cells associated with impaired endogenous elongase function (data not shown), suggesting that AtKCR2 has no KCR activity in yeast.
Figure 6.
AtKCR1 complements fatty acid elongation in yeast. Gas chromatography-flame ionization detection analysis of FAMEs prepared from wild-type (WT) or mutant (ybr159Δ) yeast coexpressing AtFAE1 (A) or IgELO9 (B) and either a vector control, yeast YBR159, AtKCR1, or AtKCR2 gene.
The fact that atkcr1 loss-of-function mutation is embryo lethal suggests that either AtKCR2 is not a functional KCR or that AtKCR2 is not expressed during embryogenesis. GUS activity assays in transgenic plants transformed with the AtKCR2promoter:GUS construct (Fig. 4, L–W) and global gene expression analyses during Arabidopsis embryogenesis (Schmid et al., 2005) indicate that AtKCR2 is transcribed at all times during embryo development, albeit at a much lower level than AtKCR1. To determine if the low expression level of AtKCR2 was the reason why AtKCR2 could not support embryogenesis in the absence of AtKCR1 or whether the AtKCR2 was indeed inactive, we expressed AtKCR2 using the AtKCR1 promoter in the AtKCR1/atkcr1 background. In contrast to AtKCR1, which complemented the atkcr1 mutant (Fig. 5B), AtKCR2 could not rescue the embryo lethality of atkcr1 even when expressed in the AtKCR1 manner behind the AtKCR1 promoter (data not shown), confirming that it is not a functional KCR isoform.
RNAi-Suppressed and Cosuppressed AtKCR1 Plants Display Fused Vegetative and Reproductive Organs and an Abnormal Root Morphology
Embryo lethality prevented further phenotypic analysis on homozygous atkcr1 lines, and no developmental differences or changes in seed VLCFA composition or cuticular wax accumulation were detected in AtKCR1/atkcr1 heterozygotes in comparison with wild-type plants. We therefore generated transgenic plants in which AtKCR1 was down-regulated by RNAi to characterize their morphological and biochemical phenotypes and determine the role of AtKCR1 in the different VLCFA metabolic pathways.
To test the specificity of our RNAi construct and ensure that the phenotypes that we describe are due to suppression of AtKCR1 alone, we performed RT-PCR analysis of the RNAi suppressed plants using mRNA prepared from flowers that showed high KCR1 and KCR2 expression in the wild type in previous assays (Fig. 3). This experiment confirmed that the RNAi construct specifically down-regulates the expression of only AtKCR1, but not AtKCR2 (Supplemental Fig. S2).
Twenty T1 RNAi transformants were chosen for further evaluation. These plants displayed a variety of phenotypes, including growth retardation and fused rosette leaves (Fig. 7, A and D). Individuals with the most severe morphological phenotype never formed rosettes, but instead produced incompletely developed leaf-like structures (Fig. 7A). They were also highly sensitive to dehydration and needed to be cultivated in a controlled environment with atmospheric humidity above 85% in wet soil at all times. Eleven T1 plants failed to grow to maturity, developed necrotic spots, and died. From the nine remaining plants, three (designated 4, 6, and 7) required mechanical organ separations to allow distorted leaves and inflorescence stems to develop (Fig. 7, C and D).
Figure 7.
Morphological characterization of AtKCR1-RNAi plants. A, C, and D, Fused rosette leaves and inflorescences of 4-week-old T1 plants. B, Eight-week-old AtKCR1-RNAi T1 plant. E, Flowers from 6-week-old glossy T2 plants opened manually. F, Mature siliques from 8-week-old glossy T2 plants. G and H, Glossy (G) and nonglossy (H), but curly, 6-week-old T2 plants. I and J, Wild-type (I) and glossy (J) AtKCR1-RNAi T2 seedlings showing changes in root morphology.
Soil-grown T2 progeny of these AtKCR-RNAi primary transformants exhibited two predominant phenotypes: glossy inflorescence stems (Fig. 7G) or twisted and curled fiddlehead (fdh)-like stems that were not obviously glossy (Fig. 7H). Both glossy and nonglossy plants were present in the progeny of the 4, 6, and 7 lines. These three lines, together with a fourth line (line 9, showing a less extreme phenotype), were selected for a detailed morphological and biochemical characterization. Examination of the flowers from glossy plants revealed an unusual bushy stigma morphology with longer papillae and stamen with shorter filaments unable to position anthers at the height of the stigma, similar to that reported for cer10 (Zheng et al., 2005; Fig. 7E). These plants also produced short and crooked siliques, containing mostly aborted seeds, suggesting a problem with pollen viability/fertility (Fig. 7F). Cross-pollination of glossy plants with wild-type pollen resulted in longer siliques and a considerably increased number of seeds produced (data not shown). Similar short and crooked siliques were also found on the nonglossy plants. Interestingly, mature dried seeds collected from both glossy and nonglossy AtKCR-RNAi plants were considerably larger than seeds collected from wild-type plants grown under identical conditions (Table II).
Table II.
Dry weight of seeds collected from glossy and nonglossy AtKCR RNAi plants compared to the wild type
Each value is the mean of three independent measurements ± sd. GL, Glossy; NGL, nonglossy.
| Line | Wild Type | 4 GL | 7 GL | 7 NGL | 9 NGL |
|---|---|---|---|---|---|
| Seed weight (μg/50 seeds) | 21.77 ± 1.0 | 36.47 ± 0.8 | 34.53 ± 1.0 | 40.47 ± 1.4 | 43.20 ± 1.1 |
An aspect of the AtKCR-RNAi phenotype that has not previously been reported in other mutants impaired in VLCFA biosynthesis is the abnormal root morphology of the glossy plants (Fig. 7J). When grown horizontally on plates, the roots of these plants ran on the surface of the gel matrix, lacked lateral branches, and had dramatically reduced root hairs (Fig. 7J, inset). Closer observation revealed that lateral root primordia were formed but did not elongate. In contrast, roots of nonglossy plants with curly inflorescences appeared wild-type.
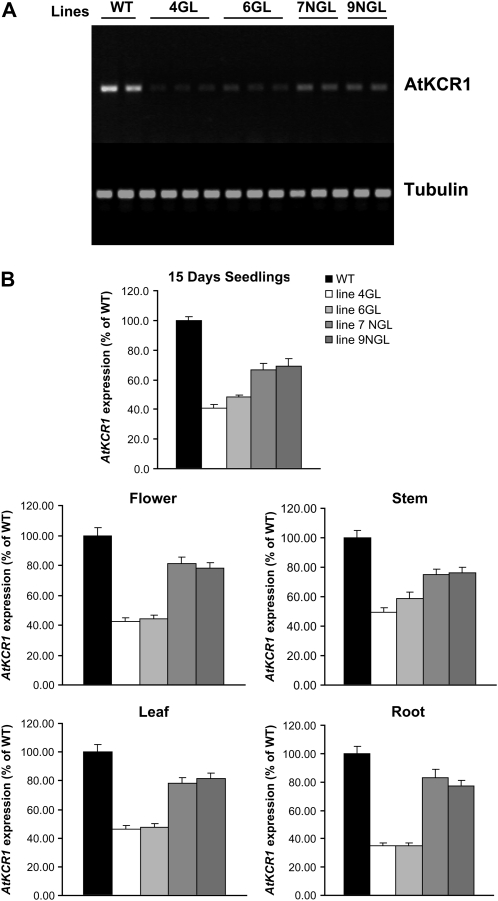
To determine if the phenotypes observed were due to AtKCR1 RNAi suppression, AtKCR1 transcript levels in transgenic lines were determined by RT-PCR in 15-d-old-seedlings and in a number of tissues from 8-week-old T2 plants. As shown in Figure 8, in the developing seedlings and in all of the tissues analyzed, the levels of AtKCR1 transcripts were lower than in the wild type. The reduction in KCR transcripts was more pronounced in the glossy plants compared with the more limited reduction observed in the nonglossy lines. Within each line and for each plant phenotype (glossy or nonglossy), levels of AtKCR1 transcripts were similar in all tissues. These results show a clear correlation between reduced levels of AtKCR1 transcripts and increased severity of the phenotype in the T2 generation.
Figure 8.
Expression of Arabidopsis KCR genes. A, RT-PCR analysis of the expression levels of AtKCR1 in 2-week-old wild-type (WT) and T2 AtKCR1-RNAi seedlings. The bottom panel shows the expression of a tubulin gene as a loading control for the corresponding lanes in the top panel. B, RT-PCR analysis of the expression levels of AtKCR1 in 15-d-old seedlings, flowers, stems, leaves, and roots of wild-type and T2 AtKCR1-RNAi plants. GL, Glossy; NGL, nonglossy.
We also overexpressed the AtKCR1 cDNA under the control of 35S promoter. About 50% of transgenic lines displayed abnormal phenotypes, similar to those of the AtKCR1-RNAi lines, including dwarfism, curly cauline leaves, fused flower buds, and bright green glossy stems (Supplemental Fig. S3, A–C). To investigate if these phenotypes were caused by AtKCR1 cosuppression, AtKCR1 transcript levels in a variety of tissues of transgenic plants were determined by RT-PCR. As shown in Supplemental Figure S3D, in cauline leaves, stems, and floral buds, the organs exhibiting the most pronounced phenotypes, AtKCR1 transcript levels were considerably lower than in the wild type.
RNAi-Suppressed AtKCR1 Plants Have Abnormal Trichome and Epidermal Cell Morphology
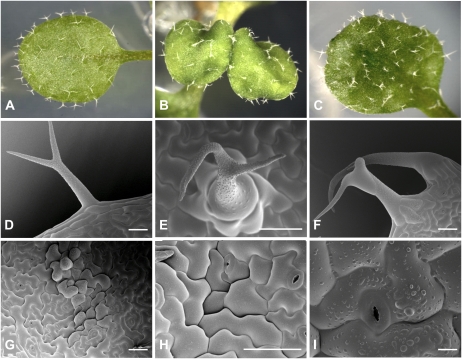
As previously reported for cer10, fdh, wbc11, and wax2/pel6, both glossy and nonglossy AtKCR-RNAi plants display clearly altered trichome morphology (Fig. 9, A–F). As shown in Figure 9B, trichomes are sparsely distributed, have fused branches in glossy plants, and have shorter or bent branches in nonglossy plants (Fig. 9C). Scanning electron microscopy examination showed that the basal cells of these trichomes are swollen and bulging out of the epidermal surface (Fig. 9, D–F). A further inspection of the surface of young leaves revealed impaired integrity of the epidermis in glossy plants. Groups of cells appeared swollen to the point were they did not adhere tightly to each other, resulting in deep intercellular grooves (Fig. 9, G–I) and probably contributing to the extreme sensitivity of glossy AtKCR1-RNAi plants to dehydration. This type of abnormal epidermal structure could not be detected in nonglossy plants.
Figure 9.
Glossy AtKCR1-RNAi plants have abnormal trichomes and epidermal structures. A to C, Primary leaves of 2-week-old wild-type (A), line 4 glossy (B), and line 9 nonglossy (C) T2 seedlings. D to F, Trichomes from the cauline leaves of wild-type (D) and glossy plants (E and F). G to I, Abnormal epidermal structure of the abaxial surface of cauline leaves in glossy plants. In the scanning electron micrographs (D–I), bars = 50 μm, except in E, where the bar = 10 μm.
Cuticular Wax Load Is Reduced in AtKCR1-RNAi Lines
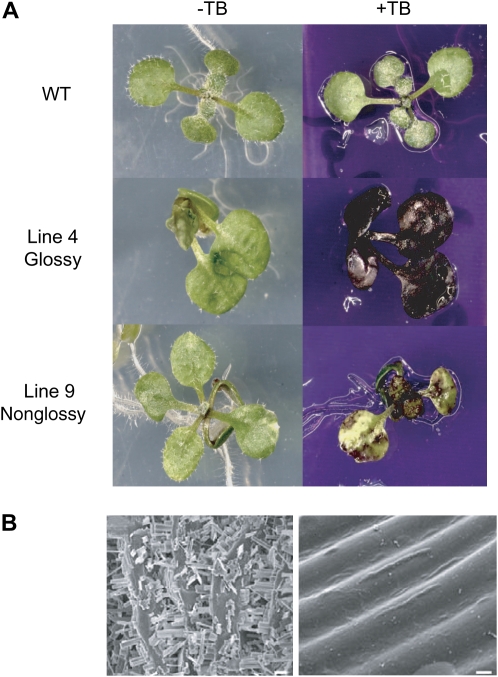
Organ fusions and stem glossiness observed in the AtKCR1-RNAi plants are typically associated with defects in plant cuticles. Cuticle abnormalities can be detected by a toluidine blue (TB) test, which results in staining of organs with permeable cuticles (Tanaka et al., 2004). We used the TB test to evaluate cuticles of glossy line 4 and nonglossy line 9 individuals (Fig. 10A). The test resulted in complete staining of line 4 glossy seedlings, indicating severe cuticular abnormalities of this RNAi line. Nonglossy line 9 seedlings displayed a range of partially stained patterns, including patchy or random staining as observed for fdh and cer14 mutants (Fig. 10A; Tanaka et al., 2004).
Figure 10.
AtKCR1-RNAi plants display cuticular defects. A, Two-week-old wild-type (WT) and AtKCR1-RNAi seedlings stained with TB. B, Epicuticular wax crystal structure on stem surfaces visualized by scanning electronic microscopy. Shown are images of the wild type (left panel, bar =10 μm) and glossy AtKCR1-RNAi line 4 (right panel, bar = 2 μm).
Because VLCFAs are precursors of all the aliphatic components of cuticular wax, we suspected that cuticle permeability of AtKCR1-RNAi lines may at least in part be due to reduced wax accumulation. We therefore examined the glossy inflorescence stem surfaces of the AtKCR1-RNAi plants by scanning electron microscopy. As shown in Figure 10B, wild-type stems are densely covered with epicuticular wax crystals. In contrast, glossy stem surfaces of AtKCR1-RNAi plants are virtually devoid of wax crystals.
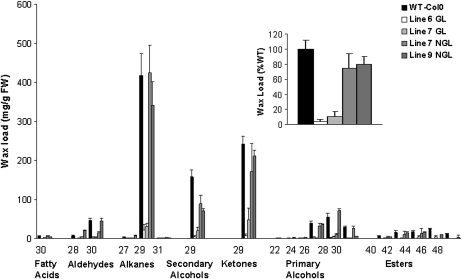
Wax analyses confirmed major reductions in wax loads in AtKCR1-RNAi plants. The total wax coverage on wild-type Columbia-0 (Col-0) inflorescence stems under the conditions used here was 1086 μg per g fresh weight. In contrast, wax loads measured in glossy AtKCR1-RNAi plants reached 124.2 μg per g fresh weight (11.4% of the wild type) in line 7 (7 GL) and 42.8 μg per g fresh weight (3.9% of the wild type) in line 6 (6 GL; Fig. 11). Quantification of wax components demonstrated that reduced wax loads were due to decreased accumulation of all compound classes (Fig. 11). In nonglossy AtKCR1-RNAi plants, the decrease in wax load was far less pronounced, and it was proportional to the levels of AtKCR1 transcripts detected in the stem of those plants (Fig. 8). Wax loads measured for nonglossy plants from lines 7 and 9 were 811 and 869 μg per g fresh weight (75% and 80% of the wild type), respectively (Fig. 11, inset). However, these data indicate that while lines 7 and 9 were phenotypically classified as nonglossy, they do have a reduction in wax load, as might be expected for plants with an impaired capacity to synthesize VLCFAs.
Figure 11.
Cuticular wax composition on stems of wild-type (WT) and AtKCR1-RNAi plants. Each bar represents the amount of a specific wax constituent, labeled on the x axis by a chemical class and a carbon chain length. Each value is the mean of four measurements. Error bars indicate sd. FW, Fresh weight; GL, glossy; NGL, nonglossy.
VLCFA Content Is Decreased in Root Lipids and Seed Triacylglycerols of AtKCR1-RNAi Lines
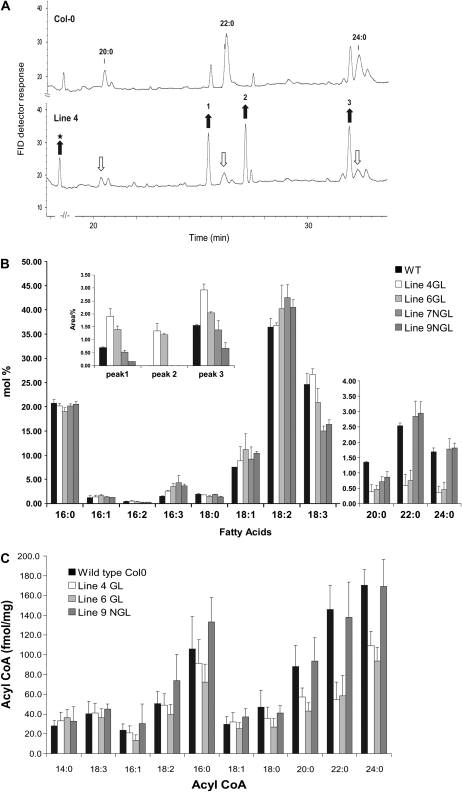
Since Arabidopsis leaves contain very low levels of VLCFAs, we analyzed the fatty acid composition in the roots of wild-type and AtKCR-RNAi plants that exhibit abnormal root morphology (Fig. 7J). A clear reduction in the content of C20, C22, and C24 fatty acids was detected in glossy lines 4 and 6 (Fig. 12, A and B). VLCFAs accounted for 5.6% of the total root fatty acid extracted from wild-type Col-0 plants grown in this study, predominantly reported in phospholipids, such as phosphatidylethanolamine and phosphatidylserine (Devaiah et al., 2006). The VLCFA content measured in glossy plants was reduced to 1.4% and 1.7% in lines 4 and 6 (24% and 29% of the wild type), respectively. The VLCFA content in nonglossy plants was similar to that of wild-type plants with 5.3% and 5.6% measured in lines 7 and 9, respectively. These results correlated well with the levels of VLCFAs measured in the acyl-CoA pool in roots of the same plants (Fig. 12C).
Figure 12.
Fatty acid composition in the root of wild-type and AtKCR1-RNAi T2 lines. Gas chromatography-flame ionization detection (FID) trace of FAMEs prepared from the roots of wild-type and AtKCR1-RNAi 15-d-old seedlings. In addition to the expected reduction in C20+ fatty acids, several elevated peaks were identified as C16:0 and C18:0 dicarboxylic acids (peaks 1 and 2, respectively) and an ω-hydroxylated fatty acid (peak 3). The star indicates a phenolic compound, most likely ferulic acid. B, Quantitation of variation in root fatty acid composition in wild-type and AtKCR1-RNAi lines. WT, Wild type. C, Acyl-CoA profiling of the roots of wild-type and AtKCR1-RNAi 15-d-old seedlings. Each value is the mean of four measurements. Error bars indicate sd. GL, Glossy; NGL, nonglossy.
Surprisingly, in the root fatty acid methyl ester (FAME) extracts prepared from glossy plants, we detected three novel peaks (Fig. 12, A and B, peaks 1–3) that were either absent or low in wild-type extracts. These peaks were tentatively identified by mass spectrometry as C16 and C18 α,ω-dicarboxylic acids and an ω-hydroxy fatty acid, compounds that are abundant in the cutin and suberin in Arabidopsis (Bonaventure et al., 2004, Franke et al., 2005, Beisson et al., 2007). The presence of these usually highly polymerized constituents in the AtKCR1-RNAi FAME samples after mild methanolic acid extraction may reflect the altered composition and/or structure of the suberin when the synthesis of longer C20, C22, and C24 compounds is reduced. The nonglossy plant extracts did not contain the three additional compounds founds in the roots of glossy plants.
Because Arabidopsis seed oil also contains VLCFAs, we examined the fatty acid composition and content of mature dry seeds. Compositional analyses of oil produced by the AtKCR1-RNAi seeds revealed that only the oil from glossy plants was depleted in C20 and C22 saturated and monounsaturated fatty acids. The total VLCFAs content in these seeds was about 30% lower compared to the wild type (Table III; Fig. 13). The larger seeds collected from both glossy and nonglossy AtKCR1-RNAi plants had a higher fatty acid content than wild-type seeds (Tables II and III). Overall, the increase in total fatty acid content was largely proportional to the increase in seed size and did not result from a higher proportion of oil as a percentage of the seed weight (data not shown).
Table III.
Fatty acid composition of seed storage lipids in wild-type and AtKCR-RNAi lines (mol %)
Each value is the mean of three independent measurements ± sd. GL, Glossy; NGL, nonglossy.
| Fatty Acid | Wild Type | Line 4 GL | Line 6 GL | Line 7 NGL | Line 9 NGL |
|---|---|---|---|---|---|
| 16:0 | 9.38 ± 0.79 | 9.28 ± 0.52 | 8.87 ± 0.76 | 8.25 ± 0.45 | 8.93 ± 0.58 |
| 16:1 | 0.52 ± 0.07 | 0.53 ± 0.14 | 0.52 ± 0.19 | 0.54 ± 0.06 | 0.52 ± 0.03 |
| 18:0 | 1.19 ± 0.12 | 0.76 ± 0.20 | 1.12 ± 0.04 | 0.99 ± 0.05 | 1.02 ± 0.00 |
| 18:1 | 16.61 ± 0.63 | 16.44 ± 0.95 | 18.19 ± 0.25 | 17.96 ± 0.52 | 17.88 ± 0.34 |
| 18:2 | 29.89 ± 1.77 | 31.32 ± 1.49 | 30.15 ± 1.88 | 29.50 ± 1.61 | 30.32 ± 1.58 |
| 18:3 | 20.55 ± 2.26 | 21.41 ± 1.16 | 20.34 ± 2.48 | 20.41 ± 1.02 | 20.48 ± 0.98 |
| 20:0 | 0.83 ± 0.02 | 0.66 ± 0.22 | 0.77 ± 0.13 | 0.63 ± 0.16 | 0.65 ± 0.19 |
| 20:1 | 17.28 ± 1.11 | 12.99 ± 1.24 | 13.66 ± 1.10 | 18.14 ± 1.34 | 16.87 ± 0.96 |
| 20:2 | 1.61 ± 0.14 | 1.71 ± 0.08 | 1.43 ± 0.10 | 1.55 ± 0.17 | 1.48 ± 0.16 |
| 20:3 | 0.49 ± 0.01 | 0.50 ± 0.01 | 0.44 ± 0.08 | 0.43 ± 0.07 | 0.42 ± 0.04 |
| 22:0 | 0.23 ± 0.09 | 0.12 ± 0.04 | 0.13 ± 0.03 | 0.17 ± 0.05 | 0.14 ± 0.04 |
| 22:1 | 1.43 ± 0.13 | 0.94 ± 0.20 | 0.98 ± 0.25 | 1.44 ± 0.33 | 1.28 ± 0.16 |
| Total fatty acids (nmole/seed) | 22.62 ± 1.04 | 45.55 ± 1.00 | 37.1 ± 1.08 | 47.25 ± 1.59 | 50.95 ± 1.31 |
| Total fatty acids (μg/seed) | 6.44 ± 0.30 | 12.89 ± 0.28 | 10.51 ± 0.31 | 13.46 ± 0.45 | 14.49 ± 0.37 |
Figure 13.
Fatty acid composition of total seed lipids from mature wild-type (WT) and AtKCR1-RNAi T2 seeds. Each value is the mean of four measurements. Error bars indicate sd.
Sphingolipid Analyses
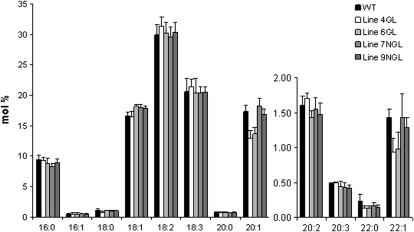
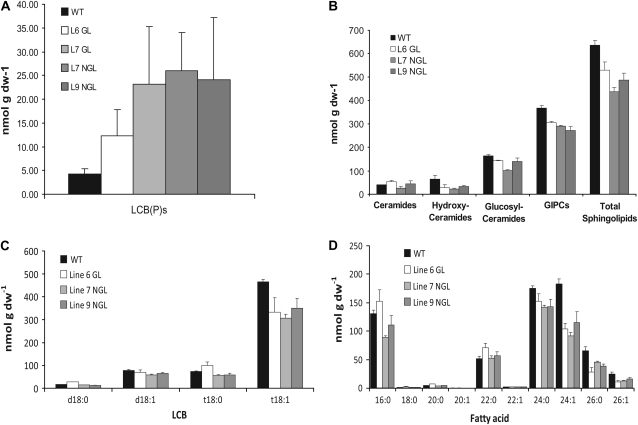
To evaluate the effect of reduced AtKCR1 expression on VLCFA incorporation into sphingolipids, we analyzed the complex sphingolipid composition in the stems of glossy and nonglossy AtKCR1-RNAi plants that all exhibit abnormal shoot morphology. All the examined lines accumulated free long-chain bases (LCBs), which are characteristic of impaired sphingolipid metabolism as reported for the yeast FAE mutants (Kohlwein et al., 2001; Fig. 14A). In addition, a moderate but statistically significant decrease in almost all the sphingolipid species was detected (Fig. 14B). Conversely, neither the accumulation of free LCBs nor the reduction in any of the sphingolipid species could be detected in the stems of morphologically normal plants (data not shown). This suggests that the twisted shoot phenotype may also be caused by altered sphingolipid structure and not only the defective cuticular lipid composition as proposed for the fdh mutant.
Figure 14.
Sphingolipid composition of the stems of wild-type (WT) and AtKCR1-RNAi plants. A, Free LCBs in the stems of wild-type, glossy (GL), and nonglossy (NGL) AtKCR1-RNAi T2 plants. B, Sphingolipid species in the stems of wild-type, glossy, and nonglossy AtKCR1-RNAi T2 plants. GIPC, Glycosylinositolphosphoceramide. C, LCB composition of the stem sphingolipids in wild-type, glossy, and nonglossy AtKCR1-RNAi T2 plants. D, Fatty acid composition of the stem sphingolipids in wild-type, glossy, and nonglossy AtKCR1-RNAi T2 plants. dw, Dry weight.
DISCUSSION
This study describes the functional characterization of the Arabidopsis orthologs of the yeast KCR YBR159w and confirms the central role of this enzyme activity in the synthesis of VLCFAs in plants, as well as the essential nature of these fatty acids. Like maize, Arabidopsis contains two KCR-like sequences (KCR1 and KCR2), but they do not share the same high degree of identity exhibited by the monocot genes. Our data indicate that only KCR1 is a bona fide KCR of the FAE in Arabidopsis, based on several independent criteria. First, KCR1 but not KCR2 is capable of rescuing fatty acid elongation in the yeast ybr159wΔ mutant. Second, the expression of the KCR2 ORF under the control of the KCR1 promoter is unable to suppress the embryo lethality of the kcr1 insertional mutant. Finally, while insertional mutations of KCR1 are embryo lethal, similar mutations of KCR2 have no obvious phenotypic effect and do not affect the composition of lipids that are known to accumulate or are derived from VLCFAs, such as seed storage lipids, cuticular waxes, and sphingolipids. It is possible that this apparently normal phenotype of the kcr2 mutant is a result of compensation by the KCR1 protein, but we have not detected increased KCR1 transcript levels in the kcr2 mutant to support this idea. The precise function of KCR2 remains to be determined, although it is still likely to be as a member of the short-chain dehydrogenases/reductase superfamily. Thus, Arabidopsis differs from maize, in which two closely related KCRs (GL8A and GL8B) both contribute to the activity of the microsomal elongase.
Since KCR1 represents the sole microsomal elongase KCR, it is not surprising that disruption of the KCR1 gene is embryo lethal. This also indicates that the capacity to synthesize VLCFAs is crucial at early stages of embryo development and is in agreement with the recent data of Bach et al. (2008) showing that the dehydratase PAS2, another core microsomal elongase component, is essential during embryogenesis. It is interesting to note that while PAS2 and KCR1 are both essential single genes in Arabidopsis, the CER10 gene is not, even though it represents the sole ortholog of the yeast TSC13 ECR. This very likely indicates the presence of structurally unrelated but functionally equivalent forms of the ECR in Arabidopsis.
To evaluate the requirement for VLCFAs during the growth and development of Arabidopsis after embryogenesis, we generated a number of transgenic lines with reduced KCR1 transcript levels via RNAi-mediated silencing. Such plants displayed a number of developmental and biochemical phenotypes that correlated with the relative reduction in transcript levels for KCR1. In particular, lines with more pronounced reduction in KCR1 transcripts showed an almost complete loss of cuticular waxes and a strong fiddlehead-like phenotype, whereas less severe reduction in KCR1 transcripts still generated an fdh phenotype, but only a small reduction in waxes. Given that the KCSs required for wax synthesis (predominantly CER6) is expressed at significantly higher levels than the FDH KCS (Joubès et al., 2008), the phenotypes that result from the knockdown of KCR1 may reflect not only the reduction in this elongase component activity, but a perturbation to the normal stoichiometry of different KCS activities. Thus, in the absence of sufficient KCR to interact with all the different KCSs present in a cell, the most abundant KCS could titrate out other less abundant KCSs. Such a model could explain the occurrence of an fdh phenotype in the absence of a glossy phenotype. Alternatively, loss of KCR activity could result in reduced activity of all KCSs, mediated through a currently undefined homeostatic process leading to a more uniform reduction in the synthesis of VLCFAs. However, in both scenarios, the outcome is a perturbation (reduction) in the normal total synthesis of VLCFAs as mediated by the activity of several different KCSs (which could include both FAE-like KCSs and ELO-like forms). Given our observations of discrete phenotypes, such as fdh, in the absence of significant reduction in wax loads, we favor the titration model.
Our data confirm the essential nature of VLCFAs in Arabidopsis development, though given the evidence that these fatty acids accumulate in virtually all classes of lipids (sphingolipids, waxes, triacylglycerols, suberin, and some phospholipids), the precise reason(s) for the observed embryo lethality in insertion mutants and gross phenotypic perturbations in the KCR1-RNAi knockdowns requires further investigation. For example, there is clear evidence for an absolute requirement for VLCFAs in Arabidopsis for the synthesis of sphingolipids (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008), similar to the situation observed in yeast (Dickson et al., 2006). However, recent studies have shown that it is possible to generate viable yeast mutants with sphingolipids entirely devoid of VLCFAs (Cerantola et al., 2007), even though the yeast double KCS mutant elo2Δ/elo3Δ, which is unable to synthesize VLCFAs, is not viable (Oh et al., 1997). Thus, the assumption that the lack of VLCFAs critically impairs the formation of essential sphingolipids may not hold true in all cases. It is also interesting to note that a suppressor screen for yeast mutants capable of overcoming an absence of sphingolipids recovered a mutant allele (slc1-1) of a phospholipid acyltransferase SLC1, resulting in the abnormal accumulation of VLCFAs in yeast phospholipids (Nagiec et al., 1993). These studies indicate that the cellular function of VLCFAs, even in extensively studied single-cell model systems, is still only poorly understood.
The results obtained in our study do provide some new insights into the role of VLCFAs in Arabidopsis. First, it is clear that the ability to make waxes is intimately linked to the capacity to synthesize VLCFAs, with some of the KCR1-RNAi knockdown lines showing an almost complete absence of waxes. As previously reported, such wax-deficient mutants are still viable, but they have to be grown at high humidity. Thus, the essential nature of VLCFAs is unlikely to relate to defects in wax biosynthesis. Our study also shows that the more severe KCR1-RNAi knockdown lines are not able to form lateral roots and root hairs, which correlates with a strong reduction in C20+ saturated fatty acids. In Arabidopsis roots, saturated VLCFAs are present in phospholipids, predominantly phosphatidylethanolamine, and are also components of suberin. Strikingly, in lines 4 and 6, which show the strongest defect in lateral root formation, we detected the presence of several suberin-derived dicarboxyclic acids in simple fatty acid methyl-ester extractions. Dicarboxyclic acids are not normally extracted under such mild conditions and may indicate a perturbation to the VLCFA-containing suberin polymer in the root. Further work is needed to determine if impaired root development is linked to phospholipid- or suberin-related VLCFA deficiency.
One final role of VLCFAs that needs to be considered relates to their potential involvement in the establishment of cellular polarity, most likely through the action of VLCFA-containing sphingolipids. Sphingolipids, in conjunction with sterols, have been shown to be enriched in Arabidopsis detergent-insoluble membrane fractions (Borner et al., 2005), consistent with their presence in so-called lipid rafts. Auxin carrier proteins, such as the PINs, have promiscuous (apolar) targeting in plants that are disrupted in sterol metabolism, and this has been hypothesized to relate to defects in endocytosis (Feraru and Friml, 2008). PIN1 has also recently been shown to be present in detergent-insoluble membranes (Titapiwatanakun et al., 2009). One scenario that cannot be excluded is that the KCR1 knockdown phenotypes result from perturbations to cell polarity of some critical cell types. This may be due to disruption of membrane composition, either the formation of plasma membrane microdomains (e.g. lipid rafts) or membrane trafficking as described for the ECR mutant (Zheng et al., 2005). Currently almost nothing is known in plants about the role of VLCFA-containing lipids in these processes, and as such, this represents a topic ripe for further research.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Ds transposon insertional line (RATH12-5282-1-G) was obtained from Plant Functional Genomics Research Group of RIKEN Genomic Sciences Center (Kuromori et al., 2004), whereas SALK_096487 and SAIL_536_H04 T-DNA insertional lines were ordered from the Arabidopsis Biological Resource Center (www.arabidopsis.org). The seeds were germinated on AT-agar plates (Somerville and Ogren, 1982) and grown in soil (Sunshine Mix 5; SunGro) at 20°C under continuous light (90–120 μE m−2 s−1 photosynthetically active radiation).
After transformation with the KCR1-RNAi construct, T1 seeds were surface sterilized, spread evenly onto agar plates containing kanamycin, stratified for 3 d in a cold room, and germinated at 22°C under continuous light (150 μE m−2 s−1). After 2 weeks, kanamycin-resistant plant were transferred to soil and grown in a controlled environment cabinet with a photoperiod of 16 h light (150 μE m−2 s−1) at 22°C and 8 h dark at 18°C, keeping the soil damp and atmospheric humidity above 80%.
Wax and Fatty Acid Extraction and Analysis
Cuticular wax was extracted and analyzed as described by Rowland et al. (2006). For the determination of the seed fatty acid composition, FAMEs were prepared by incubation with methanolic-HCl (Supelco) at 80°C for 2 h, followed by extraction with hexane and analysis by gas-liquid chromatography (Rossak et al., 2001).
Extraction and Analysis of Fatty Acids, Acyl-CoAs, and Sphingolipids
Total fatty acids from leaves and root tissues and from yeast cultures were extracted and methylated as described before (Sayanova et al., 1997; Napier et al., 1998). Total FAMEs were analyzed by gas chromatography and identified by mass spectrometry. Acyl-CoA extraction and profiling was carried out as described by Larson and Graham (2001). Sphingolipids were extracted from root and shoot tissues and quantified as detailed by Markham and Jaworski (2007).
Total RNA Isolation and Semiquantitative RT-PCR Analysis
Total RNAs from aerial tissues of 2- to 6-week-old Arabidopsis (Arabidopsis thaliana) plants were extracted using TRIZOL reagent (Invitrogen Life Technologies) following the manufacturer's instructions. Total RNA from root tissue was isolated from 14-d-old seedlings grown on AT-agar plates placed vertically. For RT-PCR analysis of AtKCR1 mRNA levels in seeds from immature siliques, total RNA (2.5 μg) from each sample, treated with RNase-free DNase (Promega), was used for reverse transcriptase reactions. First-strand cDNA was synthesized with random hexamers using a SuperScript first-strand synthesis system according to the manufacturer's manual (Invitrogen Life Technologies), and the constitutive expression gene glyceraldehyde-3-P dehydrogenase C subunit was used as a control for RT-PCR experiments. One microliter of reverse transcription reaction mixture was used as a template in a 20-μL PCR. The primer sequences used were as follows: AtKCR1 forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGAGATCTGCACTTACTTCAAAT-3′ (bold sequences = directional attB sites) and AtKCR1 reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCTTTCTTCATGGAGTCTTTTTGG-3′. The amplification conditions were 94°C (30 s), 56°C (30 s), and 72°C (30 s), and the number of cycles varied from 28 to 32 for different genes. Each RT-PCR was repeated twice. For RT-PCR analysis of AtKCR1 and AtKCR2 mRNA levels in 2- to 6-week-old plant tissues, total RNA (2.5 μg) from each sample, treated with Turbo DNA-free DNase (Ambion), was used for reverse transcriptase reactions. First-strand cDNA was synthesized using oligo(dT)12-18 primer and SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen). The constitutive expression gene tubulin (α-1 tubulin) was used as a control for RT-PCR experiments. One microliter of reverse transcription reaction mixture was used as a template in a 25-μL PCR. The primer sequences used were as follows: AtKCR1 forward primer 5′-GGCCGGTACCATGGAGATCTGCACTTAC-3′ and AtKCR1 reverse primer 5′- GGCCCTCGAGTCATTCTTTCTTCATGGA-3′ or AtKCR2 forward primer 5′-GGCCGGTACCATGCAGGGAGCATGCATCTC-3′ and AtKCR2 reverse primer 5′-GGCCCTCGAGTTAAGATAAACTTCTTCTGC-3′. The amplification conditions were 94°C (30 s), 55°C (30 s), and 72°C (60 s), and the number of cycles varied from 25 to 28 for different genes. Each RT-PCR was repeated three times.
Plasmid Construction and Plant Transformation
The full-length cDNAs of AtKCR1 and AtKCR2 were obtained by RT-PCR with primers containing Gateway (Invitrogen) recombination sites (bold sequences), AtKCR1 forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGAGATCTGCACTTACTTCAAAT-3′ and reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCTTTCTTCATGGAGTCTTTTTGG-3′; AtKCR2 forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCAGGGAGCATGCATCTCCGAGA-3′ and reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGATAAACTTCTTCTGCGAAGTCCG-3′, and subcloned into Gateway donor vector (pDONR207; Curtis and Grossniklaus, 2003). After sequencing the coding sequences of the AtKCR1 and AtKCR2 genes, the confirmed clones of AtKCR1 and AtKCR2 were transferred into the Gateway destination vectors pMDC32 (Curtis and Grossniklaus, 2003), for overexpression, or pEarleygate101 (Earley et al., 2006), for determination of subcellular localizations of AtKCR1 and AtKCR2. The destination vectors were introduced into 5-week-old relevant Arabidopsis plants by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). The first 390 bp of AtKCR1 were amplified using either primers KCR1RNAiXhoF (5′-GGCCCTCGAGATGGAGATCTGCACTTAC-3′) and KCR1RNAiKpnR (5′-GGCCGGTACCATCTAATCCTTCAATACT-3′) or primers KCR1RNAiXbaF (5′-GGCCTCTAGAATGGAGATCTGCACTTAC-3′) and KCR1RNAiClaR (5′-GGCCATCGATATCTAATCCTTCAATACT-3′). The amplified products were subcloned into the pCR4-TOPO vector (Invitrogen) and sequenced. Verified inserts were restricted from pCR4-TOPO using XhoI and KpnI or XbaI and ClaI (underlined in primer sequences) and cloned in sense or antisense orientation, respectively, into the pKannibal vector (Wesley et al., 2001). The hairpin RNA encoding cassette thus created was restricted from pKannibal using NotI and cloned into a NotI cut pART27 binary vector. This construct was introduced into Arabidopsis (ecotype Col) by Agrobacterium-mediated transformation as described in Clough and Bent (1998).
Functional Complementation of the Yeast ybr159wΔ Mutant by AtKCR1 or AtKCR2
Wild-type and ybr159wΔ mutant yeast strains expressing either AtFAE1 or IgELO9 (Isochrysis galbana; IgASE1) pYES2 constructs have been described before (Beaudoin et al., 2002, Qi et al., 2002). The AtKCR2 ORF was amplified by PCR using primers AtKCR2-Fwd (5′-GGCCGGATCCATGCAGGGAGCATGCATCTC-3′) and AtKCR2-Rev (5′-GGCCGGTACCTTAAGATAAACTTCTTCTGC-3′). The amplified sequence was restricted using BamHI and KpnI (underlined in the forward and reverse primers, respectively), purified using the Qiagen PCR purification kit, and ligated into a BamHI/KpnI cut pESC-TRP plasmid vector (Stratagene). For functional complementation studies, the ybr159wΔ strains described above were cotransformed with pESC-TRP constructs containing either YBR159w, AtKCR1 (Beaudoin et al., 2002), or AtKCR2 (this study). The empty pESC-TRP vector was used as negative control. Yeast cells were cultivated and the transgenes induced in the presence or absence of exogenously supplied fatty acid substrate as described previously (Beaudoin et al., 2002).
GUS Activity Assays
A length of 780 bp of the 5′ promoter region of AtKCR1 was amplified from genomic DNA by PCR using forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCCTTTGGACTTACCAACG-3′ and reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGGTTGAGACTTTGGAGATGAGA-3′. The amplified product was introduced into Gateway entry vector by Gateway BP Clonase enzyme mix (Invitrogen). The confirmed entry clones were transferred into destination vector pMDC163 (Curtis and Grossniklaus, 2003) carrying the GUS gene. The generated construct was introduced into wild-type Col-0 plants by Agrobacterium-mediated transformation.
For GUS activity assays, 1- or 2-week-old seedlings, leaves, flowers, and siliques from 4-week-old plants, and embryos of different developmental stages were incubated in GUS assay buffer (50 mm sodium phosphate, pH 7.0, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.1% [v/v] Triton X-100, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) at 37°C overnight. Then, samples were cleared in 70% ethanol and visualized by light microscopy.
TB Test
For rapid visualization of leaf cuticle defects, we used the TB test described by Tanaka et al. (2004) with the following differences. After surface sterilization, seeds were sown on plate containing half-concentration Murashige and Skoog (1962) medium solidified with 0.5% (w/v) phytagel (Sigma-Aldrich).
Microscopy
For the YFP or CFP fusion protein localization analysis, leaf samples of transgenic plants were examined using confocal laser scanning microscope as described by Zheng et al. (2005). Excitation light wavelength was 458 to 514 nm, the emission filter set at 505 to 530 nm, and autofluorescence was detected using an emission filter set at 600 to 650 nm.
Siliques of different developmental stages from heterozygous plants were dissected. Ovules from individual siliques were mounted on slides in Hoyer's clearing solution (chloral hydrate, water, glycerol, 8:2:1 v/v) and cleared overnight at 4°C (Liu and Meinke, 1998). The cleared ovule was observed with a LSM 510 Meta DIC microscope (Carl Zeiss).
For electron microscopy, sections of stem and leaves were mounted on Al cryo stubs using Optimal Cutting Temperature compound (Ted Pella) and plunge frozen in slushed liquid nitrogen. The samples were then transferred to the cryo chamber stage under vacuum and etched for 2 min at −95°C. The stage temperature was returned to −175°C, and the samples were coated for 90 s with Au Pd. The samples were then loaded on a JEOL 6700 FEG scanning electron microscope chamber stage maintained at −160°C for imaging using the on board software.
Bioinformatics
For routine sequence comparison, BLAST was used (Altschul et al., 1990). The amino acid sequences were aligned using the ClustalW program (Thompson et al., 1994). Transmembrane spanning helices were predicted using the DAS transmembrane prediction server (http://www.sbc.su.se/∼miklos/DAS/). Motifs were predicted using the ELPH program (http://www.cbcb.umd.edu/software/ELPH). The FASTA sequence comparison program (http://wrpmg5c.bioch.virginia.edu/fasta_www2/fasta_www.cgi) was used for sequence identity and similarity.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NY143811 (AtKCR1), NM102292 (AtKCR2), AY557868 (Ybr159), AF302098 (GL8a), and AF527771 (GL8b).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence comparisons of AtKCR1 and AtKCR2 with homologs from other species.
Supplemental Figure S2. Expression of AtKCR1 and AtKCR2 in AtKCR1-RNAi plants.
Supplemental Figure S3. The phenotypes of cosuppressed plants.
Supplementary Material
Acknowledgments
We thank the Genomic Analysis Laboratory of the Salk Institute and RIKEN Genomic Sciences Center for providing Arabidopsis T-DNA insertion mutants, the Bioimaging Facility at the University of British Columbia for providing microscopy and technical support, and Jean Devonshire and the Centre for Bioimaging at Rothamsted who helped with the scanning electron microscopy work. We are also grateful to Louise Michaelson and Teresa Dunn for helpful discussions.
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (UK) to Rothamsted Research and a grant from the Natural Sciences and Engineering Research Council of Canada to L.K.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Johnathan A. Napier (johnathan.napier@bbsrc.ac.uk) and Ljerka Kunst (kunst@interchange.ubc.ca).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Gable K, Sayanova O, Dunn T, Napier AJ (2002) A Saccharomyces cerevisiae gene required for heterologous fatty acid elongase activity encodes a microsomal β-keto-reductase. J Biol Chem 29 11481–11488 [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA (2002) Demystifying suberin. Can J Bot 80 227–240 [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M (2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J 40 920–930 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerantola V, Vionnet C, Aebischer OF, Jenny T, Knudsen J, Conzelmann A (2007) Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem J 401 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti DL, Cook L, Nagi MN, Suneja SK (1992) The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog Lipid Res 31 1–51 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Weissman JS (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Nature 130 663–677 [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dickson RC, Sumanasekera C, Lester RL (2006) Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res 45 447–465 [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Han G, Chen M, Berg RH, Dunn TM, Cahoon EB (2008) Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J 54 284–298 [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Perera MADN, Yandeau-Nelson MD, Meeley BM, Nikolau BJ, Schanable PS (2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42 844–861 [DOI] [PubMed] [Google Scholar]
- Dunn TM, Lynch DV, Michaelson LV, Napier JA (2004) A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann Bot (Lond) 93 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE (1995) Effects of plant epicuticular lipids on insect herbivores. Annu Rev Entomol 40 171–194 [Google Scholar]
- Feraru E, Friml J (2008) PIN polar targeting. Plant Physiol 147 1553–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry 66 2643–2658 [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L (2007) Suberin—a biopolyester forming apoplastic plant interfaces. Curr Opin Plant Biol 10 252–259 [DOI] [PubMed] [Google Scholar]
- Gable K, Garton S, Napier JA, Dunn TM (2004) Functional characterization of the Arabidopsis thaliana orthologue of Tsc13p, the enoyl reductase of the yeast microsomal fatty acid elongating system. J Exp Bot 55 543–545 [DOI] [PubMed] [Google Scholar]
- Han G, Gable K, Kohlwein SD, Beaudoin F, Napier JA, Dunn TM (2002) The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J Biol Chem 277 35440–35449 [DOI] [PubMed] [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EA (1994) Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol 105 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67 547–566 [DOI] [PubMed] [Google Scholar]
- Karimi R, Brumfield T, Brumfield F, Safaiyan F, Stein S (2007) Zellweger syndrome: A genetic disorder that alters lipid biosynthesis and metabolism. Internet J Pharmacol 5 1–15 [Google Scholar]
- Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T (2001) Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol 21 109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Thiel F (1989) A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J Hered 80 118–122 [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42 51–80 [DOI] [PubMed] [Google Scholar]
- Kunst L, Taylor DC, Underhill EW (1992) Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem 30 425–434 [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Tkabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37 897–905 [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA (2001) A novel technique for the sensitive quantification of acyl-CoA esters from plant tissues. Plant J 25 115–125 [DOI] [PubMed] [Google Scholar]
- Leonard AE, Kelder B, Bobik EG, Chuang LT, Lewis CJ, Kopchick JJ, Mukerji P, Huang YS (2002) Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37 733–740 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16 21–31 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reverse-phase high-performance liquid chromatography coupled to electrospray-ionization tandem mass-spectrometry. Rapid Commun Mass Spectrom 21 1304–1314 [DOI] [PubMed] [Google Scholar]
- McMahon A, Butovich I, Mata NL, Klein M, Ritter R, Richardson J, Birch DG, Edwards AO, Kedzierski W (2007) Retinal pathology and skin barrier defect in mice carrying a Stargardt disease-3 mutation in elongase of very long chain fatty acids-4. Mol Vis 13 258–272 [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Kunst L (1997) Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12 121–131 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC (1993) A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem 268 22156–22163 [PubMed] [Google Scholar]
- Napier JA, Hey SJ, Lacey DJ, Shewry PR (1998) Identification of a Caenorhabditis elegans Δ6-fatty-acid-desaturase by heterologous expression in Saccharomyces cerevisiae. Biochem J 330 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugteren DH (1965) The enzymic chain elongation of fatty acids by rat-liver microsomes. Biochim Biophys Acta 106 280–290 [DOI] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272 17376–17384 [DOI] [PubMed] [Google Scholar]
- Paul S, Gable K, Beaudoin F, Cahoon E, Jaworski J, Napier JA, Dunn TM (2006) Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J Biol Chem 281 9018–9029 [DOI] [PubMed] [Google Scholar]
- Poulos A, Beckman K, Johnson DW, Paton BC, Robinson BS, Sharp P, Usher S, Singh H (1992) Very long-chain fatty acids in peroxisomal disease. Adv Exp Med Biol 318 331–340 [DOI] [PubMed] [Google Scholar]
- Preuss D, Lemieux B, Yen G, Davis RW (1993) A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signalling during fertilization. Genes Dev 7 974–985 [DOI] [PubMed] [Google Scholar]
- Qi B, Beaudoin F, Fraser T, Stobart AK, Napier JA, Lazarus CM (2002) Identification of a cDNA encoding a novel C18-Delta(9) polyunsaturated fatty acid-specific elongating activity from the docosahexaenoic acid (DHA)-producing microalga, Isochrysis galbana. FEBS Lett 510 159–165 [DOI] [PubMed] [Google Scholar]
- Rossak M, Smith M, Kunst L (2001) Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol Biol 46 717–725 [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L (2006) CER4 encodes an alcohol-forming fatty acyl-Coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayanova O, Smith MA, Lapinskas P, Stobart AK, Dobson G, Christie WW, Shewry PR, Napier JA (1997) Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of Δ6-desaturated fatty acids in transgenic tobacco. Proc Natl Acad Sci USA 94 4211–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schneiter R, Brugger B, Amann CM, Prestwich GD, Epand RF, Zellnig G, Wieland FT, Epand RM (2004) Identification and biophysical characterization of a very-long-chain-fatty-acid-substituted phosphatidylinositol in yeast subcellular membranes. Biochem J 381 941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM (1996) A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16 7161–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Isolation of photorespiratory mutants of Arabidopsis. In RB Hallick, NH Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier, New York, pp 129–139
- Sprecher H (1974) The influence of dietary alterations, fasting and competitive interactions on the microsomal chain elongation of fatty acids. Biochim Biophys Acta 360 113–123 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y (2004) A new method for rapid visualization of defects in leaf cuticule reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37 139–146 [DOI] [PubMed] [Google Scholar]
- Tehlivets O, Scheuringer K, Kohlwein SD (2007) Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta 1771 255–270 [DOI] [PubMed] [Google Scholar]
- Teng C, Dong H, Shi L, Deng Y, Mu J, Zhang J, Yang X, Zuo J (2008) Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol 146 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Bibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, et al (2009) ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57 27–44 [DOI] [PubMed] [Google Scholar]
- Toulmay A, Schneiter R (2007) Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie 89 249–254 [DOI] [PubMed] [Google Scholar]
- Trenkamp S, Martin W, Tietjen K (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA 101 11903–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasireddy V, Uchida Y, Salem N, Kim SY, Mandal MN, Reddy GB, Bodepudi R, Alderson NL, Brown JC, Hama H, et al (2007) Loss of functional ELOVL4 depletes very long-chain fatty acids (> or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum Mol Genet 16 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein-Knowles P (1982) Elongase and epicuticular wax biosynthesis. Physiol Veg 20 797–809 [Google Scholar]
- von Wettstein-Knowles P (1993) Waxes, cutin and suberin. In T Moore, ed, Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 127–166
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–90 [DOI] [PubMed] [Google Scholar]
- Westerberg R, Tvrdik P, Undén AB, Månsson JE, Norlén L, Jakobsson A, Holleran WH, Elias PM, Asadi A, Flodby P, et al (2004) Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem 279 5621–5629 [DOI] [PubMed] [Google Scholar]
- Wertz PW, Downing DT (1983) Ceramides of pig epidermis: structure determination. J Lipid Res 24 759–765 [PubMed] [Google Scholar]
- Xu X, Dietrich CR, Delledonne M, Xia Y, Wen TJ, Robertson DJ, Nikolau BJ, Schnable PS (1997) Sequential analysis of the cloned glossy8 gene of maize suggests that it may code for a β-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant J 15 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ, et al (2001) A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet 27 89–93 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.