Abstract
Understanding the cellular differentiation of multicellular organisms requires the characterization of genes whose expression is modulated in a cell type-specific manner. The Arabidopsis (Arabidopsis thaliana) root hair cell is one model for studying cellular differentiation. In this study, root hair cell-specific genes were screened by a series of in silico and experimental filtration procedures. This process included genome-wide screening for genes with a root hair-specific cis-element in their promoters, filtering root-specific genes from the root hair-specific cis-element-containing genes, further filtering of genes that were suppressed in root hair-defective plant lines, and experimental confirmation by promoter assay. These procedures revealed 19 root hair-specific genes, including many protein kinases and cell wall-related genes, most of which have not been characterized thus far. Functional analyses of these root hair-specific genes with loss-of-function mutants and overexpressing transformants revealed that they play roles in hair growth and morphogenesis. This study demonstrates that a defined cis-element can serve as a filter to screen certain cell type-specific genes and implicates many new root hair-specific genes in root hair development.
Multicellular organisms are organized structures with diverse morphologically and functionally differentiated cells. Cellular differentiation in multicellular organisms is highly coordinated in time and space and is primarily directed by transcriptional regulation of cell type-specific genes. Where, when, and how much a gene is transcribed is mostly coded in the gene promoter region. The short nucleotide motif for this modular code is called the cis-element, which is recognized by factors for transcriptional activation or suppression (Carroll et al., 2001). Although the identification and characterization of cell type-specific genes and their cis- and trans-regulatory elements would be useful for deciphering cell differentiation mechanisms, relatively few cases are known in plants.
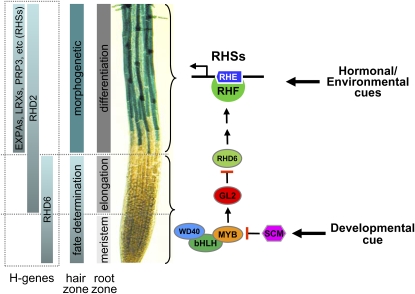
The root hair is an outgrowth of the root epidermal cell. Root hair development proceeds sequentially along the root longitudinal axis from the root tip to the basal (upper) regions: cell specification, hair initiation, and hair elongation by tip growth (Grierson and Schiefelbein, 2002). Hair/nonhair cell fate in the Arabidopsis (Arabidopsis thaliana) root is established by the position-dependent activities of a receptor-like kinase, a WD40 protein/basic helix-loop-helix transcription factor/MYB transcription factor complex, and a MYB-like protein, which are active mostly in the epidermis of the root meristem and elongation zones (fate-determination zone in Fig. 1; Schiefelbein and Lee, 2006). In contrast, root hair morphogenetic processes (hair initiation and elongation) occur in the upper mature region (morphogenetic zone in Fig. 1). Many genetic studies have shown that different gene sets are implicated in cell fate determination and hair morphogenesis (Parker et al., 2000; Grierson et al., 2001; Grierson and Schiefelbein, 2002; Jones et al., 2006). Root hair morphogenesis is ultimately attributable to the activities of hair cell (H cell or trichoblast)-specific genes, although these should collaborate with other general cell morphogenetic genes.
Figure 1.
Root hair development and expression patterns of H cell-specific genes. Root zones indicate three temporospatial root development stages, and hair zones indicate two stages for the fate determination of hair/nonhair cells and hair morphogenesis. The fate determination zone approximately covers the root region where the fate determinants (WEREWOLF [WER], CAPRICE [CPC], GLABRA3/ENHANCER OF GLABRA3 [GL3/EGL3], and GL2) are expressed. H genes are the genes that are specifically expressed in the hair-forming H cell (trichoblast cell file) of both fate determination and morphogenetic zones. The root image shows the blue expression pattern of ProEXPA7:GUS. The right part designates the signaling flow from fate determinants to morphogenetic genes. RHSs are RHE-containing genes, and RHF is a putative transcription factor that binds RHE. Hormonal/environmental cues are likely to act on the morphogenetic genes downstream of RHD6. bHLH, Basic helix-loop-helix transcription factor (GL3/EGL3); GL2, a homeodomain transcription factor; MYB, MYB transcription factor (WER and CPC); RHD6, a bHLH transcription factor; SCM, SCRAMBLED, a receptor-like kinase; WD40, WD40 domain protein (TRANSPARENT TESTA GLABRA).
Depending on their temporospatial expression pattern, three types of H cell-specific genes can be proposed: genes expressed in the fate-determination zone (meristem and elongation zones, where fate determinants are expressed), in both the fate-determination and morphogenetic zones (elongation and differentiation zones), and only in the morphogenetic zone (differentiation zone; Fig. 1). For example, ROOT HAIR DEVELOPMENT6 (RHD6) is expressed in the meristem and elongation zones (Menand et al., 2007), RHD2 in the elongation and morphogenetic zones (Takeda et al., 2008), and PROLINE-RICH PROTEIN3 (PRP3), LEUCINE-RICH REPEAT/EXTENSIN1 (LRX1), LRX2, EAXPANSIN A7 (EXPA7), and EXPA18 in the morphogenetic zone (Bernhardt and Tierney, 2000; Baumberger et al., 2001, 2003; Cho and Cosgrove, 2002). These genes are all expressed in the H cell position and are likely necessary for normal root hair formation. Loss of these genes almost completely blocks root hair formation (RHD6; Masucci and Schiefelbein, 1994), greatly reduces hair elongation (RHD2; Schiefelbein and Somerville, 1990), or deforms hair morphogenesis (LRXs; Baumberger et al., 2001, 2003). Thus, the H cell-specific genes expressed in the differentiation zone, designated hereafter as “morphogenetic H genes” (Fig. 1), seem to mediate hair morphogenesis, such as bulge formation, elongation, and morphological alterations.
The same cis-element (RHE, for root hair element) controls the root hair cell-specific expression of all known morphogenetic H genes (LRX1, LRX2, EXPA7, EXPA18, PRP3, and the orthologous genes of EXPA7; Kim et al., 2006; Cho, 2007). The core RHE consists of 16 or 17 nucleotides with an incomplete palindromic structure. Because the biological function of RHE for H cell specificity is conserved among the known root hair cell-specific genes of Arabidopsis and other angiosperms, the RHE consensus sequence could potentially be used for in silico screening for additional root hair cell-specific genes at the genome level. Coexpression patterns by certain genetic cues or external stimuli in transcriptome analyses have been used to identify coregulated genes at a large scale (Lisso et al., 2005; Zhang et al., 2005; Jones et al., 2006; Brady et al., 2007), and the defined cis-elements for abscisic acid and abiotic stress responses were used for targeted gene searches (Zhang et al., 2005). However, use of a similar approach for the targeted search of cell type-specific genes and subsequent experimental confirmation have not yet been performed.
Using the root hair cell as a model, we sought to demonstrate that a defined cis-element can aide in the genome-wide screening of cell type-specific genes. To refine the screening process and confirm root hair cell specificity, we experimentally analyzed root hair-specific transcriptomes and gene promoter activity. Analyses of loss-of-function mutants and overexpressing transformants for the screened genes demonstrated that the morphogenetic H genes genuinely function in root hair morphogenesis.
RESULTS
We applied four sequential filtering steps to isolate root hair morphogenetic H genes from the whole Arabidopsis genome: in silico screening of RHE-containing genes from the Arabidopsis genome; selection of root-specific genes; filtration of putative morphogenetic H genes; and experimental confirmation of root hair specificity (Fig. 2).
Figure 2.
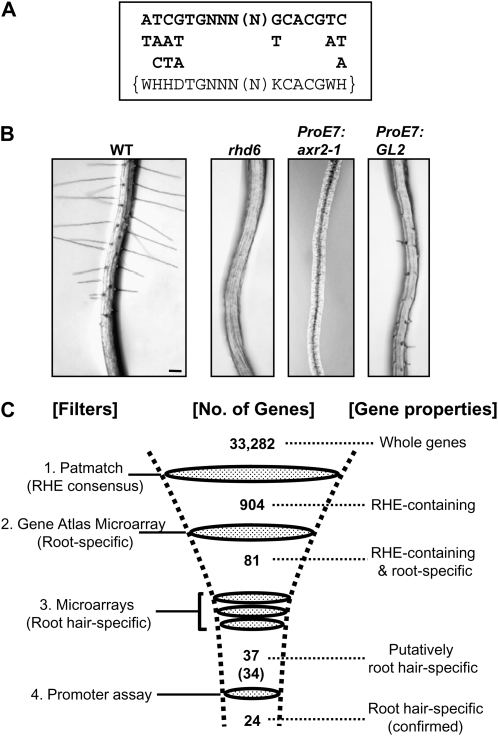
Tools for screening root hair-specific Arabidopsis genes. A, The RHE consensus. The top sequence represents majority nucleotides in the RHEs from angiosperm root hair genes. The nucleotides below the sequence are the next most frequent nucleotide species. The bottom sequence inside brackets refers to the multiple nucleotide consensus from the above sequence and was used in Patmatch to screen out RHE-containing genes from the Arabidopsis genome. B, Root hair-defective Arabidopsis mutant and transgenic lines used to produce triple microarray filters for root hair specificity. The root hair-repressing genes axr2-1 and GL2 were specifically expressed in the root hair using the root hair-specific EXPA7 promoter (ProE7). Genes down-regulated in the root hair-defective rhd6 mutant or transgenic lines were identified by microarray analysis of root-expressible RNAs. WT, Wild type. Bar = 0.1 mm for all. C, Filtering of root hair-specific genes from the whole Arabidopsis genome. Four filtering steps were used to purify the root hair-specific genes: 1, Patmatch to filter RHE-containing genes; 2, Genevestigator Gene Atlas microarray database to filter the RHE-containing genes with root specificity; 3, microarray data from root hair-defective mutant and transgenic lines to filter putative root hair-specific genes; 4, experimental promoter assay of 34 putative root hair-specific genes to confirm root hair specificity, yielding 24 root hair-specific genes, among which five were previously identified.
Screening of RHE-Containing Genes from the Whole Arabidopsis Genome
To identify genes carrying RHE in their proximal promoter regions, we used the Patmatch analysis tool with the RHE consensus sequence (Fig. 2A). As a cis-element sequence is generally flexible for transcription factor binding (Latchman, 2004) and the functional RHE includes plural syntactic variants (Kim et al., 2006), a less stringent, composite RHE consensus [WHHDTGNNN(N)KCACGWH], where W = A/T, H = A/T/C, D = G/T/A, K = G/T, and N = A/T/C/G, was used in Patmatch to obtain as many starting root hair genes as possible (Fig. 2A). The Patmatch screen for the whole 33,282 Arabidopsis genes yielded 904 nuclear genes (for the gene list, see Supplemental Table S1) with one or more RHEs within 1,000 bp upstream of the start codon. Because actual RHE positions were somewhat different from the positions indicated by the database, these were manually corrected (Table I).
Table I.
RHE-containing root hair genes in Arabidopsis
| Locus | RHS Gene No. | Gene Product | Putative Subcellular Localizationa | RHE Position from ATGb | RHE Orientationc | RHE Sequenced | RHE Linker Typee | Fold Changes of Transcript in Root Hair-Defective Linesf
|
Promoter Length from ATGg | Root Hair-Specific Expressionh | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rhd6 | ProE7:axr2 | ProE7:GL2 | ||||||||||
| AT1G05990 | RHS1 | Calcium-binding protein | Cytosol | −38 | F | TTCTTCCTCCACACGCA | 3 | −3.5 | −21.1 | −21.0 | −1,099 | Yes |
| −739 | R | AACGTGCAACCAATAT | 2 | |||||||||
| AT1G12950 | RHS2 | MATE family protein | Plasma membrane | −536 | F | CATATGGAGTCCACGAC | 3 | −4.0 | −3.5 | −2.8 | −1,147 | Yes |
| −566 | F | TAATTGCTCCGCACGTA | 3 | |||||||||
| AT1G16440 | RHS3 | Protein kinase | Chloroplast | −257 | F | AACCTAATAACACGAG | 2 | −3.0 | −7.5 | −4.6 | −882 | Yes |
| −454 | R | TACGTGGAAAACAGGCC | 3 | |||||||||
| −547 | R | TTCGTGATAACAAGTA | 2 | |||||||||
| AT1G30850 | RHS4 | Hypothetical protein | Nucleus | −111 | R | AACGTGCTTACCATTTT | 3 | −19.7 | −1.6 | −1.6 | −655 | Yes |
| −306 | R | TACGTGCAAACCATTTC | 3 | |||||||||
| −975 | F | ATCTTGGATCCACGAG | 2 | |||||||||
| AT1G34760 | RHS5 | 14-3-3 protein | Cytosol or peroxisome | −52 | R | ATCGTGACAGCAGTTT | 2 | −2.6 | −5.3 | −2.3 | −923 | Yes |
| −559 | F | TTTTTTGTTCCACGAA | 2 | |||||||||
| AT1G51880 | RHS6 | Leu-rich repeat protein kinase | Plasma membrane | −214 | F | AATGTCGTAGACACGTA | 3 | −59.7 | −32 | −34.3 | −721 | Yes |
| −373 | R | AACGTGCCTCCATAAA | 2 | |||||||||
| AT1G54970 | RHS7 | Pro-rich protein (PRP1) | Cell wall | −212 | R | ATCGTGCATAACACTAT | 3 | −4.6 | −6.5 | −5.2 | −1,162 | Yes |
| AT1G63450 | RHS8 | Exostosin | Unclear | −73 | R | GACGTGAGTGCATGTC | 2 | −5.7 | −1.3 | −1.4 | −1,408 | Yes |
| −301 | F | AACTTGGAGGCACGTT | 2 | |||||||||
| AT1G69240 | RHS9 | Hydrolase, α/β-fold family protein | Chloroplast | −91 | F | CACATGCAAGCACGAA | 2 | −5.7 | −5.7 | −3.0 | −1,446 | Yes |
| −202 | R | TTCGTGAGTCTCAAGAA | 3 | |||||||||
| AT1G70450 | Ser/Thr protein kinase | Cytosol | −311 | R | GACGTGCCTACAATTA | 2 | −3.5 | −3.2 | −3.7 | −922 | Noi | |
| −478 | F | TTGCTTATTGCACGAG | 2 | |||||||||
| AT1G70460 | RHS10 | Ser/Thr protein kinase with extensin | Nucleus | −447 | R | GACGTGCCTCCAATAA | 2 | −5.3 | −4.3 | −4.6 | −946 | Yes |
| −636 | F | GACTTACAAACACGTC | 2 | |||||||||
| −761 | R | TACGTGTGTATTACAAA | 3 | |||||||||
| AT2G03720 | Universal stress protein (USP) family protein | Cytosol | −317 | F | TTCATGGCAAGCACGTT | 3 | −8.0 | −7.0 | −4.6 | −678 | Whole epidermis | |
| AT2G45890 | RHS11 | Kinase partner protein (KPP); RopGEF4 | Nucleus | −96 | F | TTAATCTTTGTCACGTT | 3 | −3.5 | −1.9 | −4.5 | −968 | Yes |
| −220 | F | AATGTGGAAGGCACGTC | 3 | |||||||||
| −706 | R | GACGTGGACTTCATCCC | 3 | |||||||||
| −877 | R | CACGTGCCACCACCAC | 2 | |||||||||
| AT3G10710 | RHS12 | Pectinesterase | Cell wall | −166 | F | TTTGTGCCTCGCACGTA | 3 | −7.0 | −16.0 | −7.0 | −757 | Yes |
| −1,288 | F | TGCTTGGTAACACGTC | 2 | |||||||||
| AT3G12540 | Expressed protein | Nucleus | −430 | F | TGATTGCAGGCACGAC | 2 | −6.5 | −2.3 | −2.8 | −847 | Whole epidermis | |
| −515 | F | TTCATGGCTTTCACGTC | 3 | |||||||||
| AT3G47740 | ABC transporter family protein | Plasma membrane | −310 | F | ATAGTGCGCTCACGTA | 2 | −3.2 | −8.6 | −3.2 | −1,260 | Whole epidermis | |
| AT3G48940 | Remorin | Nucleus | −246 | F | TTTTTTTCTGTCACGAA | 3 | −2.5 | −3.0 | −3.7 | −1,057 | Whole epidermis and vasculature | |
| −270 | R | ATCGTGCACAACAATAA | 3 | |||||||||
| −462 | R | GTCGTGACGGTGAGCTT | 3 | |||||||||
| −1,031 | F | TGAGTATCTTTCACGTC | 3 | |||||||||
| AT4G02270 | RHS13 | Extensin family protein | Cell wall | −200 | R | ACCGTGCATACACGTT | 3 | −4.6 | −22.6 | −5.3 | −697 | Yes |
| −215 | F | ACCGTGCATACACGTT | 2 | |||||||||
| −253 | F | ATAATGCGACTCACGTC | 3 | |||||||||
| AT4G15740 | C2 domain-containing protein | Nucleus | −336 | F | AATGTTTTTACACGAG | 2 | −2.5 | −1.7 | −2.6 | −1,085 | No | |
| −532 | R | TACGTGAGCACCATGAA | 3 | |||||||||
| AT4G22080 | RHS14 | Pectate lyase family protein | Cell wall | −110 | R | ATCGTGTTTACCACACC | 3 | −8.6 | −68.6 | −78.8 | −1,307 | Yes |
| −348 | F | TGGATTGGTACACGTA | 2 | |||||||||
| −597 | F | TCCTTGATCTCACGTA | 2 | |||||||||
| AT4G25220 | RHS15 | Putative transporter (major facilitator superfamily) | Plasma membrane | −510 | R | TACGTGCGGGCACAAA | 2 | −7.0 | −59.7 | −2.5 | −1,111 | Yes |
| −879 | F | CAAATAAACTTCACGAG | 3 | |||||||||
| −1,159 | R | GTCGTGGAGAGATGAC | 2 | |||||||||
| AT4G29180 | RHS16 | Leu-rich repeat protein kinase | Plasma membrane | −155 | R | GTCGTGAGCAGCACTTA | 3 | −4.9 | −4.3 | −2.5 | −694 | Yes |
| AT4G37060 | Patatin-like protein (PLP5) | Cytosol or cell wall | −750 | R | CACGTGAAGTTTATTTT | 3 | −1.2 | −1.4 | −2.0 | −1,289 | No | |
| −838 | F | ATCATGATTAGCACGAA | 3 | |||||||||
| AT4G37070 | Patatin-like protein (PLP1) | Chloroplast or cytosol | −661 | R | ATCGTGCATGCAGAGA | 2 | −1.2 | −1.4 | −2.0 | −1,223 | Whole epidermis (stronger in hair cells) | |
| −771 | R | AGCGTGCTTGCATGCT | 2 | |||||||||
| −924 | F | ATCATGATTAGCACGAA | 3 | |||||||||
| AT4G38390 | RHS17 | Expressed protein | Plasma membrane | −591 | F | TACATGTCATCACGTA | 2 | −4.0 | −3.2 | −2.8 | −1,187 | Yes |
| AT5G19560 | Kinase partner protein (KPP); RopGEF10 | Nucleus | −402 | F | AACATGGACCGCACGTT | 3 | −22.6 | −20.0 | −34.3 | −1,270 | Whole epidermis (stronger in root hairs) | |
| −551 | F | CAATTTTCTTACACGAA | 3 | |||||||||
| AT5G22410 | RHS18 | Peroxidase | Cell wall | −129 | F | TCCATGAACGCACGTT | 2 | −5.7 | −21.0 | −6.5 | −892 | Yes |
| −485 | F | TTAGTAGTATCCACGTT | 3 | |||||||||
| −534 | F | ATGATTTTTGCACGTT | 2 | |||||||||
| AT5G41280 | Receptor-like protein kinase | Cell wall? | −272 | F | TAATTGGTTTCACGTC | 2 | −2.5 | −1.9 | +9.8 | −695 | Whole epidermis | |
| −453 | R | ATCGTGCGTAATAATTT | 3 | |||||||||
| AT5G67400 | RHS19 | Peroxidase | Cell wall | −199 | R | AACGTGGCTCCAAGTT | 2 | −6.1 | −27.9 | −4.6 | −965 | Yes |
| −538 | F | TTTTTGCTCTGCACGAA | 3 | |||||||||
| AT1G12040 | Leu-rich repeat extensin (LRX1) | Cell wall | −370 | R | ATCGTGCTATAATGTAG | 3 | −5.7 | −9.2 | −5.7 | −404 | Yesj | |
| −417 | R | GTCGTGCGTGCAGGTT | 2 | |||||||||
| AT1G12560 | Expansin (EXPA7) | Cell wall | −131 | F | ATAGTGATCCGCACGTA | 3 | −5.7 | −2.3 | −1.4 | −181 | Yesk | |
| −302 | F | AACCTGCTCGCACGTC | 2 | |||||||||
| −361 | F | ATACATAAACGCACGTA | 3 | |||||||||
| AT1G62440 | Leu-rich repeat extensin (LRX2) | Cell wall | −241 | F | ATATTGATAAGCACGTC | 3 | −1.3 | −2.1 | −3.2 | Approximately –1,500 | Yesl | |
| AT1G62980 | Expansin (EXPA18) | Cell wall | −160 | F | AACATGAACGCACGTC | 2 | −4.9 | −6.1 | −13.9 | −251 | Yesm | |
| −205 | F | ATAGTGTTTCTCACGTT | 3 | |||||||||
| AT3G62680 | Pro-rich protein (PRP3) | Cell wall | −207 | R | TAAGTGTAATGCACGAT | 3 | −3.5 | −13.0 | −9.2 | −302 | Yesn | |
| AT1G58270 | Meprin and TRAF homology domain-containing protein | Endomembrane | −186 | F | TACCTACGTGTCACGAT | 3 | −2.1 | +1.1 | −1.2 | N.D.o | ||
| −334 | F | GACGTGCGTAACACGAA | 3 | |||||||||
| AT1G64780 | Ammonium transporter | Plasma membrane | −660 | R | ATCGTGCCGTCAAAAA | 2 | −2.1 | −2.0 | +1.1 | N.D. | ||
| AT1G78260 | RNA-binding protein | Nucleus | −1,011 | F | TATATGTATCACACGAA | 3 | −1.9 | −2.6 | −2.3 | N.D. | ||
| AT3G07350 | Unknown | Unclear | −688 | F | TGAATGTATGCCACGAA | 3 | 1.0 | −1.2 | −2.1 | N.D. | ||
| −596 | F | TAATTGATATCACGTT | 2 | |||||||||
| AT5G02330 | Zinc finger protein | Nucleus | −546 | R | AACGTGTGAGATAGTCA | 3 | −2.1 | +2.5 | −3.5 | N.D. | ||
| −370 | R | GACGTGCTATGCACAGA | 3 | |||||||||
| −88 | R | TACGTGTCTTTAATAT | 2 | |||||||||
Predicted by the WoLF PSORT protein subcellular localization prediction program (http://wolfpsort.org/; Horton et al., 2006).
Position of the first nucleotide for forward RHEs and the last nucleotide for reverse RHEs.
F, Forward; R, reverse.
All of the functional RHE sequences (Kim et al., 2006) in addition to the RHE consensus for Patmatch screening.
Linker length of the RHE; two-nucleotide (2) or three-nucleotide (3) spacing.
Fold decrease (−) or fold increase (+) compared with wild-type levels in the microarray analysis.
The promoter length (bp) used for the promoter assay.
Yes indicates root hair cell-specific expression in the root; No indicates no significant GFP signal in the root.
Control (mPro35S:GFP) level expression.
From Baumberger et al. (2001) and Kim et al. (2006).
From Cho and Cosgrove (2002) and Kim et al. (2006).
From Baumberger et al. (2003).
From Cho and Cosgrove (2002) and Kim et al. (2006).
N.D., Not determined.
Isolation of Putative Root-Specific Genes from the RHE-Containing Genes
The Genevestigator Arabidopsis microarray database (www.genevestigator.com; Hruz et al., 2008), which provides information on the tissue/organ specificity of gene expression, was used to screen root-specific genes from the RHE-containing genes. We adopted the root-specific filter to focus more on root genes that contain the RHE motif and also to reduce the gene number in the following experimental steps. Eighty-one root genes from the 904 RHE-containing genes were selected (Supplemental Table S1). These genes are predominantly expressed in root tissues (Supplemental Fig. S1). In this collection, we also included some genes that show additional expression peaks in one or two other tissue types as long as their gene transcript levels are clearly high in root tissues. These genes might be expressible in both root hairs (by RHE) and other cell types (by other cell-type cis-elements). We included these additional genes because they could enrich the RHE-mediated, root hair-expressing genes. Thus, these 81 genes largely represent root-specific and potentially RHE-containing root hair-specific genes at least in the root.
Filtering Putative Morphogenetic H Genes by Microarray Analyses of Root Hair-Specific Mutant and Transgenic Plants
Root hair microarray filters were used to further screen putative morphogenetic H genes from the 81 root-specific, RHE-containing genes. To design root hair microarray filters, we performed microarray analyses with the rhd6 mutant and EXPA7 promoter (ProE7)-driven overexpression transgenic lines of axr2-1 (dominant mutant of AUXIN RESISTANT2; ProE7:axr2-1) and GL2 (for GLABRA2; ProE7:GL2). The rhd6 mutant has impaired root hair initiation and scarcely forms root hairs (Masucci and Schiefelbein, 1994; Fig. 2B). The dominant axr2-1 mutant is defective in root hair elongation and also partly in root hair initiation (Wilson et al., 1990; Masucci and Schiefelbein, 1996; Cho and Cosgrove, 2002), although it also bears many other phenotypes. Root hair cell-specific overexpression of the dominant mutant gene (ProE7:axr2-1) completely abrogated root hair formation, with no other obvious phenotypic effects (Fig. 2B). GL2 suppresses the root hair morphogenetic machinery (Masucci and Schiefelbein, 1996; Ohashi et al., 2003). Accordingly, root hair cell-specific GL2 (ProE7:GL2) overexpression greatly decreased root hair formation and growth (Fig. 2B).
The common feature of these three mutant and transgenic lines is the specific defect in root hair development without other detectable phenotypic effects. We took advantage of this property to screen for putative root hair-specific genes that showed transcript level changes in these lines compared with the wild type. Microarray analyses were performed with transcriptomes from wild-type, rhd6, ProE7:axr2-1, and ProE7:GL2 roots.
Transcriptional changes of the 81 RHE-containing root-specific genes were then identified from the microarray data. Thirty-seven of these genes were down-regulated by more than 2-fold in at least one of the three root hair-defective lines (Table I; Supplemental Table S1). This 37-gene collection included four previously known morphogenetic H genes: LRX1 (AT1G12040; Baumberger et al., 2001, 2003; Kim et al., 2006), EXPA7 (AT1G12560) and EXPA18 (AT1G62980; Cho and Cosgrove, 2002; Kim et al., 2006), and PRP3 (AT3G62680; Bernhardt and Tierney, 2000; Kim et al., 2006). One more previously known H gene, LRX2 (AT1G62440; Baumberger et al., 2001, 2003), has a slightly different RHE structure from the one we used for the Patmatch screening (Table I). The majority of these genes were down-regulated in all three root hair-defective lines (Table I), indicating that the root hair-suppressing mechanism of axr2-1 or GL2 or the loss of RHD6 could be achieved by targeting a common set of morphogenetic H genes.
Twenty-nine (78%) of the putative morphogenetic H genes carried two or more RHEs, where a forward RHE orientation was slightly dominant (58%; 46 of the 80 RHEs). Two- and three-nucleotide linker types occurred with an approximately equal frequency (Table I). Putative cell wall genes were also prominent (32%; 12 genes). Analysis of 51 RHEs from 29 root hair-specific genes (confirmed by promoter assay) showed that most (approximately 86%) RHEs were located within −600 bp of the start codon (Supplemental Fig. S2).
Efficiency of the RHE Filter in Screening Root Hair-Specific Genes
This primary screening with the moderately degenerated RHE filter revealed 904 RHE-containing genes, which we narrowed down to a final 37 putative morphogenetic H genes with the help of additional filtration steps using root-specific and root hair microarray data (Fig. 2C). This process enabled us to exclude RHE-containing genes that were probably also expressible in nonroot tissues due to additional cis-element modules. Given the stringency of the RHE sequence used in primary screening and the possible expression of RHE-containing genes in multiple cell types, the number of functional RHE-containing genes should be much greater than that obtained in our final screening step. This possibility is also implied by two previous studies. The cellulose synthase gene KOJAK (KJK)/CELLULOSE SYNTHASE-LIKE D3 (CSLD3) is expressed in various shoot tissues and root epidermal cells, but at higher levels in the root hair cells (Favery et al., 2001). The promoter of the kinesin-related gene ZWICHEL (ZWI) shows root hair specificity with a promoter fragment 460 bp from the start codon, but longer promoter fragments reveal expression patterns in multiple tissues, including root hair cells (Reddy and Reddy, 2004). Both KJK/CSLD3 and ZWI promoters contain RHE-like motifs in the proximal region (our observation), which could be operative (based on the detailed functional analysis of RHE; Kim et al., 2006) but which do not belong to the RHE consensus used for this screening process.
In order to know the efficiency of the RHE filter (Patmatch screening using RHE) in yielding final root hair-specific genes, we tested different screening orders among those three in silico screening steps: RHE filter, root-specific Genevestigator microarray (RS) filter, and three root hair mutant microarray (3RHmut) filter (Supplemental Fig. S3). As mentioned previously, the RS filter produced 81 genes from the RHE-filtered 904 genes, and a further screening with the 3RHmut filter narrowed the genes to 37 putative root hair-specific genes. A screening of the RHE-filtered 904 genes with the 3RHmut filter gave rise to 60 genes (Supplemental Table S1), which is comparable to the 81 genes given by the RS filter after RHE filtration. To evaluate the effect of the RHE filter, we administered the RHE filter at the final step, after 3RHmut and RS filters. The number of genes that are more than 2-fold down-regulated in any of three root hair-defective lines was 638 (Supplemental Table S1). Filtration of these 638 genes with the RS filter produced 247 genes (Supplemental Table S1), which is much greater than the 81 or 60 genes that were obtained using the RHE filter together with the RS or 3RHmut filter. However, administration of the RHE filter to this 247-gene collection reduced the number to 37 genes, among which 22 (approximately 60%) turned out to be root hair specific in the Promoter:reporter assay (Table I). These analyses suggest that the RHE filter was effective at enriching root hair-specific genes.
In Planta Promoter Assay of RHE-Containing Genes Reveals New Root Hair-Specific Genes in Arabidopsis
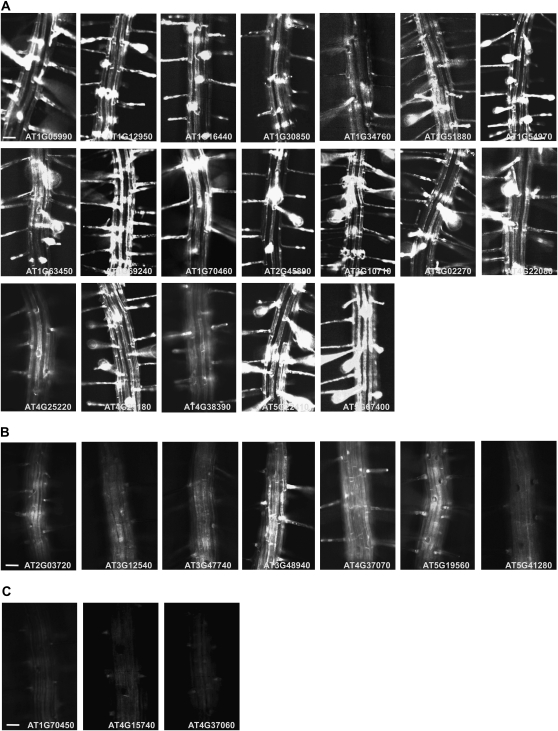
In order to experimentally confirm root hair specificity, we generated 29 Promoter:GFP constructs from those putative morphogenetic H genes, of which five were already known to be root hair-specific genes (Table I). Of the 29 tested putative morphogenetic H gene promoters, 19 Promoter:GFP constructs gave rise to H cell-specific GFP signals in the morphogenetic zone of the transformant Arabidopsis roots (Table I; Fig. 3), seven displayed GFP signals in the whole root epidermis, including the root hair cells, and three showed control (nontransgenic)-level signals. Generally, the whole epidermis-expressing transformants had relatively weaker GFP signals than the H cell-specific ones. The signal of AT4G37070 appeared stronger in the root hair cell files than in the nonhair cell (N cell) files. Thus, RHEs in these putative root hair genes are thought to be genuinely able to drive H cell-specific gene expression, and these 19 genes represent new morphogenetic H genes in Arabidopsis, except for AT1G16440, which recently was reported to be root hair specific by a Promoter:reporter assay (Zhang et al., 2009). Hereafter, the RHE-containing morphogenetic H genes are designated RHS (for root hair specific) genes. These RHE-containing RHS gene promoters began to operate during root hair bulge formation, as described in our previous study (Cho and Cosgrove, 2002).
Figure 3.
Expression patterns of root-specific RHE-containing genes in Arabidopsis root. Representative fluorescence stereomicroscope images of Promoter:GFP transformants in the Columbia background. A, Root hair-specific expression. B, Whole epidermis expression. C, Control-level expression. Bars = 50 μm for all.
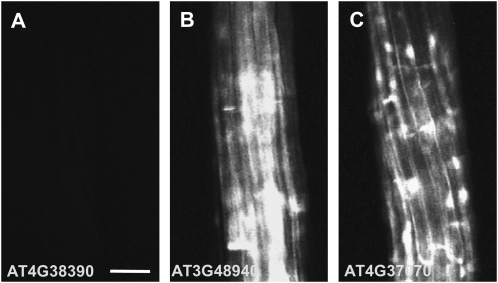
One interesting question is whether the whole epidermis-expressing gene promoters have cis-elements other than RHE for H cell expression or a completely different third element specific for whole epidermal cells. In the former case, there should be two separate cis-elements, one each for H and N cells. We introduced the Promoter:GFP constructs of two whole epidermis-expressing genes (AT3G48940 and AT4G37070) into the root hair-defective rhd6 mutant. In the rhd6 background, the root hair-specific cis-element RHE is not operational and RHS genes are not expressible (Cho and Cosgrove, 2002; Fig. 4A). Therefore, if additional cis-elements of the epidermis-expressing genes operate for H cell expression in the rhd6 root, then Promoter:GFP expression should survive in the N cell, but not the H cell, files. However, if a third cis-element contributes to whole epidermis expression, the expression pattern of the Promoter:GFP in rhd6 should not change. Our experiment showed that Promoter:GFP expression still occurred in the whole epidermis of the rhd6 root (Fig. 4, B and C), suggesting that a cis-element(s) other than RHE is functioning in the promoters of these two whole epidermis-expressing genes.
Figure 4.
Expression patterns of whole epidermis-specific RHE-containing genes in the rhd6 mutant root. Representative fluorescence stereomicroscopy images of Promoter:GFP transformants of root hair-specific (AT4G38390 as control [A]) and whole epidermis-expressing (non root hair specific; AT3G48940 [B] and AT4G37070 [C]) RHE-containing genes in the rhd6 background. Bar = 50 μm for all.
Mutant Phenotypes of RHS Genes
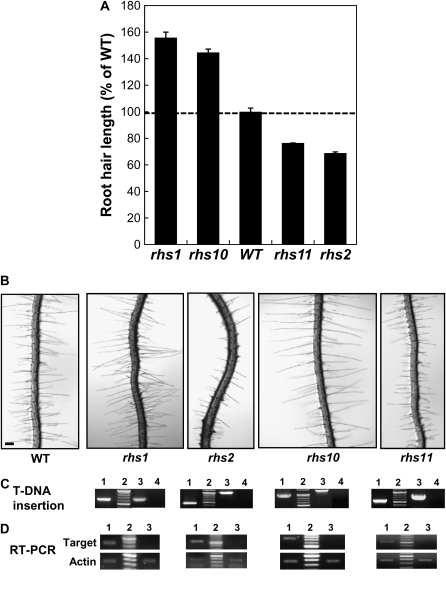
To identify the biological functions of RHS genes, we analyzed the root hair phenotypes of T-DNA insertion mutants. T-DNA insertion of eight RHS gene mutants was confirmed, and their root hair phenotypes were observed: AT1G05990 (SALK_027819; rhs1), AT1G12950 (SALK_126728; rhs2), AT1G30850 (SALK_007215), AT1G51880 (SALK_109605), AT1G54970 (SALK_061662), AT1G70460 (SALK_075892; rhs10), AT2G45890 (SALK_107520; rhs11), and AT5G67400 (SALK_093852). For AT1G70460, we tested SALK_075892, SALK_074904, SALK_079932C, and SALK_075995, but we could obtain the homozygous plants only from SALK_075892. T-DNA insertion into these eight genes showed a single hit to the corresponding gene (http://signal.salk.edu/cgi-bin/tdnaexpress), indicating that the T-DNA insertion most likely occurred once to each gene. However, further analysis with more mutant alleles and gene complementation would be helpful to completely understand the gene functions when those genes are studied in detail. Loss of AT1G05990 (rhs1), AT1G12950 (rhs2), AT1G70460 (rhs10), or AT2G45890 (rhs11) significantly changed root hair growth (for T-DNA insertion maps of these four genes, see Supplemental Fig. S4). Loss of AT1G05990 (a Ca2+-binding protein) and AT1G70460 (a Ser/Thr protein kinase) led to a longer hair phenotype than the wild type, while loss of AT1G12950 (a multidrug and toxin efflux [MATE] transporter) and AT2G45890 (RopGEF4) caused short root hairs (Fig. 5). These mutations did not have obvious phenotypic effects on root hair shape or on other tissues. As described in the next section, the mutant phenotypes are consistent with gene overexpression phenotypes. These results suggest that the RHS genes play practical roles during root hair growth.
Figure 5.
Root hair phenotypes of the loss-of-function T-DNA insertion RHS mutants. A, Comparison of the root hair lengths of wild type (WT) and mutant plants. Mutant lines were SALK_027819 (rhs1; AT1G05990), SALK_126728 (rhs2; AT1G12950), SALK_075892 (rhs10; AT1G70460), and SALK_107520 (rhs11; AT2G45890). The bars show averages ± se of 650 (wild type), 290 (rhs1), 670 (rhs2), 1,666 (rhs10), and 3,840 (rhs11) root hairs. B, Root images of 3-d-old wild-type and mutant lines. Bar = 100 μm for all. C, PCR confirmation of T-DNA insertion in the target gene. Lane 1, PCR by LBb1 and gene-specific primers with mutant genomic DNA; lane 2, molecular size marker; lane 3, PCR by both gene-specific primers with wild-type genomic DNA; lane 4, PCR by both gene-specific primers with mutant genomic DNA. D, Confirmation of null mutations of the mutants by RT-PCR. Lane 1, RT-PCR by gene-specific primers with wild-type root RNA; lane 2, molecular size marker; lane 3, RT-PCR by gene-specific primers with mutant root RNA. Actin1 gene-specific primers were used for a loading control.
Overexpression of RHSs Alters Root Hair Phenotypes
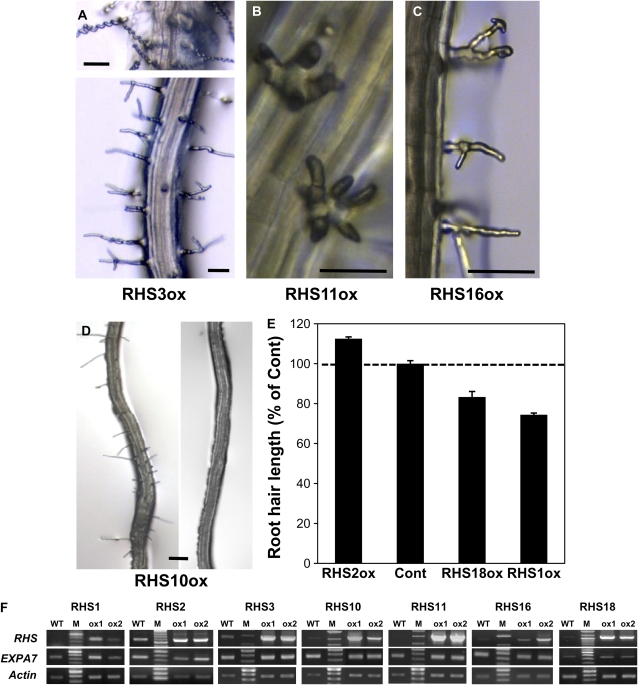
As only half of the loss-of-function RHS mutants, among the eight T-DNA insertion-confirmed genes, exhibited phenotypic changes, and as knockout mutants for many RHSs were not available, we overexpressed 17 RHSs in the root hair cell to further specify their role in root hair development. To obtain high root hair-specific expression of RHSs, we used the EXPA7 promoter (ProE7; −448 from the transcription initiation site), which includes three functional RHEs (Kim et al., 2006). The ProE7-driven expression of RHSs considerably increased their transcript levels compared with the wild type (Fig. 6F), indicating that RHSs were overexpressed in those transformants (RHSoxs). In contrast, EXPA7 levels were not much changed in RHSoxs. Seven of the 17 RHSox lines showed altered root hair phenotypes. While overexpression of RHS3, RHS11, or RHS16 caused abnormal hair morphologies (Fig. 6, A–C), overexpression of RHS1, RHS2, RHS10, or RHS18 affected hair elongation (Fig. 6, D and E). Overexpression of RHS3 (RHS3ox) caused multiple phenotypic effects on root hair morphology, such as spiral, bent, or branched hairs. These different root hair phenoytpes in RHS3ox lines appeared even in the same root. RHS16ox lines showed branched hairs at the side. In contrast, the RHS11ox lines developed multiply branched short hairs at the hair base. The RHS1ox and RHS18ox lines grew 17% and 26%, respectively, shorter root hairs than the control line (ProE7:GFP; Fig. 6E), while RHS10ox greatly or almost completely inhibited root hair elongation (Fig. 6D). RHS2ox grew slightly (13%) longer hairs than the control. The effects of RHS1ox, RHS2ox, and RHS10ox were consistent with their loss-of-function effects, as loss of function and overexpression showed opposite effects on root hair elongation. The RHSox lines did not display noticeable phenotypic changes in tissues or in organs other than root hairs.
Figure 6.
Root hair phenotypes of RHS-overexpressing lines. ProEXPA7:RHSs were introduced into wild-type Arabidopsis plants. A to D, Stereomicroscope images of RHSox lines. Bars = 50 μm (A–C) and 100 μm (D). E, Root hair length of RHSox lines. Bars show averages ± se from 694 (control [Cont]; ProEXPA7:GFP), 470 (RHS18ox), 1,704 (RHS2ox), and 1,750 (RHS1ox) root hairs. F, RT-PCR analysis of RHSox lines. Total RNA was isolated from the wild type (WT) and two ProE7-driven overexpression lines (ox1 and ox2) for each transgene. M, Molecular size marker. Transcript levels for RHS, EXPA7, and Actin were estimated by semiquantitative RT-PCR using gene-specific primers. [See online article for color version of this figure.]
DISCUSSION
Defined Cell Type-Specific cis-Elements Can Serve as Starting Points for Genome-Level Screening of Cell Type-Specific Genes
The cis-regulatory elements are likely conserved among orthologous or paralogous genes and coregulated genes (Haberer et al., 2004; Lisso et al., 2005; Zhang et al., 2005; Kim et al., 2006). Conserved cis-elements enable the in silico screening of putative coexpressed genes. However, because of its short length (5- to 9-bp core) and flexibility, the likely frequency of a cis-element sequence in the whole genome is very high (Carroll et al., 2001). Many such motifs could be syntactically correct without practical regulatory meaning; thus, putative cis-elements need to be confirmed experimentally. In contrast to stimuli-responsive genes, in which putative cis-elements can be relatively easily confirmed by examining the transcriptional activity at the organ or whole plant level, confirmation of cell type specificity requires an elaborate in planta survey of the expression pattern. This has led to a paucity of screening for cell type-specific genes using cis-elements. Here, we demonstrated the successful use of a cell type-specific cis-element to begin screening for coexpressed genes and experimentally confirmed their cell type specificity using the root hair cell as a model.
Previous approaches for identifying root hair morphogenetic genes can be divided into three categories: screening for mutants with root hair phenotypic abnormalities (Schiefelbein and Somerville, 1990; Ringli et al., 2005); analyzing root hair cell transcriptomes (Bucher et al., 1997; Covitz et al., 1998; Jones and Grierson, 2003; Brady et al., 2007); and comparing the transcriptomes of wild-type and root hair mutant plants (Jones et al., 2006). Direct mutant screening reveals the function of each gene during hair morphogenesis and has been vital for deciphering hair morphogenetic processes. In the mean time, the function of these deduced genes may not be root hair cell specific but general to the differentiation process of many cell types. Root hair transcriptome analysis allows us to test the function of many genes expressed in root hair cells, but the transcriptome necessarily includes genes that are generally expressed and function in various cell types. Although it is obvious that many essential morphogenetic genes without cell type specificity should be operative in root hair morphogenesis, we thought that root hair-specific gene products could root hair specifically cue for those general morphogenetic genes to function for hair morphogenesis. Our approach of screening for root hair-specific genes by the RHE and supplemental procedures complement the previous methodologies, provide additional insights into cell morphogenetic mechanisms, and can be applied to other cell types as well.
Putative Functions of Root Hair-Specific Genes in Root Hair Development
In our mutant analysis for the RHSs, four of eight T-DNA insertion mutants showed significant phenotypic changes in root hair growth (Fig. 5). The low frequency of phenotypic changes in the mutants could be due to overlapping gene function between homologs. Similarity in the expressional behavior of the homologs may ensure root hair morphogenesis when a gene is lost. Alternatively, root hair genes not showing obvious morphogenetic alterations upon mutation may be implicated in processes other than hair morphogenetic function, including microbe interactions and nutrient absorption. Here, we will discuss the probable function of RHE-containing root hair-specific genes in root hair development that showed phenotypic effects in loss-of-function or overexpression lines.
The RHSs can be categorized into several major functional groups, such as cell wall dynamics, protein phosphorylation, and membrane transport. Of the 24 confirmed RHSs, 11 were related to the cell wall, five to protein phosphorylation, and two to membrane transport (Table I).
Cell Wall-Related Genes
The cell wall-related RHSs identified here are Hyp-rich glycoproteins (arabinogalactan proteins, extensins, and Pro-rich proteins [PRPs]), pectin-metabolic enzymes, peroxidases, and expansins. RHS7 is a canonical PRP (PRP1), and RHS13 is a PRP-like protein with homology to the C-terminal region of typical PRPs. The pectin metabolism-related RHSs identified here included a putative pectin methyl esterase (RHS12) and a putative pectate lyase (RHS14). LRX1, LRX2, EXPA7, and EXPA18 are previously known root hair-specific extensins and expansins, respectively (Baumberger et al., 2001, 2003; Cho and Cosgrove, 2002; Table I). Although it was demonstrated that defects in LRXs weakened the integrity of root hair cell walls (Baumberger et al., 2001, 2003), in vivo roles of PRPs, pectin metabolic enzymes, peroxidases, and expansins in root hair development have been poorly understood. This study demonstrated the probable role of a peroxidase during root hair elongation. RHS18 and RHS19 are class III peroxidases that are thought to act in the cell wall. Class III peroxidases have peroxidative and hydroxylic activities, thus modulating both cell wall loosening and stiffening (Passardi et al., 2004). Root hair-specific RHS18 overexpression slightly (approximately 20%) decreased root hair elongation compared with controls (Fig. 6). We speculate that RHS18 overexpression might shift the balance of peroxidase-modulated cell wall dynamics toward wall stiffening in the root hair. For example, if cross-linking substrates such as extensins are more prominent in the root hair cell wall, increased peroxidase levels may favor wall stiffening.
Protein Kinases and Related Genes
Among the five identified kinase-related RHSs were three receptor-like kinases (RLKs), an AGC (for cAMP-dependent protein kinase A, cGMP-dependent protein kinase G, and phospholipid-dependent protein kinase C)-type protein kinase, and a guanine nucleotide-exchange factor (RopGEF)/kinase partner protein (KPP).
Two (RHS6 and RHS16) of the three identified RLK-type RHSs belonged to the Leu-rich repeat-RLK family. RHS6 (AT1G51880) is one of the closest eight tandem paralogs in Arabidopsis. RHS6 shows some homology to RHS16 (AT4G29180), sharing similarities in amino acid sequence, size, and domain order of the extracellular, transmembrane, and kinase regions. The homozygous knockout mutant for RHS16 is not yet available. Loss of RHS6 did not show significant phenotypic changes in the root hair under ordinary growth conditions, possibly due to the overlapping function of its four closest paralogs. Alternatively, RHS6 RLK may mediate specific external, rather than internal, cues for root hair development. In contrast, root hair-specific RHS16 overexpression caused dramatic morphological changes, such as branched, shortened, and kinked root hairs (Fig. 6C). Thus, root hair-specific Leu-rich repeat-RLK may be involved in internal root hair developmental processes, or its activity (or expression) may be up-regulated by some external stimuli to modulate root hair morphogenesis.
The RHS10 (AT1G70460) RLK has a Pro-rich N-terminal region and is described as a member of the PERK (for Pro-rich extensin-like receptor kinase) family (Humphrey et al., 2007). As PERKs have extensin-like motifs, the extracellular domain of RLKs might integrate with the cell wall to mediate cell wall-related signals. Loss of RHS10 enhanced root hair elongation, and its overexpression greatly inhibited hair growth, indicating that RHS10 functions as a pacemaker in root hair elongation. This role is in contrast to that of most root hair genes, whose defects largely cause hair tip growth inhibition.
RHS3/AGC1-6 (AT1G16440) is a non-RLK-type protein kinase (Bögre et al., 2003). There are six subfamilies for AGCs in Arabidopsis, of which RHS3 belongs to AGC VIIIa. Several AGC family protein kinases are involved in root hair development, including AGC2-1/OXIDATIVE SIGNAL-INDUCIBLE1 (OXI1; AGC VIIIb subfamily; Rentel et al., 2004) and INCOMPLETE ROOT HAIR ELONGATION (IRE; AGC other subfamily; Oyama et al., 2002). The oxi1 mutants develop shorter root hairs. Ectopic expression of PINOID (AGC VIIIa) in root hair cells inhibits root hair elongation by facilitating auxin efflux, resulting in insufficient cellular auxin levels for root hair elongation (Lee and Cho, 2006). Root hair-specific RHS3 overexpression leads to spiral and branched hair morphologies (Fig. 6A), suggesting that the molecular function of RHS3 might be linked to cytoskeleton-mediated cell polarity. Recently, the two closest RHS3 paralogs (AGC1-5 and AGC1-7) that are pollen specific have also been demonstrated to play critical roles in the polar growth of pollen tubes (Zhang et al., 2009).
The putative molecular function of RHS11 (AT2G45890) is assigned to RopGEF4. RhoGEFs switch GDP-bound inactive Rho to GTP-bound active Rho. At least two plant-specific RopGEFs (1 and 2) can interact with Rop, and RopGEF1 has guanine-exchange activity on Rop (Berken et al., 2005; Gu et al., 2006). Upon overexpression, RopGEF1 can cause depolarization of tobacco (Nicotiana tabacum) pollen tube growth, similar to that caused by Rop hyperactivity, indicating an in vivo role for RopGEFs for activating Rops (Gu et al., 2006). Tip-growing pollen tube and root hair cells require the massive consumption of cell wall materials, delivered by actin-mediated polar vesicle trafficking, at the growing tip. Rop GTPases may modulate tip growth by coordinating actin organization and membrane trafficking (Kost, 2008; Yalovsky et al., 2008). Hyperactivity of Arabidopsis Rop2, Rop4, or Rop6 depolarizes root hair tip growth (Molendijk et al., 2001; Jones et al., 2002). The root hair-specific RopGEF4 (RHS11) in our study might activate these Rop GTPases in the root hair, as GEF overexpression caused conspicuously depolarized root hair phenotypes, such as multiple hair initiation and branched hairs (Fig. 6B). The RopGEF AT5G19560, which is expressed in the whole root epidermis (Fig. 3), may function somewhat redundantly with RHS11 for root hair development. The moderately short root hair phenotype in the loss-of-function RHS11 mutant (Fig. 5) could be due to a partial weakening of the GEF-mediated polarity cue at the hair tip. A recent study with Arabidopsis pollen-specific RopGEF12 showed the physical interaction between RopGEF12 and PRK2a (a pollen RLK) and proposed a probable signaling pathway of RLK→RopGEF→Rop→tip growth (Zhang and McCormick, 2007). A similar pathway between RLKs, RopGEFs, and Rop GTPases, which are expressed in the root hair, is conceivable during root hair tip growth.
Other Genes
The RHS gene collection included two putative membrane transporter genes, RHS2 (AT1G12950) and RHS15 (AT4G25220). The RHS15 gene product appears to be a member of the major facilitator superfamily. RHS2 belongs to the MATE transporter family. Plant MATE transporters have been implicated in the transport of diverse substrates, such as flavonoids, alkaloids, salicylic acid, and citrate (Durrett et al., 2007). However, the function of MATE transporters has not yet been reported in root hairs. Our results suggest that the root hair-specific MATE transporter RHS2 positively mediates root hair elongation, since loss of function decreased (Fig. 5A) and overexpression increased (Fig. 6E) hair length. RHS2 might contribute to tip growth by transporting certain solutes or ions that direct tip polarity and/or sustain tip growth.
Although the role of Ca2+ in the growth of pollen tube and root hair is well studied (Gilroy and Jones, 2000; Grierson and Schiefelbein, 2002; Cole and Fowler, 2006), the function of Ca2+ sensors such as calmodulins (CaMs) and CaM-like proteins (CMLs) during tip growth is not well characterized. RHS1 (AT1G05590) is a root hair-specific CML, previously named CML7 (McCormack et al., 2005). Our results imply that RHS1 might have a unique function in Ca2+-related processes during root hair tip growth. Thus far, only positive roles for Ca2+ during tip growth have been described. However, RHS1 appears to have a negative function; its knockout mutant has longer hairs and its overexpressors grow shorter hairs than the wild type (Figs. 5 and 6). A protein microarray analysis in Arabidopsis showed that diverse protein kinases and transcription factors are the major primary targets of CaMs and CMLs, indicating their involvement in cellular signaling and transcriptional regulation (Popescu et al., 2007). Further study on the downstream targets of RHS1 should reveal how it negatively transduces a Ca2+ signal to inhibit hair tip growth.
The “RHF Regulon” for Root Hair Morphogenesis
The RHSs identified in this study represent a transcriptional regulon that includes the common regulatory cis-element and thus could be modulated by the same transcription factor. We previously proposed putative transcription factors (RHFs, for root hair factors) that recognize the RHE consensus (Kim et al., 2006). The RHE sequence is partially palindromic with a short central linker region (Kim et al., 2006). Because the linker length (2 or 3 bp) is critical for RHE function, and since its left part is more flexible than the right (Kim et al., 2006), the RHE-recognizing RHF might consist of several structurally related transcription factors belonging to the same family.
One intriguing question is how upstream factors such as GL2, RHD6, and RHD2 modulate RHF to transcribe RHSs specifically in the root hair. The homeodomain-containing transcription factor GL2, a downstream component of hair/nonhair fate determination, negatively regulates RHD6 (a basic helix-loop-helix transcription factor; Menand et al., 2007) expression and hair morphogenesis. RHD2 seems to be down-regulated in the rhd6 mutant background compared with that of the wild type (our observation), indicating that RHD2 is a downstream target of RHD6. Defects in RHD2 decrease the transcription of hair morphogenetic MRH and RHS genes (Jones et al., 2006; our observation), suggesting that RHD2-mediated signaling regulates RHF in the H cell of the maturation zone.
Another interesting question is how RHD2, an NADPH oxidase that produces reactive oxygen species (Foreman et al., 2003), regulates RHF activity. RHD2-producing reactive oxygen species activate OXI1/AGC2-1 (a Ser/Thr kinase), and in turn OXI1 activates mitogen-activated protein kinases in Arabidopsis (Rentel et al., 2004). This signaling pathway may target the RHF protein itself or close upstream regulators for RHF expression. Consistent with their probably positive effect on the hair morphogenetic RHF regulon, loss of RHD2 or OXI1 causes defects in root hair elongation (Schiefelbein and Somerville, 1990; Rentel et al., 2004). Interestingly, the promoters of RHD2 and OXI1 include RHE-like motifs (our observation), and the genes are preferentially transcribed in the H cell of the hair morphogenetic zone (Rentel et al., 2004; Takeda et al., 2008), suggesting that RHF may regulate the transcription of RHD2 and OXI1. This would lead to a positive feedback loop between RHF and the RHD2-OXI1 pathway, boosting the levels of RHSs to ensure root hair morphogenesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana Columbia ecotype) was used for transformation of Promoter:GFP constructs and for observations of root hair-specific GFP expression. T-DNA insertion mutant seeds were obtained from the Arabidopsis Biological Resource Center. The root hair-defective rhd6 mutant is as described by Masucci and Schiefelbein (1994). Arabidopsis seeds were cold treated before germination at 23°C under a 16-h-light/8-h-dark photoperiod. Arabidopsis plants were transformed using Agrobacterium tumefaciens strain C58C1 (pMP90; Bechtold and Pelletier, 1998), and transformants were selected on hygromycin-containing plates (10 μg mL−1).
Construction of Promoter:GFP and Other Transgenes
To specifically express the root hair-suppressing genes in root hair cells, the EXPA7 promoter (ProE7; Cho and Cosgrove, 2002) was used. The binary vector pGPTV-HYG (Becker et al., 1992) was the cloning vector. ProE7:GFP was as described by Kim et al. (2006). For the ProE7:axr2-1-GFP translational fusion construct, a genomic fragment of axr2-1 was obtained by PCR using primers 5′-CCCCTTCTAGACCTTTCTTCTTCCCCTCT-3′ (containing an XbaI site) and 5′-TTTATCCCGGGTAACAGATCTGTTCTTGCAGTACTTCTCCATTG-3′ (containing an XmaI site) and the genomic DNA of the axr2-1 mutant as a template. The PCR product was cloned into XbaI-XmaI sites upstream of the GFP gene. For the ProE7:GL2-GFP construct, a genomic fragment of GL2 was obtained by PCR using primers 5′-GCTAGCCCGGGACAGGATTTGTAT-3′ (containing an XmaI site) and 5′-TGTGACCCGGGTAACGCAATCTTCGATTTGTAGACTTC-3′ (containing an XmaI site) and the genomic DNA of wild-type Arabidopsis as a template. The PCR product was cloned into the XmaI site upstream of the GFP gene.
To construct the Promoter:GFP reporter systems, putative root hair-specific gene promoters were obtained from Arabidopsis (Columbia) genomic DNA by PCR using the two primers shown as a double set in the primer list (Supplemental Table S2). The PCR-amplified promoter regions were inserted into the pGPTV-HYG vector, where the uidA gene was replaced by GFP (Cho and Cosgrove, 2002; Kim et al., 2006).
Overexpression of RHSs was conducted by constructing ProE7(−448):RHSs, where the EXPA7 promoter retains its full strength (Cho and Cosgrove, 2002). ProE7 was inserted into the HindIII/SalI sites of the pCAMBIA1300 vector and followed by genomic RHS coding regions that were amplified by the primer sets listed in Supplemental Table S3.
All constructs were confirmed by nucleotide sequencing and introduced into Arabidopsis plants using Agrobacterium. Transgene insertion in the Arabidopsis transformants was confirmed by PCR analysis using the transgene-specific primers and by nucleotide sequencing if necessary.
In Silico Analyses
Arabidopsis genes containing the RHE consensus were searched using the Patmatch program (www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). RHE-containing genes with root specificity were screened using the Genevestigator Gene Atlas microarray database (www.genevestigator.com; Hruz et al., 2008). We chose genes whose transcript levels were prominently high in the root tissues (Supplemental Fig. S1).
Microarray Gene Chip Analysis for Root Hair-Defective Mutant and Transgenic Lines
Total RNA was isolated from the 3-d-old seedling roots of rhd6, ProE7:axr2-1-GFP, ProE7:GL2-GFP, and wild-type (Columbia for the transformants and Wassilewskija for rhd6) plants using the RNeasy Plant Mini kit (Qiagen). The purity and integrity of the total RNA were checked using NanoDrop (Thermo Scientific) and Experion (Bio-Rad), respectively. Five micrograms of total RNA was used for labeling. Probe synthesis from total RNA samples, hybridization, detection, and scanning were performed according to standard Affymetrix GeneChip (Affymetrix) protocols (Lockhart et al., 1996). Labeled samples were hybridized to the Arabidopsis ATH1 Genome Array GeneChip, and expression profiles were analyzed using GeneChip Operating Software programs (Affymetrix).
The one-sided Wilcoxon's signed rank test was employed to generate detection P values. The probe set was regarded as present (P) for P < 0.04 and absent (A) for P > 0.06. The detection threshold was set as the “present” call output from the GeneChip Operating Software (P < 0.05). Gene expression was regarded as increased (I) for a change P < 0.0025 and decreased (D) for a change P > 0.9975. Genes showing 2-fold or greater transcript level changes in at least one root hair-defective line, compared with the wild type, were considered to be putative root hair-specific genes and were further tested for promoter activity.
Observation of Reporter Gene Expression
For promoter activity, GFP fluorescence from the seedling root was observed in nine to 63 (average = 22.8) independent T1 or T2 transgenic lines. The cauliflower mosaic virus 35S minimal promoter:GFP (mPro-35S:GFP) rtansformant (Kim et al., 2006) was used as a control for the root basal fluorescence level. GFP fluorescence was observed using an epifluorescence stereomicroscope (MZ FLIII; Leica). GFP signals were detected using a 488-nm excitation filter and a 543-nm emission filter.
Observation of Root Hair Phenotypes
Root hair length was measured as described by Lee and Cho (2006) with modifications. The root was digitally photographed with a stereomicroscope (MZ FLIII; Leica) at 40× to 50× magnifications. The hair length from 10 consecutive hairs protruding perpendicularly from each side of the root, for a total of 20 hairs from both sides of the root, was calculated using LAS software version 2.8.1 (Leica). A total of 290 to 3,840 root hairs were used to estimate the root hair length of a genotype. Root hair phenotypes of RHS-overexpressing transformants were observed in eight to 40 (average = 19.5) independent T2 or T3 transgenic lines.
Genotyping and Reverse Transcription-PCR Analysis of T-DNA Insertion Mutants and RHSox Transformants
Homozygous T-DNA insertional mutant lines were selected by PCR analysis of the genomic DNA using gene-specific and LBb1 primers (for T-DNA insertion maps of four RHS mutant lines showing root hair phenotypes, see Supplemental Fig. S4). Null mutations were confirmed by reverse transcription (RT)-PCR with gene-specific primers (Supplemental Table S4) with Actin1 (AT2G37620) as a loading control. Overexpression of RHSs (RHSox) by ProE7 (−448) was confirmed by RT-PCR using the gene-specific primers (Supplemental Table S5). For cDNA synthesis, total RNA was extracted from the roots, and RT was performed using Moloney murine leukemia virus reverse transcriptase (Nexgen Biotech) with oligo(dT)20 and RNA. The PCR cycle numbers were 32 for the genotyping PCR and 25 to 27 for RT-PCR.
If not described in the text or tables, Arabidopsis Genome Initiative and National Center for Biotechnology Information locus identifiers for the genes and proteins mentioned in this article are AT3G23050 (AXR2/IAA7), AT1G79840 (GL2), AT5G65930 (ZWI), AT1G11130 (SCM), AT5G62310 (IRE), AT2G34650 (PINOID), AT4G38430 (RopGEF1), and AT1G01700 (RopGEF2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Tissue-specific expression profile of the genes that were screened through the RHE-Patmatch and root-specific microarray filters.
Supplemental Figure S2. Distribution of RHE in the promoters (upstream 1,000 bp from ATG) of Arabidopsis root hair-specific genes.
Supplemental Figure S3. The summary of screening processes for putative root hair-specific genes using three different filters in different orders.
Supplemental Figure S4. T-DNA insertion maps of four RHS genes whose loss-of-function mutations showed phenotypic effects on root hair growth.
Supplemental Table S1. Gene lists for each screening step.
Supplemental Table S2. Primers for constructing Promoter:GFP reporters.
Supplemental Table S3. Primers for constructing ProEXPA7:RHSs.
Supplemental Table S4. Primers for genotyping and RT-PCR analysis of T-DNA insertion mutants.
Supplemental Table S5. Primers for RT-PCR analysis of RHSox transformants.
Supplementary Material
Acknowledgments
We thank Zee-Won Lee at the Korea Basic Science Institute for help with microscopy imaging analyses and Dong-Wook Kim, Young-Im Ha, Ji-Yeun Cho, You-Kang Kim, Hye-Won Kim, and Jung-Mun Choi in our lab for help with transgene construction and transformant analyses.
This work was supported by the Crop Functional Genomics Center of the 21st Century Frontier Research Program (grant no. CG2151), the BioGreen 21 Program (grant no. 20070401034022) of the Rural Development Administration, Korea Science and Engineering Foundation (grant no. R01–2007–000–10041–0), and the Korea Science and Engineering Foundation Environmental Biotechnology Research Center (grant no. R15–2003–012–02003–0).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hyung-Taeg Cho (htcho@snu.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Steiner M, Ryser U, Keller B, Ringli C (2003) Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J 35 71–81 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In JM Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Humana, Totowa, NJ, pp 259–266 [DOI] [PubMed]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20 1195–1197 [DOI] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436 1176–1180 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Tierney ML (2000) Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol 122 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Ökrész L, Henriques R, Anthony RG (2003) Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8 424–431 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Bucher M, Schroeer B, Willmitzer L, Riesmeier JW (1997) Two genes encoding extensin-like proteins are predominantly expressed in tomato root-hair cells. Plant Mol Biol 35 497–508 [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD (2001) From DNA to Diversity. Blackwell Science, Malden, MA
- Cho H-T (2007) A cis-element for root hair specificity has been co-opted repeatedly through the divergence of upstream fate-determining machineries. Plant Signal Behav 2 117–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Fowler JE (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9 1–10 [DOI] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR (1998) Expressed sequence tags from a root-hair-enriched Medicago trunculata cDNA library. Plant Physiol 117 1325–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones DL (2000) Through form to function: root hair development and nutrient uptake. Trends Plant Sci 5 56–60 [DOI] [PubMed] [Google Scholar]
- Grierson C, Schiefelbein J (2002) Root hairs. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/www.aspb.org/publications/arabidopsis/
- Grierson CS, Parker JS, Kemp AC (2001) Arabidopsis genes with roles in root hair development. J Plant Nutr Soil Sci 164 131–140 [Google Scholar]
- Gu Y, Li S, Lord EM, Yan Z (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Nakai K (2006) Protein subcellular localization prediction with WoLF PSORT. In Proceedings of the 4th Annual Asia Pacific Bioinformatics Conference APBC06, Taipei, Taiwan, pp 39–48
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008: 420747 (doi/10.1155/2008/420747) [DOI] [PMC free article] [PubMed]
- Humphrey TV, Bonetta DT, Goring DR (2007) Sentinels at the wall: cell wall receptors and sensors. New Phytol 176 7–21 [DOI] [PubMed] [Google Scholar]
- Jones MA, Grierson CS (2003) A simple method for obtaining cell-specific cDNA from small numbers of growing root-hair cells in Arabidopsis thaliana. J Exp Bot 54 1373–1378 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45 83–100 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Hai YF, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B (2008) Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol 18 119–127 [DOI] [PubMed] [Google Scholar]
- Latchman DS (2004) Eukaryotic Transcription Factors. Elsevier, London
- Lee SH, Cho HT (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J, Steinhauser D, Altmann T, Kopka J, Mussig C (2005) Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res 33 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14 1675–1680 [DOI] [PubMed] [Google Scholar]
- Masucci J, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin and ethylene-associated process. Plant Physiol 106 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Tsai YC, Braam J (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 10 383–389 [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316 1477–1480 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CSV, Friml J, Braun M, Gilroy S, Palme K (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T (2003) Signaling by GLABRA2 in root-hair pattern formation. Science 300 1427–1430 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (2002) The IRE gene encodes a protein kinase homologue and modulates root hair growth in Arabidopsis. Plant J 30 289–299 [DOI] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 11 534–540 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Reddy AS (2004) Developmental and cell-specific expression of ZWICHEL is regulated by the intron and exon sequences of its gene. Plant Mol Biol 54 273–293 [DOI] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, et al (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 858–861 [DOI] [PubMed] [Google Scholar]
- Ringli C, Baumberger N, Keller B (2005) The Arabidopsis root hair mutants der2-der9 are affected at different stages of root hair development. Plant Cell Physiol 46 1046–1053 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J, Lee MM (2006) A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis. Physiol Plant 126 503–510 [Google Scholar]
- Schiefelbein J, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L (2008) Local positive feedback regulation determines cell shape in root hair cell. Science 319 1241–1244 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222 377–383 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho TH, You Y, Yu T, Quatrano RS (2005) Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21 3074–3081 [DOI] [PubMed] [Google Scholar]
- Zhang Y, He J, McCormick S (2009) Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes. Plant J 58 474–484 [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCormick S (2007) A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA 104 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.