Abstract
Regulation of gene expression through translational control is common in many organisms. The Arabidopsis (Arabidopsis thaliana) transcription factor bZIP11 is translational repressed in response to sucrose (Suc), resulting in Suc-regulated changes in amino acid metabolism. The 5′ leader of the bZIP11 mRNA harbors several upstream open reading frames (uORFs), of which the second uORF is well conserved among bZIP11 homologous genes. The uORF2 element encodes a Suc control peptide (SC-peptide) of 28 residues that is sufficient for imposing Suc-induced repression of translation (SIRT) on a heterologous mRNA. Detailed analysis of the SC-peptide suggests that it functions as an attenuator peptide. Results suggest that the SC-peptide inhibits bZIP11 translation in response to high Suc levels by stalling the ribosome on the mRNA. The conserved noncanonical AUG contexts of bZIP11 uORFs allow inefficient translational initiation of the uORF, resulting in translation initiation of the scanning ribosome at the AUG codon of the bZIP11 main ORF. The results presented show that Suc-dependent signaling mediates differential translation of mRNAs containing SC-peptides encoding uORFs.
Developmental and physiological processes in organisms depend on regulation of gene expression. Regulation can occur at several levels, from mRNA synthesis to the control of protein activity. The regulation of transcription has been most intensively studied and involves a plethora of transcription factors that operate through several regulatory mechanisms. However, mRNA levels often are not predictive of the levels or activities of the encoded proteins (Conrads et al., 2005; Gibon et al., 2006; Bianchini et al., 2008) and other regulatory mechanisms are of crucial importance as well.
Upstream open reading frames (uORFs) are translational reading frames present in the 5′ leaders of mRNAs. The presence of uORFs usually inhibits translation of the downstream major ORF (main ORF) as eukaryotic ribosomes generally only initiate translation once per mRNA (Luo and Sachs, 1996; Ruan et al., 1996; Morris and Geballe, 2000; Mize and Morris, 2001; Gopfert et al., 2003). Over 3,000 Arabidopsis (Arabidopsis thaliana) mRNAs contain uORFs within their 5′ leader sequences, often referred to as 5′-untranslated regions (5′-UTRs; Hayden and Jorgensen, 2007). Similar uORF frequencies are found in the genomes of other eukaryotes, such as yeast (Saccharomyces cerevisiae), mammals, and Drosophila melanogaster (Morris and Geballe, 2000; Hayden and Bosco, 2008). uORFs are widespread, but their impact on translation has been studied in detail in relatively few cases. From the examples described, several different mechanisms have been uncovered. The uORF of the Arabidopsis SAC51 gene (encoding a basic helix-loop-helix-type transcription factor) was shown to inhibit translation of the main ORF (Imai et al., 2006). Recently, this translational inhibition was shown to depend on intrinsic proteins of the ribosome (Imai et al., 2008). A uORF in the AtNMT1 mRNA is feedback-inhibiting translation in response to choline (Tabuchi et al., 2006). In mammals, a highly conserved uORF-encoded hexapeptide controls S-adenosyl-Met decarboxylase translation (Mize and Morris, 2001; Hanfrey et al., 2005). In Xenopus laevis, the Connexin41 mRNA is tightly controlled by the three uORFs through a mechanism that depends on the presence of rare codons within the uORF and results in stalled ribosomes (Meijer and Thomas, 2003). The yeast transcription factor GCN4 is translationally regulated through four uORFs within the GCN4 mRNA and the starvation-dependent phosphorylation status of eIF2 (Hinnebusch, 2005). The Arabidopsis bZIP11 gene is translationally controlled by Suc (Rook et al., 1998). Suc-induced repression of translation (SIRT) is dependent on the second uORF in the bZIP11 5′ leader (uORF2). This uORF encodes a conserved Suc control peptide (SC-peptide) present in four other Arabidopsis bZip genes (bZIP1, bZIP2, bZIP44, and bZIP53) and in many other bZip genes from other species (Wiese et al., 2004, 2005). The regulatory uORF motif is present in bZip-encoding genes from plants, but is absent from the genomes of other organisms. The molecular details of this plant-specific signaling mechanism are presently unknown.
Sugars are potent signaling molecules in plants. Suc triggers signaling pathways in all plant tissues and thereby alters gene expression (Wiese et al., 2004). Changed Suc levels within the plant affect photosynthesis, metabolism, and developmental processes. High Suc levels inhibit photosynthesis (Koch, 1996) and increase storage through induction of starch synthesis (Hendriks et al., 2003). During starvation, low levels of Suc result in increased photosynthesis and starch mobilization. Suc levels affect secondary metabolism (Teng et al., 2005; Solfanelli et al., 2006), as well as developmental processes, such as flowering and root development (Ohto et al., 2001; Takahashi et al., 2003). How plants perceive Suc is currently unknown, but Suc sensing via Suc transporters has been suggested (Chiou and Bush, 1998; Sivitz et al., 2008). Several genes seem specifically activated by Suc, including NR1, encoding nitrate reductase (Cheng et al., 1992), patatin (Wenzler et al., 1989; Jefferson et al., 1990), the phloem cell-specific rolC gene (Yokoyama et al., 1994), a UDP-Glc pyrophosphorylase gene (Ciereszko et al., 2001), and MYB75 (Teng et al., 2005). Among the genes repressed by Suc treatment are PC (for plastocyanine; Dijkwel et al., 1997) and ASN1 (Lam et al., 1998). The Suc-controlled bZIP11 transcription factor was shown to regulate genes involved in amino acid metabolism (Hanson et al., 2008), but probably has a much wider role in reprogramming metabolism (J. Hanson, M. Hanssen, and S. Smeekens, unpublished data). The transcription activation potential of bZIP11 and related proteins is enhanced by the Snf1-related kinases (SnRKs) KIN10 and KIN11 that regulate responses to stress resulting from nutrient deprivation (Baena-Gonzalez et al., 2007). Here, it is shown that the SC-peptide is required and sufficient for SIRT, and a model is proposed where Suc in combination with the SC-peptide leads to ribosome stalling and translational inhibition of the main ORF.
RESULTS
Suc-Dependent Translational Regulation of bZIP11
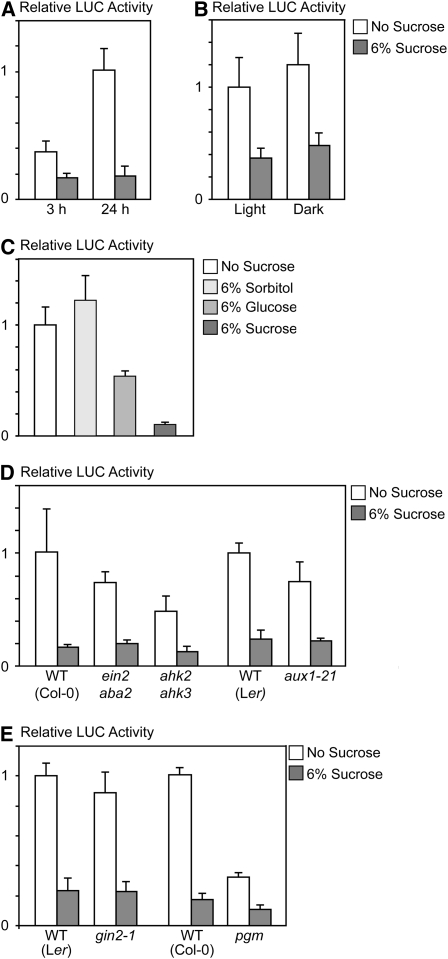
SIRT acts within 24 h of Suc addition (Wiese et al., 2004). The stability of the GUS reporter enzyme precluded detection of more rapid changes in response to Suc. A luciferase (LUC)-based transient expression system was developed based on biolistic gene transfer to investigate the kinetics of SIRT in young seedlings in more detail. Seedlings were transformed with a DNA construct expressing the 5′ leader of bZIP11 followed by the firefly LUC gene directed by the 35S promoter (35S:bZIP11 5′ leader:LUC) and relative LUC activities were determined. Suc-treated seedlings show significantly reduced LUC activities within 3 h of incubation in Suc-containing medium (Fig. 1A). After 24 h of incubation, the LUC activities of seedlings incubated in medium lacking Suc increase 2- to 3-fold, but the levels of the Suc-treated seedlings remain constant (Fig. 1A). Incubation for longer times than 24 h does not result in higher LUC levels (data not shown). Light, as well as sugars, affects the development and physiology of plants. It has been documented that light effects can be mimicked by sugar treatments (Cheng et al., 1992). However, light or darkness has no effect on SIRT (Fig. 1B).
Figure 1.
Suc-induced repression of translation is independent of hormone, light, and Glc signaling. Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct. Following transformation, seedlings were incubated in medium containing either no Suc (white bars) or Suc (gray bars). Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. A, Relative LUC activity levels of transformed seedlings (accession Col-0) incubated for 3 or 24 h. B, Relative LUC activity levels of transformed seedlings (accession Col-0) incubated for 24 h in either constant light or darkness. C, Relative LUC activity levels of transformed seedlings (accession Col-0) incubated for 24 h in medium supplemented with different sugars. Note that the molar concentration of Glc and sorbitol is close to double that of Suc. D, Relative LUC activity levels of wild-type seedlings compared with that of mutants with altered hormone biosynthesis or signal transduction. Relative LUC levels were compared to that of the corresponding wild-type accessions of the mutants used (ein2aba2 and ahk2ahk3 double mutants, Col-0; and aux1-21, Ler). E, Relative LUC activity levels of wild-type seedlings and mutants with altered sugar signal transduction or metabolism. LUC activity levels were compared with that of the corresponding accession of the mutants (gin2/hxk1, Ler; and pgm, Col-0). WT, Wild type.
Glc, at twice the molar ratio of Suc, triggers translational repression as well, but to a much lower extent compared to Suc (Fig. 1C). On the other hand, sorbitol treatment is not affecting the SIRT response. SIRT is therefore concluded to be independent of osmotic responses. Several documented examples indicate that hormone signaling and sugar signaling are tightly interconnected in plants (Smeekens, 2000; Rolland et al., 2006). Mutants in hormone signaling were tested to investigate whether hormone signaling is involved in SIRT. Mutants showing defects in ethylene and abscisic acid (ABA) signaling (ein2aba2), cytokinin signaling (ahk2ahk3), and auxin signaling (aux1-21) all show normal Suc-mediated repression of LUC activity (Fig. 1D). Hormone application to wild-type plants (auxin, GA, cytokinin, ABA, 1-aminocyclopropane-1-carboxylic acid) confirmed that SIRT does not depend on hormone signaling (data not shown).
Suc is readily hydrolyzed to Glc and Fru in planta. The major Glc receptor in plants is HEXOKINASE1 (HXK1; Smeekens, 2000; Rolland and Sheen, 2005; Rolland et al., 2006). Whether SIRT depends on Glc signaling through HXK1 was tested. No difference in relative LUC activities is detected between the wild-type and gin2-1 (HXK1-null mutant) seedlings (Fig. 1E), indicating that HXK1 does not affect SIRT. Suc treatments promote starch accumulation in plants. The pgm mutant is deficient in starch synthesis and accumulates excess Suc during the day (Periappuram et al., 2000). Relatively low LUC activities are observed in untreated pgm seedlings, possibly due to elevated Suc levels in the pgm mutant. However, Suc-treated pgm seedlings exhibit SIRT (Fig. 1E) and changed starch levels therefore seem not to be involved in SIRT.
Sequences Downstream of uORF2 Are Dispensable for Suc Repression
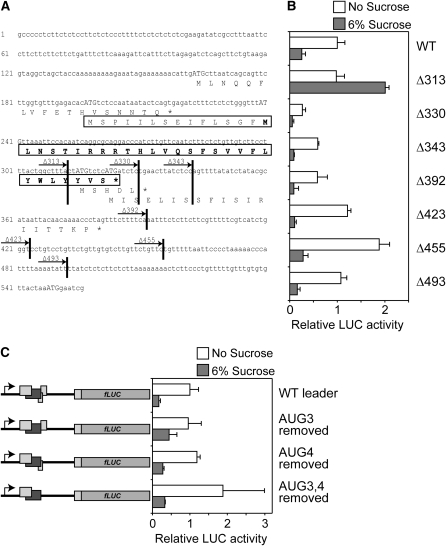
The length of the bZIP11 5′ leader is 547 nucleotides. The sequence includes four uORFs followed by an intercistronic region of 169 nucleotides before the start codon of the main bZIP11 ORF (Fig. 2A). SIRT depends on a AUG start codon within uORF2 (Wiese et al., 2004). To determine whether other regions of the bZIP11 5′ leader were important for repression, a series of deletions of the bZIP11 5′ leader were tested for repression activity. Suc-dependent repression of LUC levels is detected in seedlings transformed with all constructs containing 330 nucleotides of the leader or more (Δ330, Δ343, Δ392, Δ423, Δ455, and Δ493). In contrast, no repression of LUC levels is detected in seedlings transformed with the shorter construct (Δ313). This indicates that a critical region is located between the Δ313 and Δ330 deletions. This region includes the end of uORF2 and the start codons of uORF3 and uORF4. The possible involvement of uORF3 and uORF4 in SIRT was tested by mutations in which the start codons of these two uORFs were removed, while leaving the amino acid sequence of the uORF2 peptide unaltered. These modified leaders impose SIRT (Fig. 2C), demonstrating that the start codons of uORF3 and uORF4 are dispensable for SIRT. The results further show the importance of the peptide sequence encoded by uORF2, as the mRNA nucleotide sequence in the critical region could be mutated without affecting SIRT. The importance of the peptide sequence is also indicated by the limited conservation of the third codon position compared to the first two positions of the codons of homologous uORFs (Supplemental Fig. S1).
Figure 2.
Suc-induced repression of translation is uORF2 dependent. Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or mutant versions thereof. Following transformation, seedlings were incubated in medium containing either no Suc (white bars) or 6% Suc (gray bars) for 24 h. Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. A, Nucleotide sequence of the bZIP11 5′ leader. uORFs and the translated peptide sequences are shown below the nucleotide sequence in capital letters. The start codons (ATG) of the respective uORFs are indicated in capital letters. uORF2 is encoding a 42-amino-acid-long peptide (boxed) and the minimal peptide sufficient for Suc-induced repression is indicated in bold. The ATG at position 548 represents the start codon of the bZIP11 main ORF. The vertical lines illustrate the position of deletions in the 5′ leader. The deletion constructs harbor the 5′ leader from start of transcription to the respective vertical line fused to the LUC gene. Names of the deletion constructs are indicated above the arrows. B, Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or indicated deletions. C, Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or constructs lacking start codons of uORFs. Rectangles in schematic drawings (left) represent ORFs. Dark gray rectangles represent uORF2 of the bZIP11 5′ leader. WT, Wild type.
uORF2 Confers SIRT on an Independent mRNA
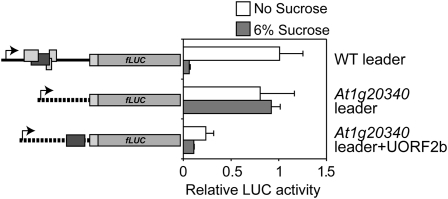
As previously shown, only the second part of uORF2 starting with the second AUG codon is needed for SIRT (uORF2b; Wiese et al., 2004). This 82-nucleotide-long uORF2 element was transplanted to the unrelated leader of the At1g20340 (encoding a plastocyanin protein) gene of Arabidopsis, which lacks uORFs. The element encodes the carboxy-terminal 28 amino acids encoded by uORF2. The At1g20340 leader does not affect the translation of the downstream LUC reporter in response to Suc treatments (Fig. 3). In contrast, Suc-induced repression of LUC activity was detected in the At1g20340:uORF2 seedlings, indicating that the bZIP11 element used is sufficient to impose SIRT on the 5′ leader of an unrelated mRNA. Non-Suc-treated seedlings transformed with the At1g20340:uORF2 construct show approximately 3-fold less LUC activity compared to seedlings transformed with the At1g20340 construct. This demonstrates the general inhibitory effect of uORF2 on translation of the main ORF.
Figure 3.
The SC-peptide is sufficient for Suc-induced translational repression. Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:5′ leader:LUC constructs. After transformation, seedlings were incubated for 24 h in medium containing either no Suc (white bars) or 6% Suc (gray bars). Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. Schematic drawings of constructs used for transient expression experiments are indicated to the left. Rectangles represent ORFs. Dark gray rectangles represent uORF2 of the bZIP11 5′ leader. The 5′ leader of the At1g20340 gene is indicated using a dashed line. WT, Wild type.
Suc Repression Depends on Conserved Amino Acids of the SC-Peptide
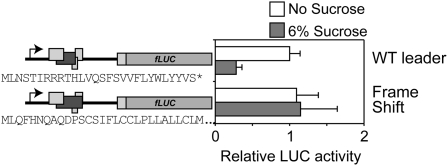
The amino acid sequence of the peptide encoded by uORF2b is highly conserved, especially in the carboxy-terminal part of the peptide (Supplemental Fig. S1; Wiese et al., 2005; Hayden and Jorgensen, 2007). This might be a consequence of the conserved nucleotide sequence of the mRNA or reflect an evolutionarily important selection pressure on the peptide sequence. To address the question of whether the mRNA sequence or the peptide sequence is important for SIRT, an extra nucleotide at position 245 in the bZIP11 5′ leader was introduced. The resulting frame shift changes the peptide sequence, whereas the nucleotide sequence of the uORF2 region shown to be sufficient for SIRT remains unaltered. The frame shift mutation totally abolished SIRT as shown by relative LUC activity levels upon transient expression of the modified 5′ leader (Fig. 4). Thus, the peptide encoded by uORF2 mediates SIRT, as previously proposed by Wiese et al. (2004).
Figure 4.
The mRNA sequence of the bZIP11 5′ leader is not sufficient to trigger Suc-induced repression of translation. Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or a mutated version. Following transformation, seedlings were incubated in medium containing either no Suc (white bars) or 6% Suc (gray bars). Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. Schematic drawings of constructs used for transient expression experiments are indicated on the left. Rectangles represent ORFs. Dark gray rectangles represent uORF2 of the bZIP11 5′ leader. The frame shift mutated 5′ leader harbors a single base pair insertion giving rise to a frame shift and therefore a total change of the sequence of the Suc control peptide, as indicated. The nucleotide insertion is situated in the 5′ part of uORF2, which is not required for SIRT (Fig. 3). WT, Wild type.
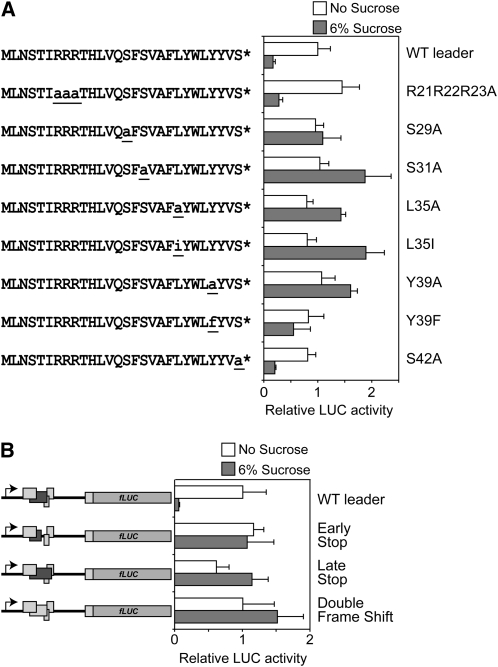
To test the relative importance of well-conserved amino acids in uORF2, a series of mutations were introduced in uORF2, all resulting in amino acid substitutions of conserved amino acids in the peptide (Fig. 5). Ser-29, Ser-31, Leu-35, Tyr-39, Ser-42, Arg-21, Arg-22, and Arg-23 were substituted to Ala residues. Leu-35 and Tyr-39 were also changed to their structurally similar amino acids Ile and Phe, respectively. The mutated 5′ leader sequences were tested for SIRT. Mutating the Arg-rich stretch or the Ser-42 residue did not affect SIRT because the resulting normalized expression values do not differ from the values of the wild-type 5′ leader (Fig. 5A). However, mutations affecting either Ser-29 or Ser-31, Leu-35, or Tyr-39 result in loss of SIRT (Fig. 5A). The LUC activity levels of the non-Suc-treated seedlings were not affected significantly by the point mutations. Interestingly, the substitution of Tyr-39 to Ala (Y39A) abolishes Suc repression, whereas replacing it with the structurally related amino acid Phe (Y39F) resulted in a reduced repression (Fig. 5A).
Figure 5.
Conserved amino acids and the stop codon position of the Suc control peptide are necessary for Suc-induced repression of translation. Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or versions with mutated 5′ leaders. Following transformation, seedlings were incubated for 24 h in medium containing either no Suc (white bars) or 6% Suc (gray bars). Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. A, The amino acid sequence of the 28 C-terminal amino acids of the SC-peptide or mutated versions used for transformation experiments are indicated on the left. Changed amino acids are indicated in underlined lowercase letters. B, Schematic drawings of constructs used for transient expression experiments are indicated to the left. Rectangles represent ORFs. Dark gray rectangles represent uORF2. The mutated 5′ leaders encode either a shorter uORF2, early stop (the last two residues missing); a longer uORF2, late stop (13 residues added after the intact uORF2 sequence); or a combinatorial mutant, double frame shift, in which a frame shift mutation (Fig. 3) is combined with another frame shift mutation at the end of uORF2. This latter construct encodes a uORF2 peptide in which only the first two and last two residues are the same as in the wild-type counterpart. WT, Wild type.
The position of the stop codon within uORF2 is also well conserved (Supplemental Fig. S1). The importance of the stop codon position for SIRT was tested by introducing three mutations in the bZIP11 5′ leader that affected the length and sequence of the uORF2. One mutation lengthened the peptide by 13 residues, late stop, and the other shortened the peptide by two residues, early stop. In another mutated 5′ leader, two frame shift mutations in opposite directions were introduced in the beginning of the sequence and in the end, respectively. These mutations give rise to a uORF2 sequence encoding a peptide of wild-type length, but with an unrelated sequence, except for the first two and last two residues (double frame shift). The mutated 5′ leaders are not able to confer SIRT as determined by LUC activity measurements (Fig. 5B). Thus, both the amino acid sequence and stop codon position of the SC-peptide are important for SIRT. Mutations in the mRNA sequence might affect their steady-state levels and therefore mRNA levels were quantified. The results show that the absence of SIRT in the mutants cannot be explained by changes in mRNA levels, in agreement with earlier observations (Supplemental Fig. S2; Rook et al., 1998; Wiese et al., 2004).
Leaky Scanning Is Important for bZIP11 Protein Translation
In eukaryotes, uORFs generally inhibit translation of the downstream main ORF (Vilela and McCarthy, 2003). The SC-peptide encoded by uORF2 of the bZIP11 5′ leader efficiently represses bZIP11 translation in response to Suc. Sequences surrounding the AUG codon (the AUG context) affect the efficiency of translational initiation by scanning translational preinitiation complexes (PICs). The AUG contexts of uORF2 differ from the consensus sequence of plants, indicating that initiation of translation of uORF2 is inefficient. In contrast, the AUG context of the main ORF is more similar to the consensus sequence. This AUG context pattern also holds for bZIP11 homologous genes of Arabidopsis, as well as from other species (Fig. 6A). Probably, the scanning PICs are prone to skipping the uORF start codon while efficiently initiating translation of the main ORF. To test this hypothesis, the internal AUG context of uORF2 (uuuAUGuu) was optimized to the aaaAUGgc sequence (Fig. 6B). This consensus sequence has been shown to yield maximal initiation frequency in higher eukaryotes (Joshi et al., 1997) and corresponds to the dicot consensus sequences of initiation context (Rangan et al., 2008). Scanning PICs that pass the weak first start codon of uORF2 should be efficiently captured by an optimized internal AUG context and initiate translation of uORF2. The second start codon was chosen for modification as the first part of the peptide was shown to be dispensable for SIRT (Fig. 3) and due to the fact that only one AUG is needed to cause SIRT (Wiese et al., 2004). Moreover, the second AUG is not fully conserved among SC-peptide homologs; therefore, minimal secondary effects of the change are expected (data not shown). The mutated 5′ leader was tested for SIRT using the transient expression assay and compared to the activity of the wild-type 5′ leader. The results of transient expression assays show that improving the AUG context of uORF2 retains SIRT, but dramatically reduces translation of the main ORF (Fig. 6B), providing support for a leaky scanning-dependent mechanism operating at the uORF initiation codons.
Figure 6.
The main ORF is translated through a leaky scanning-dependent mechanism. A, AUG codon contexts of bZIP11 and homologous genes from other dicotyledonous species and Arabidopsis. The conserved positions are indicated in green if the sequence matches the consensus sequence and in red if the nucleotide does not match the consensus sequence. The indicated consensus sequence is that of dicots (Rangan et al., 2008). B, Relative normalized LUC activity levels of 10-d-old seedlings transformed with the 35S:bZIP11 5′ leader:LUC construct or mutated versions. Following transformation, seedlings were incubated for 24 in medium containing either no Suc (white bars) or 6% Suc (gray bars). The constructs were either transformed into wild-type seedlings (accession Wassilewskija-0) or stv1 seedlings, as indicated. Means of at least three biological replicates are presented. Error bars represent sd from the mean. Two or more independent experiments were performed with essentially similar results. Schematic drawings of constructs used for transient expression experiments are indicated on the left. Rectangles represent ORFs. Dark gray rectangles represent uORF2 of the bZIP11 5′ leader. The AUG contexts are indicated below the schematic drawings and colored in accordance with similarity to the consensus sequence (A). The positions of the mutations are indicated with a white stripe in the boxes. WT, Wild type.
Ribosomes probably dissociate after translating uORF2. To test whether translational reinitiation occurs after termination of uORF2 translation, the short valve1 (stv1) mutant was analyzed for SIRT. This mutant was shown to be deficient in translational reinitiation (Nishimura et al., 2005), but was not found to affect SIRT (Fig. 6B). The relative LUC activity of an At1g20340 construct lacking uORFs was not affected by Suc in the stv1 mutant, confirming the equal translatability of mRNAs in the mutant treated with or without Suc.
DISCUSSION
Diurnal changes in sugar levels affect the expression levels of approximately one-third of the Arabidopsis genes, highlighting the central role of sugar signaling in plants (Bläsing et al., 2005). The Arabidopsis transcription factor bZIP11 is translationally regulated by Suc (Rook et al., 1998; Wiese et al., 2004) and was shown to regulate a subset of sugar-regulated genes in Arabidopsis (Hanson et al., 2008). bZIP11 activation results in extensive changes in gene expression and appears involved in metabolic reprogramming (Hanson et al., 2008; J. Hanson, M. Hanssen, and S. Smeekens, unpublished data). Likely, bZIP11 is part of a stress-dependent adaptation system that involves the SnRK1 (KIN10, KIN11) protein kinase pathway. In protoplast transfection assays, KIN10 activates the transactivation activity of bZIP11 and other Suc-regulated bZIP proteins (Baena-Gonzalez et al., 2007). It appears that bZIP11 is a node in a signaling network that activates genes in response to stress or starvation. Importantly, bZIP11 translational repression in response to Suc abrogates the downstream bZIP11 response pathway. Here, the mechanism of SIRT has been investigated and a novel mechanistic model is proposed.
Translational Repression of bZIP11 Is Independent of Light and Hormone Signaling
SIRT has previously been shown to act within 24 h of Suc addition. The stability of the GUS reporter protein prohibited more accurate testing of the response (Wiese et al., 2004). A LUC-based transient transformation system showed that SIRT acts within a few hours (Fig. 1A); likely, SIRT acts in a much shorter time frame, but this could not be tested due to initial low LUC activity levels.
Sugar signaling has been shown to be tightly interconnected to light and hormone-signaling pathways (Smeekens, 2000; Rolland and Sheen, 2005; Rolland et al., 2006). For example, the light effect on expression of the NR gene is replicated by sugar treatments (Cheng et al., 1992; Ciereszko et al., 2001). Several Glc-insensitive mutants identified were shown to be allelic to ABA-insensitive biosynthetic or signaling mutants and the effects of Glc were suggested to be dependent on increased levels of ABA compared to untreated seedlings (Arenas-Huertero et al., 2000; Huijser et al., 2000; Rook et al., 2001). Sugar signaling is also connected to ethylene signaling (Gibson et al., 2001; Arroyo et al., 2003; Moore et al., 2003; Yanagisawa et al., 2003), cytokinin, and auxin signaling (Ohkama et al., 2002; Ohto et al., 2006). Assaying SIRT in plants subjected to darkness or in various mutant backgrounds impaired in hormonal signaling demonstrated normal SIRT to occur. This indicates the independence of SIRT signaling of these signaling pathways.
HXK1 is an important Glc sensor in plants and was shown to be involved in several sugar responses (Rolland et al., 2006). Suc is readily metabolized to Glc and Fru in plants, but SIRT is not dependent on the HXK1 pathway as determined by the analysis in gin2 (HXK1-null) mutants. Recently, a G protein-dependent Glc-signaling pathway was identified (Grigston et al., 2008). However, the stronger effect of Suc compared to Glc (Fig.1C; Wiese et al., 2004) indicates that SIRT depends on a novel, Suc-dependent signaling pathway. The possibility that starch levels affect SIRT is excluded because the starchless pgm mutant shows SIRT. The generally lower LUC levels in pgm seedlings compared to wild-type seedlings could be due to the higher sugar levels of the pgm mutant during the day (Periappuram et al., 2000).
Suc Repression Is Mediated by a 28-Amino-Acid-Long Peptide
The bZIP11 5′ leader contains four partly overlapping uORFs, followed by a 169-nucleotide-long intercistronic region lacking ORFs. Systematic shortening of the bZIP11 5′ leader from the 3′ direction and testing for functionality in planta showed that the intercistronic region was dispensable for SIRT. The Δ330 construct displays SIRT, whereas the 17-nucleotide shorter Δ313 construct lacks SIRT activity. The 313 to 330 region of the bZIP11 mRNA includes the start codons of uORF3 and uORF4, but mutational analysis showed that uORF3 or uORF4 are not involved in the repression mechanism.
Translation of the 28 C-terminal amino acid residues of uORF2 are sufficient to impose SIRT (Wiese et al., 2004). Transplantation of this region to the 5′ leader of the At1g20340 gene is sufficient to impose SIRT (Fig. 3). This 3′ part of uORF2 sequence shows pronounced conservation at both amino acid and nucleotide levels (Supplemental Fig. S1). The importance of the peptide sequence was demonstrated by insertion of a single base pair in the 5′ nonessential part of the 5′ leader, which abolishes SIRT (Fig. 4). Mutations in the 5′ leader that did not result in changed amino acid sequence of the SC-peptide did not affect SIRT (Fig. 2). Therefore, nucleotide sequence conservation of uORF2 is most likely due to selection pressure on the peptide sequence, as shown by limited conservation at the third position of the codon of the uORF2 sequence (Supplemental Fig. S1). Together, this indicates the importance for SIRT of the 28-amino acid SC-peptide encoded by the 3′ part of uORF2. This SC-peptide has also been shown to be translated in vitro (Wiese et al., 2004). Mutational analysis has shown that inhibiting translation of uORF2 abolishes SIRT (Wiese et al., 2004). Mutagenesis showed the highly conserved Ser-29, Ser-31, Leu-35, and Tyr-39 to be essential for SIRT. These single amino acid substitutions disrupt functionality of the SC-peptide. Interestingly, changing Tyr-39 to the structurally similar amino acid Phe conferred some repression activity. This highlights the sequence dependence of the repression system and indicates that the SC-peptide interacts with other molecules to repress translation. Such proposed interaction occurs only in Suc-treated plants because SC-peptide amino acid changes specifically affect the translation in plants treated with Suc.
The stop codon position of uORF2 is evolutionarily conserved. Changing the stop position by lengthening the uORF2 abolishes SIRT. uORFs are common within mRNAs of plants and other eukaryotes and most uORFs act in a sequence-independent way (Hayden and Jorgensen, 2007). However, several examples exist in which the protein sequence of the uORF is conserved and therefore is believed to be of regulatory importance. The AdoMetDC and the SAC51 genes in Arabidopsis harbor uORFs with conserved protein sequences (Hanfrey et al., 2005; Hu et al., 2005; Imai et al., 2006). In these cases, the uORFs repress translation like the uORF2 in bZIP11.
Translation of the bZIP11 Main ORF
The eukaryotic translational apparatus first interacts with the 5′ cap structure of the mRNA and then scans the mRNA for the first available AUG codon within an appropriate sequence context to initiate translation. Once the ORF translation is terminated, the ribosomal subunits separate and dissociate from the mRNA. As a consequence, eukaryotic mRNAs generally only encode one ORF. However, several translational mechanisms have been documented that allow translation of ORFs preceded by uORFs (Kozak, 2002). For example, an internal ribosome entry site (IRES) in the mRNA allows the ribosome to directly associate with a sequence within the mRNA. Several plant viral RNAs are translated through IRES sequences (Jaag et al., 2003; Dorokhov et al., 2006; Karetnikov and Lehto, 2007), but so far only one cellular plant mRNA was shown to be translated through an IRES-dependent mechanism (Dinkova et al., 2005). Translation of bZIP11 does not depend on IRES as all sequences 3′ of the uORF2 can be deleted without affecting translation of the main ORF (Fig. 2A). This excludes the possibility of secondary structures in the mRNA leading to ribosome shunting as documented for several virus RNAs (Xi et al., 2005; Pooggin et al., 2006). Ribosomes are able to reinitiate translation after translating short uORFs (Kozak, 2002), but this seems unlikely for bZIP11 because, in that case, the AUG context-dependent improvement of uORF2 translational initiation would lead to increased main ORF translation, whereas the opposite is observed. This conclusion is supported by the results from the analysis of the stv1 mutant, which is impaired in translation reinitiation (Nishimura et al., 2005), but shows normal SIRT. Moreover, reinitiation after translation of a peptide longer than 35 amino acids is impossible in yeast (Vilela and McCarthy, 2003). The complete uORF2 is 42 amino acids long and thus to long for reinitiation. The distance between the uORF and the main ORF can be important for reinitiation (Kozak, 2001). Changing the distance between the stop codon of the SC-peptide and the AUG of the main ORF does not affect SIRT (Fig. 2A). Taken together, we conclude that a mechanism dependent on reinitiation appears unlikely.
Inefficient translation initiation of uORF2 is important for translation of the main ORF (Fig. 6) as the optimization of the AUG2b context strongly represses translation of the main ORF. This observation suggests that ribosomes failing to initiate translation of the weak context of uORF2 continue scanning and translate the main ORF. This notion is also supported by the experiments using the At1g20340 5′ leader:uORF2 construct with an improved, but not perfect, context, translation of the main ORF is strongly reduced, further supporting the conclusion that the weak AUG contexts of the uORF are important for efficient main ORF translation (Fig. 3). Most of the known bZIP11 homologs have noncanonical AUG context sequences of the uORFs, whereas sequences that allow efficient translation initiation surround the start codons of the main ORFs (Fig. 6). This indicates that the main ORF is translated by leaky scanning ribosomes and that this mechanism is evolutionary conserved. bZIP11 translation requires eIF3h (Kim et al., 2004). eIF3h was suggested to increase the processivity of the scanning ribosomes (Kim et al., 2007), which is in agreement with our model of bZIP11 translation. Leaky scanning was also found to be required for the translation of other plant genes, such as AtLIG1 (Sunderland et al., 2006) and THI1 (Chabregas et al., 2003). Inserting a strong uORF2b AUG context in uORF2 reduces main ORF translation, but Suc repression is retained (Fig. 6). This important observation excludes a regulatory model in which main ORF translation is regulated by differential initiation of translation of the repressing SC-peptide in response to Suc. Such regulation of translation by uORFs was observed in the translational regulation of AdoMetDC (Hanfrey et al., 2005).
We conclude that bZIP11 is translated by leaky scanning ribosomes and that translation of the SC-peptide is inhibiting translation of bZIP11 in the presence of Suc. Suc concentrations as low as 20 mm already reduce main ORF translation; thus SIRT acts at physiologically relevant Suc concentrations (Wiese et al., 2004, 2005). SIRT is lost in mutants with changed amino acid sequences of the uORF2 encoding SC-peptides, indicating that specific interaction with other biomolecules is important. When translated, nascent peptides move from the peptidyl transfer center through the exit tunnel of the ribosome. The exit tunnel covers approximately 40 amino acids (Matlack and Walter, 1995). Thus, during translation, the SC-peptide interacts with ribosome-associated molecules. The different parts of the translational apparatus are fixed in position relative to the peptidyl transfer center of the ribosome during translation elongation. The stop codon position of the SC-peptide is important for SIRT as shown experimentally (Fig. 5) and by essentially total evolutionary conservation of the stop codon position (Supplemental Fig. S1; Hayden and Jorgensen, 2007). Changes in stop codon position will move the relative position of the SC-peptide in the exit tunnel at termination of translation. Thus, positioning of the SC-peptide is important for SIRT, possibly as the SC-peptide interacts with a component of the translational apparatus, which is located in the exit tunnel. Changing the Tyr-39 residue abolished SIRT, which indicates that the SC-peptide must be translated entirely for biological activity and even the last residue of the SC-peptide is evolutionarily conserved. This implies that SIRT occurs at translational termination of the SC-peptide. In mammalian and cytomegalovirus systems, ribosome stalling depending on the position of the stop codon of the uORF has been documented (Cao and Geballe, 1995; Janzen et al., 2002). Likely the nascent SC-peptide stalls the translating ribosome in the presence of Suc and thereby halts the translation of bZIP11. The presence of a stalled ribosome at the mRNA will efficiently block scanning PICs from reaching the bZIP11 AUG codon and thus inhibit translation of the transcription factor. The nature of the molecule interacting with the SC-peptide remains to be elucidated.
The bZIP11 transcription factor was shown to specifically activate Suc-repressed genes in Arabidopsis (Hanson et al., 2008). Four other closely related bZip transcription factors in Arabidopsis harbor the SC-peptide encoding uORF in the 5′ leader sequence and show SIRT as well (Weltmeier et al., 2009). SIRT thus represents a general molecular mechanism of Suc-controlled gene expression in plants. In this study, the molecular nature of this Suc control was investigated and a ribosomal stalling mechanism suggested for SIRT. This novel translational control mechanism provides further understanding of how Suc regulates gene expression in plants and is important with respect to the proposed biological function of bZip transcription factors in reprogramming metabolism in response to stress and starvation (Hanson et al., 2008).
MATERIALS AND METHODS
Plant Growth Conditions
Wild-type or mutant Arabidopsis (Arabidopsis thaliana) seedlings (var Columbia-0 [Col-0], Wassilewkija-0, or Landsberg erecta [Ler]) were grown for 10 d on one-half-strength Murashige and Skoog medium, pH 5.7 (Duchefa), supplemented with 0.2% Suc, 0.5 mg/mL MES (Sigma-Aldrich), and 14 g/L plant agar (Duchefa). The seedlings were grown in growth chambers (Snijders Scientific) at 22°C under constant fluorescent light (100 μmol m−2 s−1).
Construction of bZIP11 5′ Leader Vectors Used for Transient Expression Experiments
A Gateway-compatible destination vector, p35S-ccdB-LUC, containing the 35S promoter in front of firefly LUC (fLUC) was constructed using the XbaI/SacI restriction fragment of pGWB35 (provided by Dr. T. Nakagawa, Shimane University, Matsue, Japan) containing the fLUC gene with Gateway sites for translational fusion. This was ligated into the 35S promoter-containing vector (pUC19 based). The wild-type bZIP11 5′-leader was amplified using primers with Gateway sites (bZIP11 5′-UTR FWD, bZIP11 5′-UTR REV) and transferred to pDONR-Zeo vector. The resulting plasmids were sequenced and the inserts were transferred to the p35S-ccdB-LUC destination vector by LR reaction, according to the manufacturer's instructions (Invitrogen). The deletion constructs were amplified by using primer bZIP11 5′-UTR FWD and bZIP11Δ313, Δ330, Δ342, Δ392, Δ423, Δ455, and Δ493 (Supplemental Table S1). For mutated versions of the bZIP11 5′ leader, the whole 5′ leader fragment, including Gateway sites, was translocated to the pALTER vector (Promega) using traditional methods (Sambrook et al., 1989). The resulting clone was used as a mutagenesis template using the Altered Sites II in vitro mutagenesis system kit (Promega), according to the manufacturer's instructions and indicated primers listed (Supplemental Table S1). Mutated 5′ leaders were moved to p35S-ccdB-LUC using Gateway technology.
To fuse the 84-nucleotide-long fragment of the uORF2 to the At1g20340 5′ leader, this region was amplified using uORF2FWDBamHI and uORF2REV primers (Supplemental Table S1). The PCR fragment was subcloned into pGemT-easy vector (Promega). The At1g20340 5′ leader was amplified using the At1g20340-leader FWD and At1g20340-leader REV BamHI primers and the amplified fragment was subcloned into the pGemT-easy vector (Promega). The uORF2 plasmid was restricted by BamHI/NcoI and the resulting fragment was ligated into the BamHI/NcoI sites of the At1g20340 5′ leader vector. The resulting insert was transferred to the p35S-ccdB-LUC vector using Gateway technology. The At1g20340 5′ leader for the control plasmid was amplified using the At1g20340-leader FWD and At1g20340 Gateway REV primers and transferred to the p35S-ccdB-LUC vector using Gateway technology. The integrity of the final constructs was confirmed by sequencing (Macrogen).
Transient Transformation of Arabidopsis Seedlings
Gold particles (1 μm diameter; Bio-Rad) were coated with plasmid DNA according to Giovanna et al. (1998). DNA for coating was premixed in a concentration equivalent of 1.2 mg fLUC vector and 0.4 mg Renilla LUC (rLUC) vector per transient expression experiment. The rLUC vector includes the rLUC gene driven by the constitutive 35S promoter. Seedlings were transformed using the Biolistic particle delivery system, model PDS-1000 He (Bio-Rad), according to the manufacturer's instructions using a vacuum adjusted to 28 Pa and 900-psi rupture discs. Prior to particle bombardment, selected seedlings (approximately 50) were transplanted to fresh plates. Following particle bombardment, one-half of the transformed seedlings were transferred into 250-mL flasks containing 50 mL of liquid one-half-strength Murashige and Skoog medium supplemented with 6% Suc and the other half were transferred to flasks containing medium without Suc. The seedlings were incubated in growth chambers, rotary shaking (45 rpm) under constant light for 24 h, and harvested in liquid nitrogen. All experiments were independently replicated and yielded similar results as the ones presented in this article. More than 90% of the seedlings were transformed as determined in parallel experiments using a plasmid harboring the GUS reporter gene and histochemical staining (data not shown).
LUC Activity Assays
Relative LUC levels were determined by the ratio of fLUC activity to rLUC activity. The LUC activities were measured by the Dual-Luciferase reporter assay system kit, according to the manufacturer's instructions (Promega). Approximately 25 mg ground Arabidopsis tissue were lysed using 100 μL passive lysis buffer supplemented with 2 mm DDT and incubated for 15 min at room temperature, followed by 2-min centrifugation (rcf 16,000). Twenty microliters of the supernatant were transferred into a new 2-mL tube and processed according to the Dual-Luciferase reporter assay system kit manual (Promega). LUC activity was measured using the TD-20/20 Glomax luminometer (Promega). All experimental series included transient expression of the wild-type bZIP11 5′ leader. The normalized fLUC activity levels were adjusted to the value of the plants transformed with the wild-type 5′ leader treated with medium lacking Suc. All activity levels were measured using three replicates minimum, averaged, and sd from the mean was calculated.
Quantitative PCR Analysis
Total RNA was isolated from 10-d-old seedlings (var Col-0) subjected to transient transformation. Plant tissues were homogenized by grinding in liquid nitrogen and total RNA was isolated using the RNeasy kit (Qiagen). DNA was removed from the preparations using RNase-free DNase I (Fermentas). cDNA was synthesized using M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. Due to low RNA yield, RNA preparations from three independent preparations were pooled and used as substrate for cDNA synthesis. Real-time quantitative PCR was performed using an ABI7900HT sequence detector using SYBR Green II master mix (Applied Biosystems). The ACTIN2 gene (At3g18780) was used as an internal reference. Relative RNA levels were calculated by the ΔΔCt method (Pfaffl, 2001). Primer efficiency was determined as described by Rasmussen (2001). Sequences of primers used in quantitative real-time PCR reactions are listed in Supplemental Table S2.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EE466106, AF430372, DQ787056, BG446720, AJ292744, EF147315, EV526128, and BQ519273.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evolutionary conservation of uORF2b sequences in Arabidopsis and other plant species. Amino acid sequence of uORF2b is more conserved than nucleotide sequence.
Supplemental Figure S2. Different LUC mRNA levels in transiently transformed Arabidopsis seedlings do not correlate with LUC activity levels.
Supplemental Table S1. Primers used to construct wild-type and mutated forms of the bZIP11 5′ leaders used for transient expression.
Supplemental Table S2. Primers used for quantitative real-time PCR.
Supplementary Material
Acknowledgments
We are grateful to Dr. A.G. von Arnim (The University of Tennessee, Knoxville, TN) for the kind gift of the rLUC vector used for normalizing transient expression levels and for providing the transient transformation protocol ahead of publication. We also acknowledge Dr. T. Nakagawa (Shimane University, Matsue, Japan) for providing plant vectors, and Dr. K. Okada (Kyoto University, Kyoto) for providing stv1 mutant seeds. Mutant seeds were obtained from the Nottingham Arabidopsis Stock Centre. The hxk1/gin2 mutant was provided by Dr. J. Sheen (Massachusetts General Hospital, Boston).
This work was supported by the Centre for BioSystems Genomics and by the Dutch Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Johannes Hanson (s.j.hanson@uu.nl).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, Leon P (2003) Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol 133 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942 [DOI] [PubMed] [Google Scholar]
- Bianchini C, Pastore A, Pelucchi S, Torreggiani E, Lambertini E, Marchesi E, Magri E, Frasson C, Querzoli P, Piva R (2008) Sex hormone receptor levels in laryngeal carcinoma: a comparison between protein and RNA evaluations. Eur Arch Otorhinolaryngol 265 1089–1094 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Geballe AP (1995) Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J Virol 69 1030–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, Van Sluys MA, Menck CF, Silva-Filho MC (2003) Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J Cell Sci 116 285–291 [DOI] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Cristinsin M, Conkling MA (1992) Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci USA 89 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA (2001) Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem J 354 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrads KA, Yi M, Simpson KA, Lucas DA, Camalier CE, Yu LR, Veenstra TD, Stephens RM, Conrads TP, Beck GR Jr (2005) A combined proteome and microarray investigation of inorganic phosphate-induced pre-osteoblast cells. Mol Cell Proteomics 4 1284–1296 [DOI] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, Chua NH, Smeekens SCM (1997) Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Zepeda H, Martinez-Salas E, Martinez LM, Nieto-Sotelo J, de Jimenez ES (2005) Cap-independent translation of maize Hsp101. Plant J 41 722–731 [DOI] [PubMed] [Google Scholar]
- Dorokhov YL, Ivanov PA, Komarova TV, Skulachev MV, Atabekov JG (2006) An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. J Gen Virol 87 2693–2697 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7 R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280 196–203 [DOI] [PubMed] [Google Scholar]
- Giovanna S, Ugo B, Giorgio M, Ida R (1998) A transient assay for rapid functional analysis of transcription factors in Arabidopsis. Plant Mol Biol Rep 16 191–197 [Google Scholar]
- Gopfert U, Kullmann M, Hengst L (2003) Cell cycle-dependent translation of p27 involves a responsive element in its 5′-UTR that overlaps with a uORF. Hum Mol Genet 12 1767–1779 [DOI] [PubMed] [Google Scholar]
- Grigston JC, Osuna D, Scheible WR, Liu C, Stitt M, Jones AM (2008) d-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett 582 3577–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ (2005) A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem 280 39229–39237 [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53 935–949 [DOI] [PubMed] [Google Scholar]
- Hayden CA, Bosco G (2008) Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics 9 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA (2007) Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59 407–450 [DOI] [PubMed] [Google Scholar]
- Hu WW, Gong H, Pua EC (2005) The pivotal roles of the plant S-adenosylmethionine decarboxylase 5′ untranslated leader sequence in regulation of gene expression at the transcriptional and posttranscriptional levels. Plant Physiol 138 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23 577–585 [DOI] [PubMed] [Google Scholar]
- Imai A, Hanzawa Y, Komura M, Yamamoto KT, Komeda Y, Takahashi T (2006) The dwarf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORF of a bHLH gene. Development 133 3575–3585 [DOI] [PubMed] [Google Scholar]
- Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T (2008) A semi-dominant mutation in a ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56 881–890 [DOI] [PubMed] [Google Scholar]
- Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prufer D (2003) An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc Natl Acad Sci USA 100 8939–8944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DM, Frolova L, Geballe AP (2002) Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol Cell Biol 22 8562–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R, Goldsbrough A, Bevan M (1990) Transcriptional regulation of a patatin-1 gene in potato. Plant Mol Biol 14 995–1006 [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35 993–1001 [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K (2007) The RNA2 5′ leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J Gen Virol 88 286–297 [DOI] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8 R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47 509–540 [DOI] [PubMed] [Google Scholar]
- Kozak M (2001) Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16 345–353 [DOI] [PubMed] [Google Scholar]
- Luo Z, Sachs MS (1996) Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol 178 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Walter P (1995) The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J Biol Chem 270 6170–6180 [DOI] [PubMed] [Google Scholar]
- Meijer HA, Thomas AA (2003) Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res 31 3174–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mize GJ, Morris DR (2001) A mammalian sequence-dependent upstream open reading frame mediates polyamine-regulated translation in yeast. RNA 7 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T (2002) Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol 43 1493–1501 [DOI] [PubMed] [Google Scholar]
- Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K (2006) Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol 47 1603–1611 [DOI] [PubMed] [Google Scholar]
- Periappuram C, Steinhauer L, Barton DL, Taylor DC, Chatson B, Zou J (2000) The plastidic phosphoglucomutase from Arabidopsis. A reversible enzyme reaction with an important role in metabolic control. Plant Physiol 122 1193–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin MM, Ryabova LA, He X, Futterer J, Hohn T (2006) Mechanism of ribosome shunting in Rice tungro bacilliform pararetrovirus. RNA 12 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan L, Vogel C, Srivastava A (2008) Analysis of context sequence surrounding translation initiation site from complete genome of model plants. Mol Biotechnol 39 207–213 [DOI] [PubMed] [Google Scholar]
- Rasmussen R (2001) Quantification on the LightCycler. In S Meuer, C Wittwer, K Nakagawara, eds, Rapid Cycle Real-Time PCR, Methods and Applications. Springer Press, Heidelberg, pp 21–34
- Rolland F, Baena-Gonzalez E, Sheen J (2006) SUGAR SENSING AND SIGNALING IN PLANTS: conserved and Novel Mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Sheen J (2005) Sugar sensing and signalling networks in plants. Biochem Soc Trans 33 269–271 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26 421–433 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15 253–263 [DOI] [PubMed] [Google Scholar]
- Ruan H, Shantz LM, Pegg AE, Morris DR (1996) The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271 29576–29582 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sivitz AB, Reinders A, Ward JM (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51 49–81 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland PA, West CE, Waterworth WM, Bray CM (2006) An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J 47 356–367 [DOI] [PubMed] [Google Scholar]
- Tabuchi T, Okada T, Azuma T, Nanmori T, Yasuda T (2006) Posttranscriptional regulation by the upstream open reading frame of the phosphoethanolamine N-methyltransferase gene. Biosci Biotechnol Biochem 70 2330–2334 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Sato-Nara K, Kobayashi K, Suzuki M, Suzuki H (2003) Sugar-induced adventitious roots in Arabidopsis seedlings. J Plant Res 116 83–91 [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C, McCarthy JE (2003) Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol Microbiol 49 859–867 [DOI] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schutze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, et al (2009) Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol 69 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzler H, Mignery G, Fisher L, Park W (1989) Sucrose-regulated expression of a chimeric potato tuber gene in leaves of transgenic tobacco plants. Plant Mol Biol 13 347–354 [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2005) Sucrose-induced translational repression of plant bZIP-type transcription factors. Biochem Soc Trans 33 272–275 [DOI] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ (2005) Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J Virol 79 5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425 521–525 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Hirose T, Fujii N, Aspuria ET, Kato A, Uchimiya H (1994) The rolC promoter of Agrobacterium rhizogenes Ri plasmid is activated by sucrose in transgenic tobacco plants. Mol Gen Genet 244 15–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.