Abstract
The process of transcription initiation in eukaryotic cells is complex and our view of how the transcription apparatus is assembled at the promoter has changed over the last decade. For most genes transcribed by RNA polymerase II (Pol II) general transcription factors and Pol II assemble at the core promoter and initiate transcription at specific sites. The number of distinct core promoter elements is increasing and so is the number of specific proteins that act through these elements. Core promoter elements include the TATA box, TFIIB recognition element (BRE), the initiator (INR), and the downstream promoter element (DPE). Transcription factors that act through these elements are TATA binding protein (TBP) and its associated factors (TAFs), Negative Cofactor 2 (NC2), Mot1p, TFIIB, and TFIIA. Recent observations suggest that TBP, Mot1p, and NC2 exert positive and negative effects on transcription complex assembly depending on the presence or absence of specific core promoter elements. It is proposed here that Mot1p and NC2 together function as a remodeling complex that positions TFIID and perhaps other protein complexes at specific core promoter elements.
Faithful transcription initiation by Pol II requires a set of general transcription factors that guide the polymerase to the promoter region of genes and aide in the formation of elongation competent transcription complexes.1 The minimal set of proteins or protein complexes required for transcription of a TATA-box containing promoter are TBP, TFIIB, TFIIE, TFIIF, TFIIH, and Pol II.2,3 Most of our knowledge of transcription complex assembly on RNA polymerase II transcribed genes stems from experiments with promoters containing a TATA box.1 The first step is considered to be the binding of TBP to the TATA box, which is stabilized by TFIIA and leads to the recruitment of TFIIB. TFIIB establishes a bridge between the core promoter binding factors and Pol II. TFIIB is a single protein that contains different domains for protein/protein and protein/DNA interactions. Elegant biochemical studies by the Hahn group have shown that the B-finger of TFIIB reaches into the active site of Pol II and in doing so determines the start site of transcription in TATA box containing genes.4 Some promoters contain BRE elements, which can be located either upstream (BREu) or downstream (BREd) of the TATA-box.2,3 BREs can exert both positive and negative effects on transcription. The positive effect may be due to enhancing the efficiency of TFIIB recruitment. How BREs inhibit transcription is currently not known.
The INR was the first DNA element shown to mediate transcription complex assembly on TATA-less genes.5 In addition to the minimal set of transcription factors sufficient for TATA-box directed transcription, INR promoters utilize additional components including TAFs.2,3 TAF1 and TAF2 can be crosslinked to the INR and likely establish first contacts with the promoter DNA.2 However, it should be pointed out that transcription from INR promoters can not be reconstituted with a defined set of basal transcription factors leaving open the possibility that other factors first bind to the INR and recruit the TFIID complex to the promoter.2,3
The DPE, originally discovered in Drosophila by Burke and Kadonaga, is an additional common promoter element in higher eukaryotes.6 Whereas the TATA-box and INR can recruit transcription complexes independent from other DNA elements, the DPE is unable to do so and only appears to function in combination with other core elements.2,3 It is interesting to note in this respect that the spacing between INR and DPE is highly conserved between distantly related Drosophila species.7 This suggests that the DPE and INR cooperate to recruit and position transcription complexes for specific transcription initiation. Like TATA and INR, the DPE also interacts with subunits of the TFIID complex, namely TAF6 and TAF9. Lewis et al. demonstrated that DPE dependent transcription requires casein kinase II α (CKIIα) and mediator, a large Pol II associated co-regulator complex.8
The transcription co-regulator NC2 has previously been shown to stimulate transcription from DPE containing promoters and to repress transcription from TATA-box containing promoters.9 NC2 is a heterodimer composed of NC2α and NC2β and was originally isolated from a larger co-regulator complex (USA, upstream stimulatory factor, USF, stimulatory activity) involved in transcription activation of the Adenovirus 2 major late promoter (Ad2MLP).10 NC2 directly interacts with TBP and prevents the association of TBP with TFIIB.11 There are conflicting data regarding the effect of NC2 on the association of TFIIA. While Xie et al.11 found that NC2 inhibits interactions between TBP and TFIIA, Masson et al.12 showed that NC2 does not significantly affect the recruitment of TFIIA. The different results could be explained by the possibility that differences in promoter structure, for example presence or absence of INR or BRE elements, could affect the ability of NC2 to inhibit association of TFIIA with the TBP-TATA box complex. In this respect it is noteworthy that the INR is capable of attenuating NC2 mediated repression of TATA-box containing promoters.13
NC2 associates with TATA-containing as well as TATA-less promoters.14 A recent genome wide promoter ChIP on chip analysis revealed that NC2 occupies a large number of promoters and that this occupancy positively correlates with mRNA levels.15 Furthermore the same study also found a negative correlation between NC2 binding and the presence of a BRE. Geisberg et al. have previously demonstrated a positive correlation between NC2 occupancy and transcriptional activity of yeast promoters.16 The results from the global association analysis suggest that NC2 function is not restricted to DPE-containing promoters but that it also plays a positive role in transcription directed by INR and TATA. Recently, Schluesche et al.17 demonstrated quite elegantly that NC2 mobilizes TBP on DNA and induces conformational changes in the TBP-DNA complex, which regulate the ability of TBP to interact with the TATA-box.
Another protein regulating the assembly of the preinitiation complex is Mot1p, which was originally identified in yeast.18 Mot1p contains a Snf2/Swi2-related ATPase domain and is able to mobilize TBP on or to dissociate TBP from DNA.19 Interestingly, the human ortholog of Mot1p, BTAF1 was shown to interact with NC2 via its α-subunit.20 NC2 stimulates the ability of BTAF1 to interact with TBP in an ATP but not ATP-hydrolysis dependent manner.20 Like NC2, yeast Mot1p is involved in both activation and repression of transcription.21 However, many more genes appear to be repressed by Mot1p than activated. The repression function is likely due to the ability of Mot1p to displace TBP from the TATA box in an ATP hydrolysis dependent manner.22
The data regarding the function of NC2 and Mot1p in transcription complex assembly, particularly with regard to their function at TATA-box containing promoters, have not led to a conclusive consensus on how these proteins function during transcription complex assembly. Three recent publications from the Kadonaga and Timmers laboratories shed light on the function of these proteins and suggest that NC2 and Mot1p may function as an ATPase dependent TFIID/core promoter remodeling complex that positions the TFIID complex at different core promoter elements (TATA, INR, or DPE).7,23,24
Recently, Juven-Gershon et al.7 analyzed the role of core promoter elements in transcription regulation by Caudal, a key regulator of homeotic (Hox) gene expression in Drosophila. The authors show that most Hox gene promoters lack a TATA-box but contain INR and DPE elements. They further demonstrate that Caudal preferentially activates promoters containing a DPE motif. Interestingly, however, TATA-box containing promoters were activated as well, but only if they lack an upstream BRE. Addition of a BRE to the TATA-box promoter abolished activation by Caudal. These results demonstrate that the absence or presence of specific core promoter sequences restrict the regulation of transcription by specific activator proteins. Similar results have been shown previously with regard to enhancer mediated transcription activation.2,3 Some enhancers only activate transcription of TATA-driven promoters, while others only stimulate DPE-driven transcription.
In another study van Werven et al.23 demonstrated that TBP, NC2, and Mot1p co-localize at active Pol II promoters across the genome in yeast. NC2α and Mot1p appear to prefer TATA-box containing promoters while NC2β occupancy is higher at TATA-less promoters. The authors further show that NC2, Mot1p, and TBP form a stable protein/DNA complex and that NC2 dissociates from this complex in an ATP hydrolysis dependent manner. The authors suggest that the function of NC2 and Mot1p on TATA-box containing promoters is to restrict TBP activity in order to limit transcription levels.
Hsu et al.24 reduced expression of proteins associated with core promoter function by RNA interference in a Drosophila cell line. The effect on transcription was examined using reporter gene constructs that were driven either by TATA/INR or DPE/INR promoters. Diminished TBP and TFIIA activity reduced transcription of the TATA/INR promoter but had no effect on the DPE/INR promoter. The reduction of TAF4, TFIIB, CkIIα, PC4 (positive co-factor 4, another component of USA), and components of the mediator diminished transcription from both promoters. Depletion of NC2α, NC2β, and Mot1p resulted in a strong reduction of DPE/INR promoters but also reduced transcription from TATA/INR promoters. This result is consistent with a positive role of NC2 and Mot1p in transcription of TATA-box containing genes. However, the effect on endogenous gene expression was different in that transcription of DPE containing ecdyson induced genes was elevated in cells with diminished TBP levels, while transcription of ecdyson induced TATA-box containing promoters was elevated in cells with reduced levels of NC2. The authors suggest that NC2 antagonizes the function of TBP, which acts positively on TATA-box containing promoters but negatively on DPE-containing promoters.
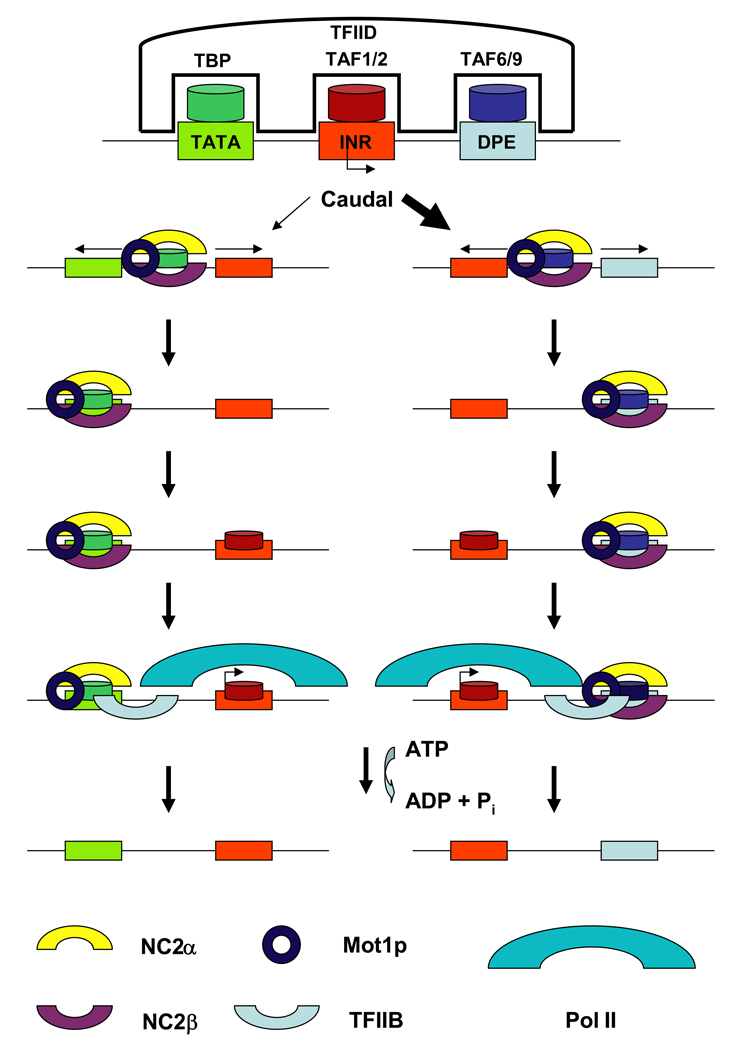
Taken all the data together it appears that NC2 and Mot1p could function as a TFIID remodeling complex that positions the DNA binding proteins (TBP, TAF1/2, or TAF6/9) at different core promoter elements (TATA, INR, DPE) as illustrated in Fig. 1. For TATA-box containing promoters NC2 and Mot1p mobilize the TFIID complex until TBP is positioned at the TATA box. This allows TAF1/2 to contact the INR, which in turn leads to a conformational change of the TFIID/NC2/Mot1p/DNA complex that results in the removal of NC2β. Dissociation of NC2β allows association of TFIIB with TBP and formation of transcription competent complexes. Basehoar et al. recently demonstrated that in yeast the SAGA complex, but not the TFIID complex, preferentially associates with TATA-box containing promoters.25 Therefore, an alternative pathway of transcription complex assembly on TATA-containing promoters begins with Mot1p and NC2 mediated mobilization of TBP and positioning of TBP at the TATA box. This would then recruit the SAGA complex leading to the dissociation of NC2β and the subsequent binding of TFIIB and Pol II.
Fig. 1.
Depiction of a model of NC2/Mot1p mediated positioning of TFIID at either the TATA-box or the DPE element. The TFIID complex via its TBP, TAF1/2, and TAF6/9 subunits interacts with the three major core promoter elements TATA, INR, and DPE. Transcription activators like Caudal preferentially enhance transcription of DPE-containing promoters (thick black arrow). On TATA/INR promoters (depicted on the left side) NC2 and Mot1p mobilize TFIID (shown here is only the TBP subunit) until TBP stably interacts with the TATA box. Subsequently, TAF1/2 will contact the INR inducing a conformational change in the TFIID complex that results in the dissociation of NC2β. Dissociation of NC2β allows association of TFIIB and formation of initiation competent transcription complexes. Alternatively, positioning of TBP at the TATA-box leads to the recruitment of the SAGA complex and subsequent dissociation of NC2β (not shown). On DPE/INR containing promoters (shown on the right) the NC2/Mot1p protein complex mobilizes TFIID until TAF6/9 stably associates with the DPE. Subsequently, TAF1/2 will contact the INR and TFIIB associates with this complex in a TBP independent manner. Association of Pol II and other general transcription factors will form initiation competent transcription complexes. To restrict transcription activation, ATP hydrolysis dependent conformational changes mediated by Mot1p will dissociate TBP, NC2, and associated proteins from TATA- and DPE containing promoters.
A similar situation is proposed for DPE initiated transcription. The NC2/Mot1p complex mobilizes the TFIID complex and positions TAF6/9 at the DPE. This allows TAF1/2 to bind the initiator, which is followed by the recruitment of TFIIB and Pol II. The difference between TATA- and DPE-containing promoters is (1) that there is a higher dependency on NC2/Mot1p at DPE-driven promoters perhaps because TAF6/9 interactions with the DPE are weaker than TBP TATA-box interactions and (2) that NC2β does not have to dissociate from DPE-containing promoters because transcription complex formation at these promoters does not require TFIIB/TBP interactions. According to this model, the NC2/Mot1p protein complex plays a similar positive role in transcription of TATA-box containing and TATA-less promoters. The difference is that on TATA box-containing promoters NC2 exerts a negative effect because it prevents association of TFIIB with TBP. The model is consistent with and explains previous observations showing that (1) NC2β occupancy is lower at TATA-box containing promoters compared to TATA-less promoters because it dissociates from these templates to allow the entry of TFIIB, that (2) the INR reduces the negative effect of NC2 on TATA-dependent transcription initiation because the binding of TAF1/2 to the INR induces a conformational change that dissociates NC2β, and that (3) NC2 facilitates the recruitment of TFIIB because NC2/Mot1p positions TFIID at the promoter and subsequent steps would allow its interaction with TFIIB.12,13,23
It has previously been proposed that TFIID functions as a specialized nucleosome that marks promoters.26 In this regard the role of NC2/Mot1p may be analogous to the function of ATP-dependent nucleosome remodeling complexes. NC2/Mot1p could lock in the TFIID complex at distinct core promoter elements to allow efficient recruitment and accurate positioning of Pol II. Mot1p mediated conformational changes during pre-initiation complex formation have previously been discussed by Dasgupta et al.21
Transcription re-initiation could be inhibited by ATP-hydrolysis dependent and Mot1p mediated dissociation of TBP or TFIID from DPE- or TATA-containing promoters. This could attenuate transcription levels as discussed by van Werven et al.23, and would be consistent with previous studies showing that Mot1p largely functions as a negative transcription factor.21
The diversity of core promoter elements and interacting proteins contribute to changes in expression programs during cell differentiation. This is best illustrated by a recent finding demonstrating that TBP is replaced by the TBP related factor TRF3 during differentiation of muscle cells, which is required for the expression of muscle specific genes.23 A switch from TATA-dependent to TATA-independent modes of transcription complex formation have been discussed.9 It is this context that renders the recent findings concerning NC2, Mot1p and TBP function highly significant. There are many open questions that will likely be addressed in the future. For example, what is the role of the BRE in restricting transcription initiation? There are a number of promoters that do not initiate transcription from a single start site but rather initiate transcription from multiple sites. Does NC2 or Mot1p regulate these genes? Finally, how do activators like Caudal enhance transcription? Do these proteins interact with NC2, or do they interfere with proteins acting negatively via the BRE? Addressing these questions and other issues will likely shed light on the repertoire of mechanisms that regulate transcription initiation.
Acknowledgements
This work was supported by the National Institutes of Health (DK052356).
Abbreviations
- BRE
TFIIB recognition element
- INR
initiator
- DPE
downstream (core) promoter element
- NC2
negative cofactor 2
- Mot1p
modifier of transcription 1p
- PIC
pre-initiation complex
- Pol II
RNA polymerase II
- SAGA
Spt-Ada-GCN5 acetyltransferase complex
- TAF
TBP associated protein
- TBP
TATA binding protein
- TFII
Transcription factor of Pol II
References
- 1.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 2.Gross P, Oelgeschläger T. Core promoter-selective RNA polymerase II transcription. Biochem Soc Symp. 2006;73:225–236. doi: 10.1042/bss0730225. [DOI] [PubMed] [Google Scholar]
- 3.Müller F, Demény MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoter with a variety of core promoter recognition factors. J Biol Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 4.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 6.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 7.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008 October 16; doi: 10.1101/gad.1698108. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BA, Sims RJ, III, Lane WS, Reinberg D. Functional characterization of core promoter elements:DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 10.Meisterernst M, Roeder RG. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 11.Xie J, Collart M, Lemaire M, Stelzer G, Meisterernst M. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 2000;19:672–682. doi: 10.1093/emboj/19.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masson P, Leimgruber E, Creton S, Collart MA. The dual role of TFIIB recruitment by NC2 is gene specific. Nucl Acids Res. 2008;36:539–549. doi: 10.1093/nar/gkm1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malecová B, Gross P, Boyer-Guittaut, Yavuz S, Oelgeschläger T. The initiator core element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem. 2007;282:24767–24776. doi: 10.1074/jbc.M702776200. [DOI] [PubMed] [Google Scholar]
- 14.Gilfillan S, Stelzer G, Piaia, Hofmann MG, Meisterernst M. Efficient binding of NC2.TATA-binding protein to DNA in the absence of TATA. J Biol Chem. 280:6222–6230. doi: 10.1074/jbc.M406343200. [DOI] [PubMed] [Google Scholar]
- 15.Albert TK, Grote K, Boeing S, Stelzer G, Schepers A, Meisterernst M. Global distribution of negative cofactor 2 subunit α on human promoters. Proc Natl Acad Sci USA. 2007;104:10000–10005. doi: 10.1073/pnas.0703490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisberg JV, Holstege FC, Young RA, Struhl K. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schluesche P, Stelzer G, Piaia, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- 18.Pereira LA, Kleijman MP, Timmers TM. Roles of BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription. Gene. 2003;315:1–13. doi: 10.1016/s0378-1119(03)00714-5. [DOI] [PubMed] [Google Scholar]
- 19.Sprouse RO, Brenowitz M, Auble DT. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleijman MP, Pereira LA, van Zeeburg HJ, Gilfillan S, Meisterernst M, Timmers HT. NC2alpha interacts with BTAF1 and stimulates its ATP-dependent association with TATA-binding protein. Mol Cell Biol. 2004;24:10072–10082. doi: 10.1128/MCB.24.22.10072-10082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darst RP, Dasgupta A, Zhu C, Hsu JY, Vroom A, Muldrow T, Auble DT. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J Biol Chem. 2003;278:13216–13226. doi: 10.1074/jbc.M211445200. [DOI] [PubMed] [Google Scholar]
- 23.Van Werven FJ, van Bakel H, van Teeffelen HAAM, Altelaar AFM, Koerkamp MG, Heck AJR, Holstege FCP, Timmers HTM. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu JY, Juven-Gershon T, Marr MT, II, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann A, Oelgeschläger T, Roeder RG. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deato MDE, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]