Abstract
AIM: To analyze the metallochaperone antioxidant-1 (Atox1) gene sequence in Wilson disease patients.
METHODS: Mutation analysis of the four exons of the Atox1 gene including the intron- exon boundaries was performed in 63 Wilson disease patients by direct sequencing.
RESULTS: From 63 selected patients no mutations were identified after the entire coding region including the intron- exon boundaries of Atox1 were sequenced. One known polymorphism within the Atox1 gene (5’UTR -99 T>C) in 31 (49%) of the Wilson patients as well as one previously undescribed variation (5’UTR -68 C>T) in 2 of the Wilson patients could be detected. Statistical analyses revealed that the existence of a variation within the Atox1- gene showed a tendency towards an earlier onset of the disease.
CONCLUSION: Based on the data of this study, no major role can be attributed to Atox1 in the pathophysiology or clinical variation of Wilson disease.
Keywords: Antioxidant-1, Wilson disease, Wilson’s disease protein, Mutation analysis
INTRODUCTION
Copper is an essential trace element for prokaryotes and eukaryotes. It acts as a cofactor for a number of proteins such as cytochrome c oxidase, dopamine β-hydroxylase (production of catecholamines), superoxide dismutase (free radical detoxification), lysyl oxidase (cross-linking collagen and elastin) and ceruloplasmin[1]. At the same time, excess or free copper is toxic to the cell.
The inherited disorders of copper metabolism Menkes and Wilson disease (WD) result from a disturbance of copper balance, resulting in either a deficiency (Menkes) or an accumulation of copper (WD) in the body[2]. WD occurs in about one of 30 000 people[3–5] and is characterized by the accumulation of copper primarily in the liver but also in the brain, kidney, cornea (Kayser-Fleischer-Rings), and spleen. WD is caused by a genetic defect in the ATP7B gene, located on chromosome 13q14.3[6]. This gene encodes a polytopic membrane protein containing several motifs characteristic of P-type ATPases, highly expressed in the liver[7]. Under steady-state conditions the gene product of ATP7B WNDP (Wilson’s disease protein) resides in the trans-Golgi network (TGN)[8] where it delivers copper to such secreted copper-dependent enzymes as ceruloplasmin. As the copper content of the hepatocytes increases, this ATPase moves from the TGN to a cytoplasmic vesicular compartment near the canalicular membrane. As copper is transported into this compartment, the intracellular copper concentration falls and the WNDP is recycled back to the TGN while copper is exported from the cell[9].
There are more than 230 mutations in ATP7B[10] accounting for Wilson Disease, and no mutation is predominant[11]. Patients present, typically between the ages of 5 and 40 years, with quite various hepatic (40%), neurological (40%), psychiatric (14%-18%) or other symptoms[12,13]. Although the specific type of mutation might have in part influence on disease severity, even patients with identical mutations show high clinical variability regarding the age of onset, signs and syndromes, ceruloplasmin levels, hepatic copper levels and presence of Kayser-Fleischer-rings[14,15]. The following factors are known to influence the disease: There are certain sex-specific differences: female patients for example show a higher prevalence of acute liver failure than male patients. Schiefermeier et al reported that an APO E epsilon3/3 genotype delays the onset of signs and symptoms[16]. Merle et al described influence on onset of symptoms of Wilson disease depending on prion protein status[17]. It has been proposed that genes influencing human copper metabolism might modify the clinical picture caused by a mutated Wilson disease gene.
As noted above copper is not free within the cell[18]. The trafficking of copper from donor and acceptor proteins is mediated by a unique class of proteins termed copper chaperones[19,20] that were first identified in the yeast Saccharomyces cerevisiae. The yeast ATX1p encodes a small cytosolic copper- binding protein that binds copper via the copper- binding MxCxxC motif and delivers this metal to CCC2[21] - the yeast homologue of WNDP - for subsequent transport into the secretory pathway and incorporation into the ceruloplasmin homologue FET3[22]. The human homologue Antioxidant-1 (Atox1) is an 8 kDa cytosolic protein that contains a single copy of the highly conserved MxCxxC motif[23] in the amino terminus that is repeated 6-fold in WNDP. This metallochaperone interacts directly with the Wilson ATPase[2] and can regulate its copper occupancy[24]. By modulating the amount of copper bound to the protein Atox1 can regulate the intracellular localization[25], the posttranslational modification[26,27] and the enzymatic activity of WNDP[24].
MATERIALS AND METHODS
Patients
The Genomic DNA of 63 WD patients was sequenced, which include 42 female and 21 male patients. The average age was 34 years (19-56). Twenty eight (44%) patients presented primarily with hepatic symptoms (including elevated liver enzyme test, ascites, liver cirrhosis, and acute liver failure), 20 (32%) patients primarily with neurological symptoms, 7 (11%) patient with both, 8 (13%) patients were diagnosed preclinically by family screening.
In all patients the definitive diagnosis of WD was established either by DNA analysis or by typical clinical and laboratory constellations [reduced copper levels (< 10 μmol/L) and ceruloplasmin levels (< 0.2 g/L) in the serum, raised free plasma copper levels, increased urinary copper excretion (≥ 2 μmol/24 h), detection of Kayser- Fleischer- rings, elevated hepatic copper concentrations (> 250 μg/g dry tissue)] or both. The ATP7B gene has been sequenced in most patients in part so far in cooperation with Professor Ferenci, Department of Gastroenterology and Hepatology, Vienna, including the analysis of the H1096Q mutation status in all patients and sequencing of exons 8, 13, 14, 15 and 18 in most patients not homozygous for H1096Q. In 36 patients two mutations could be detected within the Wilson disease gene. In 16 patients one mutation and in 11 patients no mutations could be detected within the Wilson gene so far.
Methods
Genomic DNA was isolated from peripheral EDTA- blood using the QIAamp® DNA Blood mini Kit (Quiagen, Hilden/Germany) according to the manufacturer’s instructions.
The four exons of the gene including the intron- exon boundaries were amplified by PCR, using the Oligonucleotide primers shown in Table 1, designed from the published cDNA sequence of the human homologue of Atox1 gene[23].
Table 1.
Oligonucleotide primers used to amplify Atox1
| Exon | Sequence | Fragment size |
| Exon 1-Forward primer | 5’-GGAGTGGGAGGGGCCTCCGGGACC-3’ | 273 bp |
| Exon 1-Reverse primer | 5’-GTAAGCTAGGGGACAACAGCGGCTC-3’ | 273 bp |
| Exon 2 - Forward primer | 5’-GCACTGTGTGGGGGTCACTCTACAG-3’ | 320 bp |
| Exon 2-Reverse primer | 5’-GTGAGGATTAAATGATGTGATTCAC-3’ | 320 bp |
| Exon 3-Forward primer | 5’-GAACTCTTTCTTGCTGTAACTGGGAG-3’ | 344 bp |
| Exon 3-Reverse primer | 5’-GAGGGCTCTCCCGCTCCACTCAAG-3’ | 344 bp |
| Exon 4-Forward primer | 5’-TGCAATGTCGCTATGTCCACACCA-3’ | 326 bp |
| Exon 4-Reverse primer | 5’-GATCACACAGCAAAGAATCAGAATC-3’ | 326 bp |
The 50 μL volume of each PCR reaction contained 100 ng of template DNA, 500 μmol/L of each oligonu-cleotide primer, 200 μmol/L each dNTP, 2.5 units of Stratagene Pfu-DNA Polymerase (Qiagen, Hilden, Germany) in 10 × QIAGEN PCR Buffer (Qiagen, Hilden).
PCR reactions were performed in a Progene FPROG050 cycler (Techne, Cambridge/ UK) starting with the initial denaturation of the DNA at 95°C for 10 min; followed by 45 cycles (exon 1, 3, 4)/40 cycles (exon 2) of: 45 s denaturation at 95°C, 45 s annealing at 60°C (Exon 1)/56.5°C (Exon 2)/50°C (Exon 3 + Exon 4) and 1 min extension at 72°C, then with a final extension of 72°C for 10 min.
PCR products were purified with the MinEluteTM Purification Kit (Quiagen, Hilden/Germany). The sequencing reaction was performed by SEQLAB (Sequence Laboratories Göttingen, Göttingen/Germany).
Sequences obtained by sample sequencing were compared with http://www.ncbi.nlm.nih.gov/BLAST.
Statistical analyses
Statistical analyses were performed with the SPSS, version 13.0 (Statistical Package for the Social Science, SPSS Inc., Chicago, IL, USA.). P < 0.05 was taken as significant.
RESULTS
We analysed Atox1 in 63 WD patients, diagnosed either by DNA analysis or by typical clinical constellations.
There could be no alterations in the Atox1 coding exon sequence or splice junction sequence be detected in any of these individuals.
Direct sequencing of the Atox1 gene within the 5’-UTR region located before exon 1 of the 63 Wilson disease patients examined revealed one known polymorphism within the Atox1 gene in 31 (49%) of the Wilson patients (Table 2). Thirty one of 63 (49%) Wilson patients had detectable Atox1 gene changes, with the heterozygous T/C at 5’UTR -99 being the most common with 22 (35%) patients.
Table 2.
Atox1 gene analysis in Wilson disease patients
|
Distribution of the variations detected in the Atox1-gene among the ATP7B-genotypes |
||||||
| Mutations in the ATP7B-gene | No. of patients |
5’UTR -99 T>C |
5’UTR -68 C>T |
No variation within the sequence | ||
| Heterozygous | Homozygous | Heterozygous | Homozygous | |||
| H1096Q/H1096Q | 22 | 6 | 4 | - | - | 12 |
| R1041W/R1041W | 2 | - | 2 | |||
| G12810N/G12810N | 1 | - | 1 | |||
| G1266R/G1266R | 1 | 1 | - | |||
| G710A/G710A | 1 | 1 | ||||
| K844k-fs/K844k-fs | 1 | 1 | ||||
| W779X/W779X | 1 | 1 | ||||
| D765N/Y741X | 2 | 1 | 1 | |||
| 3400 Del C/2299InsC | 1 | 1 | ||||
| 3400 Del C/H1096Q | 1 | 1 | ||||
| 3400 Del C/G982V | 1 | 1 | ||||
| 3400 Del C/W779X | 1 | 1 | ||||
| H1096Q/M769V | 1 | 1 | ||||
| m.n.d./m.n.d. | 11 | 7 | 2 | 4 | ||
| H1096Q/m.n.d. | 8 | 3 | 1 | 4 | ||
| W 778 X/m.n.d. | 1 | 1 | ||||
| W779X/m.n.d. | 2 | 1 | 1 | |||
| D765N/m.n.d. | 2 | 2 | ||||
| G710S/m.n.d. | 1 | 1 | ||||
| M769V/m.n.d. | 1 | 1 | ||||
| 2299 InsC/m.n.d. | 1 | 1 | ||||
| Total | 63 | 22 | 9 | 2 | 0 | 32 |
m.n.d.: Mutation not detected.
In 2 Wilson patients the previously undescribed 5’UTR -68 C>T (heterozygote) genetic variation could be detected.
Seven of the 11 patients where no Wilson disease gene mutation could be found so far and 8 of the 16 patients with a single identified Wilson disease gene mutation showed one of the variations in Atox1.
5’UTR -99 T>C
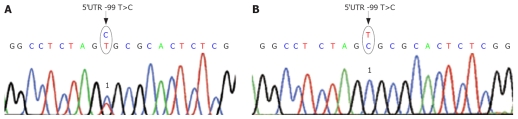
This polymorphism 5’UTR -99 T>C (Figure 1A) is already described in the database (NCBI Sequence Viewer: NM_004045: “dbSNP: 1 549 921”). 35% (22 patients) showed this substitution of the base T by C heterozygous, the other nine patients (14%) were homozygous for this variation.
Figure 1.
Sequence analysis of genomic DNA of the Atox1- gene 5`UTR -99T>C site. A: Heterozygote in this site. 1Heterozygote substitution of the base T by C on position -99 in front of the beginning of the start codon located in exon 1; B: Homozygote in this site. 1Homozygote substitution of the base T by C on position -99 in front of the beginning of the start codon located in exon 1.
Among these 22 patients with the heterozygous variation 7 persons presented with primarily hepatic symptoms, 7 patients with primarily neurological symptoms, and 3 patients with hepatic and neurological symptoms. Five were presymptomatic at diagnosis.
The nine patients with homozygous variation are composed of 3 patients with primarily hepatic symptoms, 3 persons with primarily neurological symptoms, 1 patient with hepatic and neurological symptoms, and 2 patients diagnosed preclinically by family screening (Figure 1B).
The heterozygote variation could also be found in one healthy control person, homozygote variation could be found in two persons without WD. The healthy control person, [ALT 24 U/L, AST 31 U/L (-35), copper 21 μmol/L (12-35), ceruloplasmin 0.27 g/L (0.2-0.6)] and two persons had no signs of Wilson disease (based on clinical and basic laboratory testing). They had normal levels of serum ceruloplasmin, copper and liver function tests.
5’UTR -68 C>T (heterozygot)
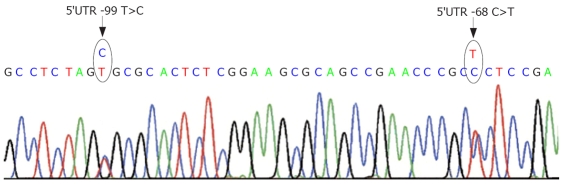
This additional variation of the 5’UTR could be found in two patients. One of them presented with the hepatic presentation, the other one had a mixed presentation (Figure 2).
Figure 2.
Localization of the 5'UTR -68 C>T gene variation in relation to the 5'UTR -99 T>C variation.
The detected nucleotide changes in the 5’UTR region of the Atox1 gene did not have significant association with or influence on the average age of initial manifestation of Wilson disease (Table 3). Statistical analyses revealed that the existence of a variation within the Atox1- gene showed no significance towards an earlier onset of the disease (Table 4).
Table 3.
Analysis of average ages of initial manifestation with regard to the variations detected in the Atox1 gene
| Atox1-variation | Average age of initial manifestation | n of patiens | Standard deviation |
| No variation within the sequence | 23.68 | 31 | 12.993 |
| Variation within the sequence detected | 18.88 | 25 | 9.909 |
| Total | 21.54 | 56 | 11.863 |
Table 4.
Statistical data for Analysis of age of initial manifestation
|
Not standardized coefficients |
Standardized coefficients |
||||
| Model | B | Standard error | Beta | T | Significance |
| ATP7B | 4.042 | 1.970 | 0.272 | 2.052 | 0.045 |
| Atox1 | -5.312 | 3.079 | -0.225 | -1.725 | 0.091 |
DISCUSSION
When sequencing the exons of the Wilson disease gene in clinically proven Wilson disease patients, only a single or even no mutation can be detected in 5%-10% of all patients depending on the population examined and the analyzing laboratory. In our study, a single Wilson disease gene mutation could only be identified in 16 patients and no Wilson disease gene mutation could be found in 11 patients so far. However complete sequencing is still under way in most of these patients.
Sixty three patients were analyzed as potential candidates of an Atox1 caused Wilson disease like disease. 31 out of these 63 patients had variations within the Atox1 gene. In terms of clinical presentation (age of onset, hepatic/neurologic onset), laboratory tests (serum ceruloplasmin, serum copper, 24 h urinary copper excretion) and clinical course (improvements, drug reactions, side effects, initial neurological worsening) there were no significant differences between the 31 WD patients with Atox1 changes compared to the remaining 32 patients without Atox1 changes. As these sequence changes were in the 5’UTL region leaving the translated regions as well as the splicing sites and the classical translation initiation complex site of the gene intact, it seemed unlikely, that these detected changes could cause a Wilson disease like disease by themselves or be able to significantly influence the clinical course of Wilson disease. On the other hand, the highly conserved region of the translated Atox1 gene in humans can be evidence for a vital role of Atox1 protein in human metabolism or embryonic development. This also might explain the still unknown phenotype of Atox1-mutation associated diseases[2], even though different roles and regulatory factors for Atox1 in human metabolism are emerging out of recent studies[28–31]. One might speculate about a Menkes disease like phenotype resulting from a complete disruption of both functional alleles of Atox1 as suggested by Atox1 knockout mice data[32].
The association between the Atox1 variations and the changes in the age of onset was weak in this study similar to data published before[2].
Taken together, Atox-1 associated modification of Wilson disease or Menkes disease are still not seen so far and the absence of mutations in the coding regions of the Atox1 gene speak for an essential role of wild type Atox1 in human metabolism.
COMMENTS
Background
Cytoplasmic copper has to be transferred into the cellular excretory pathway by copper transport pumps. In patients with the copper storage disease “Wilson disease” the copper transporter ATP7B is defective. This transmembranous ATPase receives cytoplasmic copper from the copper chaperone antioxidant-1 (Atox1).
Research frontiers
Patients with Wilson disease show a wide variation in their clinical presentation. Proteins interacting with the ATP7B copper transporter, such as Atox1, COMMD1 or chemical such as Platinum-complexes are one important area in explaining this phenomenon.
Related publications
Lutsenko S, LeShane ES, Shinde U. Biochemical basis of regulation of human copper transporting ATPases. Arch Biochem Biophys 2007; 463: 134-148 [PMID: 17562324]; Singleton C, Le Brun NE. Atx-1 like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals 2007; 20: 275-289 [PMID: 17225061]; Merle U, Schaefer M, Ferenci P, Stremmel W. Clinical presentation, diagnosis and long-term outcome of Wilson’s disease: a cohort study. Gut 2007; 56: 115-120, 314 [PMID: 16709660].
Innovations and breakthroughs
Proper ATP7B function is regulated by cytoplasmic factors (e.g. COMMD1, Atox1), cytoskeletal factors (e.g. dynactin subunit p62) and by hormones (e.g. prolactin, oestrogens). ATP7B and its sister protein ATP7A (Menkes protein) are regulated by tissue specific factors and by hormones and can be present simultaneously within one cell.
Applications
Research on modification factors of Wilson disease are aimed to identify protective factors within the clinical course of Wilson disease and providing them to affected patients.
Terminology
Atox1: antioxidant-1, small cytoplasmic protein with copper binding sides; ATP7B:copper transporting ATPase encoded by the Wilson disease gene; Wilson disease: autosomal recessive copper storage disease due to malfunction of ATP7B.
Peer review
This paper was well designed and analysed in large-scale patients. Although the results are in part negative, they are important to the scientific community.
Acknowledgments
We sincerely thank the patients for their help and willingness to participate in this study. We thank Professor. Ferenci, Department of Gastroenterology and Hepatology, Vienna for Wilson disease gene analysis and Cathrin Thunert and Uta Merle for collecting clinical data and patient specimens. There is no conflict of interest for all authors of this study.
Supported by German Research Foundation DFG; Junior-Grant Faculty of Medicine, University of Heidelberg
Peer reviewer: Valerio Nobili, Dr, Liver Unit, Research Institute, Bambino Gesù Children’s Hospital, S. Onofrio 4 Square, Rome 00165, Italy
S- Editor Sun YL L- Editor Alpini GD E- Editor Lu W
References
- 1.Linder MC, Hazegh-Azam M. Copper biochemistry and molecular biology. Am J Clin Nutr. 1996;63:797S–811S. doi: 10.1093/ajcn/63.5.797. [DOI] [PubMed] [Google Scholar]
- 2.Hamza I, Schaefer M, Klomp LW, Gitlin JD. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci USA. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheinberg IH, Sternlieb I. Major Problems in International Medicine. Philadelphia: W.B. In: Jr Smith LH., editor. Wilson‘s disease. WB Saunders Company: Saunders Company, Philadelphia; 1988. pp. 188–213. [Google Scholar]
- 4.Merle U, Schaefer M, Ferenci P, Stremmel W. Clinical presentation, diagnosis and long-term outcome of Wilson's disease: a cohort study. Gut. 2007;56:115–120. doi: 10.1136/gut.2005.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das SK, Ray K. Wilson's disease: an update. Nat Clin Pract Neurol. 2006;2:482–493. doi: 10.1038/ncpneuro0291. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi Y, Heiny ME, Gitlin JD. Isolation and characterization of a human liver cDNA as a candidate gene for Wilson disease. Biochem Biophys Res Commun. 1993;197:271–277. doi: 10.1006/bbrc.1993.2471. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer M, Gitlin JD. Genetic disorders of membrane transport. IV. Wilson's disease and Menkes disease. Am J Physiol. 1999;276:G311–G314. doi: 10.1152/ajpgi.1999.276.2.G311. [DOI] [PubMed] [Google Scholar]
- 8.Yang XL, Miura N, Kawarada Y, Terada K, Petrukhin K, Gilliam T, Sugiyama T. Two forms of Wilson disease protein produced by alternative splicing are localized in distinct cellular compartments. Biochem J. 1997;326(Pt 3):897–902. doi: 10.1042/bj3260897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutsenko S, LeShane ES, Shinde U. Biochemical basis of regulation of human copper-transporting ATPases. Arch Biochem Biophys. 2007;463:134–148. doi: 10.1016/j.abb.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenney S, Cox DW. (Since 2005) Open access data base: Wilson disease mutation database. Available from: URL: http://www.uofa-medical-genetics.org/wilson/index.php. [Google Scholar]
- 11.Cox DW. Review: molecular approaches to inherited liver disease. Focus on Wilson disease. J Gastroenterol Hepatol. 1997;12:S251–S255. doi: 10.1111/j.1440-1746.1997.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 12.Brewer GJ. Recognition, diagnosis, and management of Wilson's disease. Proc Soc Exp Biol Med. 2000;223:39–46. doi: 10.1046/j.1525-1373.2000.22305.x. [DOI] [PubMed] [Google Scholar]
- 13.Gollan JL, Gollan TJ. Wilson disease in 1998: genetic, diagnostic and therapeutic aspects. J Hepatol. 1998;28 Suppl 1:28–36. doi: 10.1016/s0168-8278(98)80373-5. [DOI] [PubMed] [Google Scholar]
- 14.Riordan SM, Williams R. The Wilson's disease gene and phenotypic diversity. J Hepatol. 2001;34:165–171. doi: 10.1016/s0168-8278(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 15.Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- 16.Schiefermeier M, Kollegger H, Madl C, Polli C, Oder W, Kuhn H, Berr F, Ferenci P. The impact of apolipoprotein E genotypes on age at onset of symptoms and phenotypic expression in Wilson's disease. Brain. 2000;123 Pt 3:585–590. doi: 10.1093/brain/123.3.585. [DOI] [PubMed] [Google Scholar]
- 17.Merle U, Stremmel W, Gessner R. Influence of homozygosity for methionine at codon 129 of the human prion gene on the onset of neurological and hepatic symptoms in Wilson disease. Arch Neurol. 2006;63:982–985. doi: 10.1001/archneur.63.7.982. [DOI] [PubMed] [Google Scholar]
- 18.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 19.Culotta VC, Lin SJ, Schmidt P, Klomp LW, Casareno RL, Gitlin J. Intracellular pathways of copper trafficking in yeast and humans. Adv Exp Med Biol. 1999;448:247–254. doi: 10.1007/978-1-4615-4859-1_22. [DOI] [PubMed] [Google Scholar]
- 20.Valentine JS, Gralla EB. Delivering copper inside yeast and human cells. Science. 1997;278:817–818. doi: 10.1126/science.278.5339.817. [DOI] [PubMed] [Google Scholar]
- 21.Lin SJ, Pufahl RA, Dancis A, O'Halloran TV, Culotta VC. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem. 1997;272:9215–9220. [PubMed] [Google Scholar]
- 22.de Silva D, Davis-Kaplan S, Fergestad J, Kaplan J. Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J Biol Chem. 1997;272:14208–14213. doi: 10.1074/jbc.272.22.14208. [DOI] [PubMed] [Google Scholar]
- 23.Klomp LW, Lin SJ, Yuan DS, Klausner RD, Culotta VC, Gitlin JD. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J Biol Chem. 1997;272:9221–9226. doi: 10.1074/jbc.272.14.9221. [DOI] [PubMed] [Google Scholar]
- 24.Walker JM, Tsivkovskii R, Lutsenko S. Metallochaperone Atox1 transfers copper to the NH2-terminal domain of the Wilson's disease protein and regulates its catalytic activity. J Biol Chem. 2002;277:27953–27959. doi: 10.1074/jbc.M203845200. [DOI] [PubMed] [Google Scholar]
- 25.DiDonato M, Hsu HF, Narindrasorasak S, Que L Jr, Sarkar B. Copper-induced conformational changes in the N-terminal domain of the Wilson disease copper-transporting ATPase. Biochemistry. 2000;39:1890–1896. doi: 10.1021/bi992222j. [DOI] [PubMed] [Google Scholar]
- 26.Vanderwerf SM, Cooper MJ, Stetsenko IV, Lutsenko S. Copper specifically regulates intracellular phosphorylation of the Wilson's disease protein, a human copper-transporting ATPase. J Biol Chem. 2001;276:36289–36294. doi: 10.1074/jbc.M102055200. [DOI] [PubMed] [Google Scholar]
- 27.Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. Novel role of antioxidant-1 (atox1) as a copper dependent transcription factor involved in cell proliferation. J Biol Chem. 2008;283:9157–9167. doi: 10.1074/jbc.M709463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singleton C, Le Brun NE. Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals. 2007;20:275–289. doi: 10.1007/s10534-006-9068-1. [DOI] [PubMed] [Google Scholar]
- 29.de Bie P, van de Sluis B, Burstein E, van de Berghe PV, Muller P, Berger R, Gitlin JD, Wijmenga C, Klomp LW. Distinct Wilson's disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology. 2007;133:1316–1326. doi: 10.1053/j.gastro.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain F, Wittung-Stafshede P. Impact of cofactor on stability of bacterial (CopZ) and human (Atox1) copper chaperones. Biochim Biophys Acta. 2007;1774:1316–1322. doi: 10.1016/j.bbapap.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Arnesano F, Scintilla S, Natile G. Interaction between platinum complexes and a methionine motif found in copper transport proteins. Angew Chem Int Ed Engl. 2007;46:9062–9064. doi: 10.1002/anie.200703271. [DOI] [PubMed] [Google Scholar]
- 32.Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin JD. The metallochaperone Atox1 plays a critical role in perinatal copper homeostasis. Proc Natl Acad Sci USA. 2001;98:6848–6852. doi: 10.1073/pnas.111058498. [DOI] [PMC free article] [PubMed] [Google Scholar]