Abstract
Use of an NF-κB-dependent selectable marker facilitated the isolation of a cell line containing a cDNA encoding Act1, an NF-κB activator. Act1 associates with and activates IκB kinase (IKK), leading to the liberation of NF-κB from its complex with IκB. Many signaling pathways that liberate NF-κB also activate activating transcription factor (ATF) and activator protein 1 (AP-1) through Jun kinase (JNK). Act1 also activates JNK, suggesting that it might be part of a multifunctional complex involved in the activation of both NF-κB and JNK. Act1 fails to activate NF-κB in an IL-1-unresponsive mutant cell line in which all known signaling components are present, suggesting that it interacts with an unknown component in IL-1 signaling.
Members of the Rel family of transcription factors, and especially NF-κB, play important roles in immune and inflammatory responses, cell survival, and stress responses by regulating the expression of many cellular and viral genes (1, 2). In most cell types, NF-κB is kept inactive in the cytoplasm through interaction with the IκB inhibitory proteins. On stimulation with certain cytokines (e.g., IL-1 and tumor necrosis factor α [TNF-α]) or other agents (e.g., phorbol ester or double-stranded RNA) or in response to the activation of Toll receptors by lipopolysaccharide or other ligands, the IκB proteins are phosphorylated on specific serine residues, followed by their ubiquitination and rapid degradation. The liberated NF-κB translocates to the nucleus and activates transcription. In addition, phosphorylation of the transactivation domain of the p65 subunit of the major p65/p50 NF-κB heterodimer in response to many or all of the same signals greatly enhances the transcriptional response (3, 4)
Many of the signals that lead to the activation of NF-κB converge on a high molecular weight oligomeric protein, the serine-specific IκB kinase (IKK) (5–10). IKK is composed of at least three subunits, the catalytic subunits IKKα and -β, and the regulatory subunit, IKKγ. IKKα and -β have very similar primary structures (52% identity), with protein kinase domains near their N termini and leucine zipper and helix–loop–helix motifs near their C termini. IKKγ lacks a kinase domain but contains long stretches of coiled-coil sequence, which function in protein–protein binding, including a C-terminal leucine zipper motif. Precisely how IKKγ participates in IKK signaling is unknown, but this regulatory protein has been proposed to interact with upstream components in response to cellular activation signals. Although IKK can be activated by members of the mitogen-activated protein kinase kinase kinase (MAP3K) family (11 and 12), including extracellular signal-regulated kinase kinase kinase 1 (MEKK1) and NF-κB inducing kinase (NIK), the identity of the upstream IKK activators remains to be identified. IL-1, TNF-α, and other related signaling pathways also activate the transcription factors activating transcription factor (ATF) and activator protein 1 (AP-1), through the activation of Jun kinase (JNK) (13, 14). Signal-induced activation of JNK may diverge from NF-κB activation at the level of a group of adapter proteins, the tumor necrosis factor receptor-associated factors (TRAFs). Activated TRAFs activate MEKK1, which in turn activates JNK (15, 16).
In this report, we present a strategy for cloning new components of NF-κB-dependent signaling pathways. In the human embryonic kidney cell line 293, an NF-κB-containing upstream region of the human IL-1-responsive gene E-selectin was used to drive the expression of proteins providing resistance to zeocin (Zeo) or sensitivity to herpes simplex virus thymidine kinase (TK) (17). 293-TK/Zeo cells, providing negative selection against the expression of TK in response to IL-1 have been used to isolate IL-1-unresponsive mutant cell lines (18). Here, we show that the same cells can be used to clone cDNAs whose expression leads to the constitutive activation of NF-κB, by using Zeo for selection. One such cDNA encodes NF-κB activator 1 (Act1), a protein that activates both NF-kB and JNK constitutively.
Materials and Methods
Biological Reagents and Cell Culture.
Recombinant human IL-1β was provided by the National Cancer Institute. An Ab against IKKγ was a kind gift from Dr. Zhaodan Cao at Tularik (South San Francisco, CA), anti-pJNK was from New England Biolabs, anti-IKKα from Santa Cruz Biotechnology, and anti-M2 from Sigma. 293-TK/Zeo cells (18) were maintained in DMEM, supplemented with 10% FCS, penicillin G (100 μg/ml), and streptomycin (100 μg/ml).
Retroviral cDNA Library and Infection of Cells.
A HaCaT retroviral cDNA library, obtained from laboratory of Harvey Lodish (Whitehead Institute, Cambridge, MA; ref. 19), was amplified in Escherichia coli and then introduced into the packaging cell line Bosc23 (20) to obtain a high-titer retroviral cDNA library (19). We produced viral supernatant suspensions by transfecting approximately 2 × 106 Bosc23 cells per 60-mm dish, plated 24 h earlier, with 5 μg of library DNA. Supernatant suspensions, recovered 48 h after transfection, were used immediately for infection or snap frozen in dry ice and stored at −80°C. Retroviruses carrying the green fluorescent protein (pMX-GFP; ref. 19) were produced in parallel with the retroviral cDNA library and used to infect NIH 3T3 cells, which were then scanned for the expression of GFP. The titer of the retroviral cDNA library was estimated from that of the pMX-GFP retroviruses produced in parallel. Titers of virus 2- to 5 × 105/ml were typical. Supernatant suspensions containing the recombinant retroviruses were incubated with 293-TK/Zeo cells for 6–9 h in normal medium containing 4 μg/ml of Polybrene (Sigma). Nine hours after infection, the supernatant suspension was removed, and the cells were cultured for 36 h more in normal medium. The infected cells were then selected with Zeo (100 μg/ml) and IL-1 (100 units/ml). Selection medium was replaced at least every 5 days, and clones were isolated after about 20 days.
Recombinant Plasmids.
The Act1 cDNA insert was obtained by reverse transcription PCR from Zeo-selected clones by using primers (5′ primer, XH15, GGCATCGCAGCTTGGATACA; 3′ primer, XH18, GCTAGCTTGCCAAACCTACAGG) derived from the retroviral vector pMX (19) and cloned into the pCR 2.1 PCR vector (Invitrogen). The cytomegalovirus (CMV)-Act1-flag plasmid was constructed with the Act1 cDNA insert (obtained through PCR) with a 5′ primer that encodes the flag-tag sequence Met-Asp-Tyr-Lys-Asp-Asp-Asp-Lys. The product was cloned in the pRcCMV mammalian expression vector (Invitrogen). Act1 deletion fragments were generated by PCR and also cloned in pRcCMV. The NIK dominant negative mutant KK429–430AA was obtained as a kind gift from Zhaodan Cao (Tularik). pE-selectin-Luc, an NF-κB-dependent E-selectin-Luc reporter plasmid, was described by Schindler and Baichwal (21).
Transfection and Reporter Assays.
For stable transfections, 2 × 105 cells were seeded onto a 10-cm plate and cotransfected the following day by the calcium phosphate method with 10 μg of each expression vector and 1 μg of pBabePuro. After 48 h, the cells were selected with 1 μg/ml of puromycin until clones appeared. For reporter assays, 2 × 105 cells were transfected by the same procedure with 1 μg of pE-selectin-Luc, 1 μg of pSV2-β-galactosidase and 100 ng of each expression construct. After 48 h, the cells were split onto two 35-mm plates and, the next day, stimulated with IL-1 for 4 h before harvest. Luciferase and β-galactosidase activities were determined by using the Promega luciferase assay system and chemiluminescent reagents, respectively.
Gel Shift Assays.
An NF-κB binding site (5′-GAGCAGAGGGAAATTCCGTAACTT-3′) from the IFN-inducible protein 10 (IP-10) gene (22) was used as a probe. Complementary oligonucleotides, end-labeled with polynucleotide kinase (Boehringer Mannheim) and [γ-32P]ATP, were annealed by slow cooling. Approximately 20,000 cpm of probe were used per assay. Cytoplasmic extracts were prepared as described by Kessler et al. (23). The binding reaction was carried out at room temperature for 20 min in a total volume of 20 μl containing 20 mM Hepes buffer, pH 7.0/10 mM KCl/0.1% Nonidet P-40/0.5 mM DTT/0.25 mM PMSF/10% glycerol.
Coimmunoprecipitation, Immunoblotting, and Kinase Assays.
For coimmunoprecipitation, cell extracts were incubated with 1 μl of anti-IKKγ polyclonal Ab (Zhaodan Cao; Tularik) or preimmune serum for 4 h, followed by a 1-h incubation with 50 μl of protein A-Sepharose beads (prewashed and resuspended in PBS at a 1:1 ratio). After incubation, the beads were washed 5 times with lysis buffer (24), followed by immunoblotting with anti-M2 Ab (Sigma). For IκB kinase assays, cell lysates were immunoprecipitated with anti-IKKα and collected on protein A-Sepharose beads. Kinase reactions were performed in 50 μl of buffer containing 20 mM Hepes, pH 7.0/20 mM MgCl2/1 mM ATP/10 mCi of [γ-32P]ATP at 30°C for 30 min. The substrate was 2 μg of glutathione S-transferase (GST)-IκB, residues (1–54a) (Joseph DiDonato, Cleveland Clinic Foundation, Cleveland, OH). Samples were analyzed by 10% SDS/PAGE, followed by autoradiography.
Northern Analyses.
Total RNA was isolated with the TRIzol Reagent (GIBCO BRL). Gene-specific probes were made with a random priming kit (Amersham). Transfers to Hybond-N membrane were performed according to the procedure provided by Amersham. A human tissue blot was obtained from CLONTECH.
Results
Act1 Contains a Helix–Loop–Helix Domain.
TK and Zeo constructs were used in which the −730 to +52 fragment of the E-selectin promoter drives reporter gene expression (18). Induction of the E-selectin promoter depends mainly on the κB sites (21). The E-selectin-TK and E-selectin Zeo constructs were cotransfected into 293 cells, and a clone was selected that survives in gancyclovir, dies completely in gancyclovir plus IL-1, dies in Zeo, and survives in Zeo plus IL-1. Previously, the IL-1-regulated negative selection provided by 293-TK/Zeo cells was used successfully to generate IL-1-unresponsive mutants (18). Here, we report that the Zeo-positive selection built into the same cells can be used to clone cDNAs whose expression leads to constitutive activation of NF-κB. We introduced a retroviral cDNA library made from human keratinocyte HaCaT cells into 293-TK/Zeo cells and then selected the infected cells in Zeo (100 μg/ml). A 2.6-kb cDNA insert was recovered by reverse transcription-PCR from one of the obtained clones. A database search showed that the sequence of the cDNA clone matched that of a previously cloned partial cDNA with no known function. The 2.6-kb insert contains an ORF predicted to encode 574 aa. This 60-kDa polypeptide (Fig. 1A) is named Act1 (NF-κB activator 1). Searches of databases showed that Act1 matches a predicted protein sequence from the human genomic clone PAC 487J7.
Figure 1.
Protein sequences. (A) The sequence of Act1. (B) Comparison of the helix–loop–helix domains of Act1 and USF-1. Identical, conserved, and homologous residues are indicated as solid lines, two-dotted bars, or one-dotted bars, respectively. Positions of the residues of Act1 are numbered.
Act1 lacks any known catalytic domain. Through a sequence similarity search (25), we found that residues 135 to 190 share significant homology with the helix–loop–helix domain of upstream stimulatory factor-1 (USF-1) (26). Within this region, the two proteins are 35% identical and share 60% homology (Fig. 1B). Furthermore, secondary structure prediction identified residues 135–155 and 182–190 to be helical, in agreement with the helix–loop–helix region in the crystal structure of USF-1 (26). USF-1 has a basic domain upstream of the helix–loop–helix region, which is required for DNA binding and its activity as a transcription factor. The basic region is not found in Act1, suggesting that Act1 is not a USF-1-like transcription factor. We also found that, using coils (27), residues 470–500 are likely to form a coiled-coil domain.
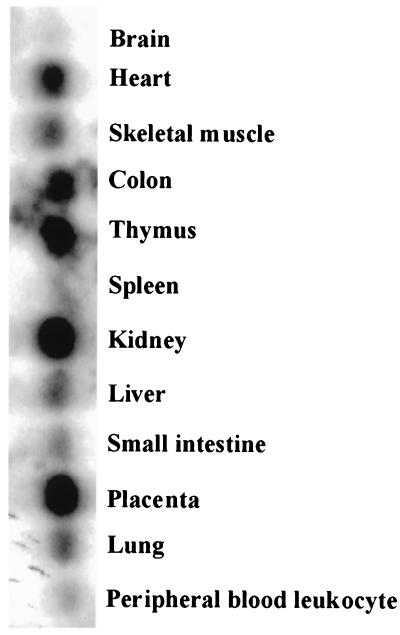
Act1 is expressed highly in thymus, kidney and placenta; moderately in heart, skeletal muscle, colon, liver, lung and small intestine; and expressed at a very low level in brain, spleen, and peripheral blood leukocytes (Fig. 2).
Figure 2.
Northern analyses. RNA from various human tissues were probed with Act1 cDNA.
Act1 Constitutively Activates Both NF-κB and JNK.
The Act1 cDNA was subcloned into a CMV expression vector and cotransfected with E-selectin-Luc into 293 cells. The E-selectin promoter activity was drastically increased without stimulation in cells cotransfected with Act1, as compared with cells cotransfected with vector only (see below in Fig. 7). Gel-shift analysis showed that NF-κB was indeed constitutively activated in 293 cells stably transfected with CMV-Act1 (Fig. 3A). This constitutive activation explains how the clone expressing Act1 cDNA survived Zeo selection. Similarly, signaling components in the IL-1, TNFα, and related pathways often constitutively activate NF-κB when overexpressed. Therefore, Act1 is likely to participate in pathway(s) involving NF-κB activation.
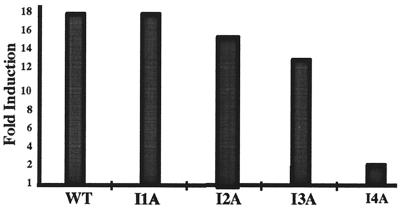
Figure 7.
Act1 function in mutant cells unresponsive to IL-1. E-selectin-Luc was cotransfected with CMV-Act1. The data are presented as fold induction of luciferase activity of cells transfected with CMV-Act1, compared with cells transfected with the control vector.
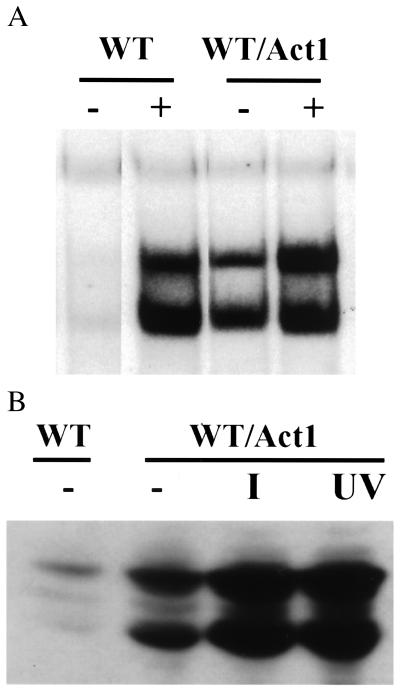
Figure 3.
Act1-dependent functions. (A) Activation of NF-κB. 293-TK/Zeo cells with and without Act1 were either untreated or treated with IL-1 for 15 min. Cell extracts were used for the gel-shift assays. (B) Activation of JNK. 293-TK/Zeo cells transfected with CMV-Act1 were untreated or treated with IL-1 (100 units/ml) or UV radiation (40 J/m) (2). Cell extracts were analyzed after Western transfer by using anti-phospho-JNK. Untransfected wild-type cells were used as controls.
Some signaling pathways that lead to NF-κB activation also activate MAP kinases and JNK, which in turn phosphorylate and activate ATF and AP-1 (13–16). Interestingly, JNK was constitutively activated in 293 cells transfected with CMV-Act1 (Fig. 3B). The fact that Act1 can activate both NF-κB and JNK suggests that it lies upstream of the branch point that separates these two pathways.
Act1 Activates NF-κB Through the NIK–IKK Complex.
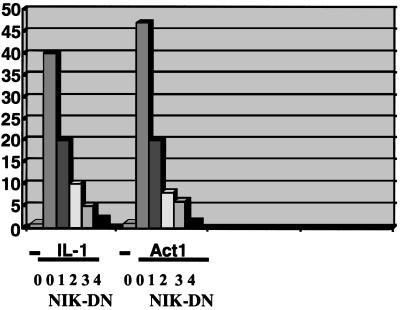
It has been suggested that a MAP3K family member, NIK, can activate IKK directly by forming a NIK-IKK complex (11 and 12). A kinase-inactive NIK-dominant negative mutant inhibited Act1 induction of the E-selectin promoter as efficiently as it did IL-1-dependent induction (Fig. 4), suggesting that NIK or a similar protein does function in the Act1-mediated NF-κB activation pathway.
Figure 4.
Effect of a NIK dominant-negative mutant on the activation of NF-κB by Act1. Increasing amounts (50–500 ng) of NIK-DN were cotransfected with Act1 and E-selectin-Luc into 293-TK/Zeo cells. The effect of NIK-DN on Act1-induced E-selectin promoter activity was compared with its effect on IL-1-induced promoter activity.
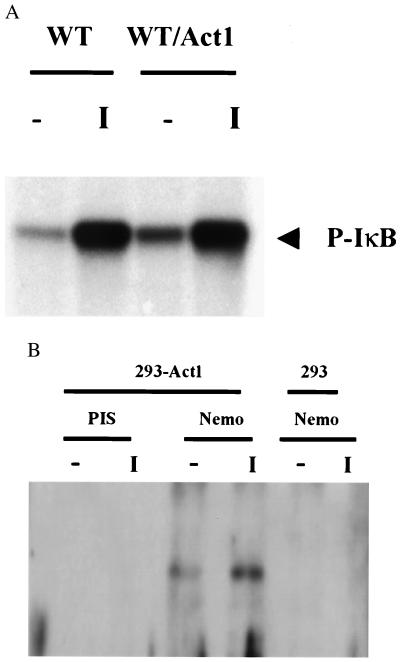
Many signaling pathways involving NF-κB activation converge on IKK. This enzyme was activated constitutively in cells transfected with CMV-Act1 (Fig. 5A), indicating that the activation of NF-κB by Act1 probably runs through IKK. To examine whether Act1 forms a complex with IKK directly, anti-IKK was used to immunoprecipitate the IKK complex in cells stably transfected with flag-tagged Act1 (CMV-Act1-flag) and then probed with anti-flag Ab (M2). Act1 was detected in the IKK complex pulled down by the IKKγ Ab, strongly suggesting that it does form a complex with IKK (Fig. 5B).
Figure 5.
Interaction of Act1 with IKK. (A) IκB kinase assay. 293-TK/Zeo cells with or without Act1 transfection were untreated or treated with IL-1 for 15 min. Cell extracts were immunoprecipitated with anti-IKKγ, followed by a kinase assay using GST-IκB (amino acids 1–55) as substrate. (B) Interaction of Act1 with IKKγ. 293-TK/Zeo cells with or without expression of Act1-flag were untreated or treated with IL-1 for 3 min. Cell extracts were immunoprecipitated with anti-IKKγ and probed with an Ab directed against the flag tag.
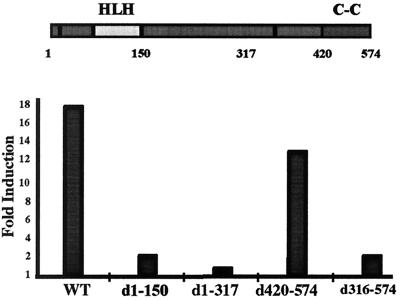
Act1 Activates IKK Through Its Helix–Loop–Helix Domain.
Act1 contains a helix–loop–helix domain in its N-terminal portion and a coiled-coil structure at the C terminus (Fig. 2B). To examine the roles of these two domains, deletion constructs were generated (Fig. 6). Disruption of the helix–loop–helix domain at the N terminus (d1–150) completely abolished the ability of Act1 to activate NF-κB. This deletion also abolished the interaction of Act1 with the IKK complex (data not shown), suggesting that this N-terminal domain is important for protein–protein interactions. Deletion of the coiled-coil region had no effect on Act1-induced NF-κB activation. Further deletion from the C terminus (d316–573) abolished this activity, suggesting that the region between residues 316 to 420 is also important for function.
Figure 6.
Deletion analysis of Act1. The constructs shown were cotransfected into 293-TK/Zeo cells with E-selectin-Luc. The data are presented as fold induction of luciferase activity of the cells transfected with Act1 constructs, compared with cells transfected with the control vector.
Act1 Fails To Activate NF-κB in IL-1-Unresponsive I4A Cells.
We previously generated several IL-1-unresponsive cell lines by selecting mutagenized 293-TK/Zeo cells in IL-1 and gancyclovir (18). Mutant clone I1A lacks IRAK (IL-1 receptor-associated kinase), and mutants I2A and I3A are defective in components upstream of IRAK (18). IL-1-induced NF-κB and JNK activation were both abolished in all three mutants. Mutant I4A is defective only in IL-1-induced NF-κB activation, with normal JNK activation (unpublished data). All known components in the IL-1-signaling pathway are expressed normally in mutant I4A, suggesting that it is defective in an unknown component specific for NF-κB activation (unpublished data). We tested the Act1-mediated activation of NF-κB in all of these mutants by cotransfection with E-selectin-Luc. Interestingly, the E-selectin promoter was activated constitutively in I1A, I2A, and I3A but not in I4A cells (Fig. 7), suggesting that Act1 fails to activate NF-κB in the latter cells. Act1 was also stably cotransfected with pBabepuro into both 293-TK/Zeo and I4A cells. The transfected cells were first selected in puromycin (1 μg/ml), followed by selection in Zeo (100 μg/ml). Similar numbers of puromycin-resistant clones were obtained from the transfected 293-TK/Zeo and I4A cells, suggesting that the transfection efficiency was about the same for the two cell lines. Western analysis showed that Act1 protein was expressed in puromycin-resistant clones from either the transfected 293-TK/Zeo or I4A cells (data not shown). However, although many of the puromycin-resistant clones from Act1-transfected 293-TK/Zeo cells also survived Zeo selection, those from the transfected I4A cells were all Zeo sensitive. Taken together, the data clearly reveal that Act1 is unable to activate NF-κB in I4A cells, indicating that the lost or defective protein in these mutant cells, necessary for IL-1-induced NF-kB activation, is also required for Act1-mediated NF-κB activation.
Discussion
We report the identification and initial characterization of an NF-κB-activating protein. The activation of NF-κB by Act1 appears to be through the NIK-IKK complex, because a NIK dominant-negative mutant protein inhibited Act1-induced NF-κB activation. Act1 coimmunoprecipitated with IKKγ and Act1 activated IKK. Interestingly, Act1-mediated NF-κB activation was abolished in the IL-1-unresponsive mutant I4A, which is defective in an unknown component specific for IL-1-induced NF-κB activation, suggesting that Act1 is part of either the IL-1 signaling pathway or a related pathway. Further biochemical analysis of how Act1 responds to various proinflammatory stimuli, including IL-1, TNF, and lipopolysaccharide (LPS), and identification of Act1-interacting proteins should help to reveal the Act1-mediated pathway(s). These studies will be greatly facilitated by Act1-specific Abs. Also, the generation and analysis of Act1 knock-out mice will provide information about its physiological function(s).
Act1 also activates JNK. Most of the ligands that lead to NF-κB activation, including LPS, IL-1, and TNF, also activate MAP kinases and JNK, which in turn phosphorylate and activate the transcription factors ATF and AP-1 (13–16). The signaling components proximal to each class of receptors are unique. The TNF pathway uses TRAF2, TRAF5 and the receptor-interacting protein (RIP) kinase, whereas the Toll and IL-1 pathways use TRAF6 and IRAK. These different signals then converge on IKK to activate NF-κB (5–10). Signal-induced activation of the MAP kinases and JNK may diverge from NF-κB activation at the level of the TRAF proteins (12, 15, 16, 28). The activated TRAFs activate MEKK1, which in turn activates MAP kinases and JNK. On the other hand, both MEKK1 and NIK have been implicated in the activation of IKK (11, 12). Future research is required to clarify the exact functions of these MAP3Ks in different signaling pathways. Nevertheless, it is clear that each pathway bifurcates at a certain point to activate NF-κB and ATF/AP-1. The fact that Act1 activates both NF-κB and JNK indicates that it functions before this point. Because Act1 does not have any known catalytic domain, it is likely that it functions as a structural component through protein–protein interactions. Act1 forms a complex with IKK through its helix–loop–helix domain and leads to the activation of NF-κB. It is possible that it forms another complex to activate the pathway leading to the activation of JNK. Alternatively, Act1 might be part of a multifunctional complex that leads to the activation of both NF-κB and JNK.
The transcription factor NF-κB is a central mediator of mammalian immune responses. Many stimuli activate NF-κB, including cytokines, a wide variety of bacteria or bacterial products, viruses, and also various environmental insults (29). The signaling pathways activated by most of these stimuli are still largely unknown. In this paper, we present a strategy for cloning new components of NF-κB-dependent signaling. The cloning of Act1 represents an example in which 293-TK/Zeo cells are used to clone cDNAs whose expression leads to constitutive activation of NF-κB, by using Zeo for selection. The same cells, providing selection against the expression of TK, can also be used to clone cDNAs whose expression leads to inhibition of signal-induced NF-κB activation. Isolation of both positive and negative regulators of NF-κB activation provides tools for enumerating and elucidating the various NF-κB signaling pathways. The isolated components can also be used as bait to identify interacting proteins in biochemical or yeast two-hybrid experiments. Targeted disruption of the corresponding genes in mice will then provide information about the physiological functions of these components and the signaling pathways mediated through them.
Acknowledgments
We thank Focco van den Akker for analyzing the Act1 protein sequence and Das Kingshuk for technical assistance. We also thank Harvey Lodish for the HaCaT retroviral cDNA library, Zhaodan Cao for anti-IKKγ, and Joseph DiDonato for the GST-IκB construct. We thank Ulrich Siebenlist for the exchange of information on Act1. This work was supported by Grants GM60020 and CA62220 from the National Institutes of Health.
Abbreviations
- IKK
IκB kinase
- JNK
Jun kinase
- MAP3K
mitogen-activated protein kinase kinase kinase
- MEKK1
extracellular signal-regulated kinase kinase kinase 1
- NIK
NF-κB-inducing kinase
- Zeo
zeocin
- TK
thymidine kinase
- ATF
activating transcription factor
- AP-1
activating protein 1
- TRAF
tumor necrosis factor receptor-associated factor
- CMV
cytomegalovirus
- Act1
NF-κB activator 1
- USF-1
upstream stimulatory factor 1
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF274303).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160265197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160265197
References
- 1.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Barnes P J, Karin M. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 3.Reddy S A, Huang J A, Liao W S. J Biol Chem. 1997;272:29167–29173. doi: 10.1074/jbc.272.46.29167. [DOI] [PubMed] [Google Scholar]
- 4.Sizemore N, Leung S, Stark G R. Mol Cell Biol. 1999;19:4798–4805. doi: 10.1128/mcb.19.7.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 6.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 7.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 8.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 9.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 10.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 11.Ling L, Cao Z, Goeddel D V. Proc Natl Acad Sci USA. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner D A, O'Hara M, Angel P, Chojkier M, Karin M. Nature (London) 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 14.O'Neill L A. Biochem Soc Trans. 1997;25:295–302. doi: 10.1042/bst0250295. [DOI] [PubMed] [Google Scholar]
- 15.Lee S Y, Reichlin A, Santana A, Sokol K A, Nussenzweig M C, Choi Y. Immunity. 1997;7:703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z G, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 17.Stark G R, Gudkov A V. Hum Mol Genet. 1999;8:1925–1938. doi: 10.1093/hmg/8.10.1925. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark G R. Mol Cell Biol. 1999;19:4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X, Liu X, Ansari D O, Lodish H F. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindler U, Baichwal V R. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majumder S, Zhou L Z, Chaturvedi P, Babcock G, Aras S, Ransohoff R M. J Neurosci Res. 1998;54:169–180. doi: 10.1002/(SICI)1097-4547(19981015)54:2<169::AID-JNR5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.Kessler D S, Veals S A, Fu X Y, Levy D E. Genes Dev. 1990;4:1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- 24.Wesche H, Henzel W J, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 25.Worley K C, Wiese B A, Smith R F. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- 26.Ferre-D'Amare A R, Pognonec P, Roeder R G, Burley S K. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupas A. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 28.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pahl H L. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]