Abstract
Background
The Huntington disease (HD) CAG repeat exhibits dramatic instability when transmitted to subsequent generations. The instability of the HD disease allele in male intergenerational transmissions is reflected in the variability of the CAG repeat in DNA from the sperm of male carriers of the HD gene.
Results
In this study, we used a collection of 112 sperm DNAs from male HD gene‐positive members of a large Venezuelan cohort to investigate the factors associated with repeat instability. We confirm previous observations that CAG repeat length is the strongest predictor of repeat‐length variability in sperm, but we did not find any correlation between CAG repeat instability and either age at the time of sperm donation or affectedness status. We also investigated transmission instability for 184 father–offspring and 311 mother–offspring pairs in this Venezuelan pedigree. Repeat‐length changes were dependent upon the sex of the transmitting parent and parental CAG repeat length but not parental age or birth order. Unexpectedly, in maternal transmissions, repeat‐length changes were also dependent upon the sex of the offspring, with a tendency for expansion in male offspring and contraction in female offspring.
Conclusion
Significant sibling–sibling correlation for repeat instability suggests that genetic factors play a role in intergenerational CAG repeat instability.
Keywords: Huntington disease, CAG repeat instability, spermatogenesis, genetic factors
Huntington disease (HD) is an autosomal dominant neurodegenerative disorder of variable onset, which results from the expansion of a polymorphic CAG repeat in exon 1 of the HD gene encoding huntingtin.1 Normal alleles at the HD locus possess up to 35 CAG repeats, while disease alleles have 36– >100 CAG repeats, with repeat lengths of 36–39 CAGs considered to be reduced‐penetrance alleles.2 Repeats in the normal range are stably inherited; in contrast, disease alleles change in length (either increase or decrease) for 70–80% of intergenerational transmissions.3,4,5 Repeat‐length changes occur in transmissions from parents of either sex, although large increases are seen predominantly in paternal transmissions.3,4,6,7,8 As the length of the CAG repeat is inversely correlated with age at onset,9,10 most cases of juvenile onset are a product of large expansions occurring with paternal transmission.
We and others have previously shown that the instability of the HD disease allele in paternal intergenerational transmissions is reflected in the variability of the CAG repeat in DNA from the sperm of male carriers of the HD gene, indicating that the instability occurs in spermatogenesis.3,6,8,11 In this study, we performed a detailed investigation of the factors that contribute to the instability of the HD CAG repeat, using an extensive collection of sperm samples and transmission data from a large Venezuelan HD pedigree.12
Subjects and methods
Samples
All subjects in this study either had diagnosed HD or were asymptomatic gene‐positive members of families with HD from the US–Venezuela Collaborative HD project12 with the exception of eight people evaluated through the New England HD Research Center who donated the sperm/lymphoblastoid samples. The study was approved by the institutional review board and appropriate informed consent was obtained from all participants.
All subjects had expanded HD CAG repeat lengths ⩾36. DNA was prepared from sperm, lymphoblastoid cell lines or fresh blood.10 Sperm samples were a crude preparation pelleted from semen and washed. We tested sperm and lymphoblastoid cell DNA from 112 HD gene‐positive subjects (42 affected with HD and 70 asymptomatic at the time of sperm donation). For the lymphoblastoid DNAs, the mean lengths of the expanded CAG repeat and normal CAG repeat were 45 (range 36–62) and 18 (range 9–35), respectively. The mean age at sperm collection was 33 years (range 14–70). In the Venezuelan pedigree, we analysed 495 transmissions of the HD allele from 206 parents. Of these, 184 transmissions were from 83 fathers and 311 transmissions were from 123 mothers.
Measures of repeat variation
We used PCR amplification of the HD CAG repeat1 to examine the range of length variation represented in total DNA from lymphoblastoid cells and sperm samples. To capture the spread in HD CAG alleles, we determined the modal HD CAG repeat length and the range of repeat lengths by densitometeric analysis as described previously.3
Statistical methods
Correlation between the quantitative traits of constitutive HD CAG repeat length, ΔCAG, in sperm, range of CAG repeat length in sperm, and age at sperm donation was performed using the Pearson correlation coefficient. Models evaluating the relationship of sperm instability to age and controlling for parental constitutive HD CAG repeat size or age were performed by linear regression analyses using the PROC GLM procedure in the SAS program (SAS Inc., Cary, North Carolina, USA).13 Similarly, models evaluating whether repeat variation in sperm was associated with affected status, and controlling for constitutive HD CAG repeat size and age, were performed by the PROC GLM procedure.
For analyses of the Venezuelan pedigree, differences in the mean repeat‐length change were assessed by the t test and differences in the proportion of expansions and contractions analysed by χ2 test. Analyses of the effects of parental CAG repeat length, age and birth order were determined using the generalised estimating equations (GENMOD) procedure in SAS, adjusting for repeated observations within each sibship. For age and birth order analyses, models were used that adjusted for parental CAG repeat and for offspring sex, adjusting for repeated observations within each sibship. For the analyses of sibling correlation, an equation to determine the “expected repeat size” was estimated by linear regression models predicting the offspring expanded CAG repeat size from the parental expanded repeat size. Separate models were run for maternal and paternal transmission. Models that included the sex of the offspring and adjusted for repeated observations within the sibship were also evaluated. The deviation from expected change was computed as the observed minus the predicted offspring CAG. The correlation for deviation from expected CAG for sibling pairs was computed by interclass correlation controlling for multiple siblings per sibship.
To control for testing of multiple hypotheses (three comparisons in the sperm dataset, five for maternal transmissions and five for paternal transmissions in the Venezuelan pedigree) a Bonferroni correction was applied. Uncorrected p values are given in the text. Corrected p values were significant after Bonferroni correction as indicated in the text.
Results
Effect of constitutive CAG repeat length on CAG repeat‐length variation in sperm
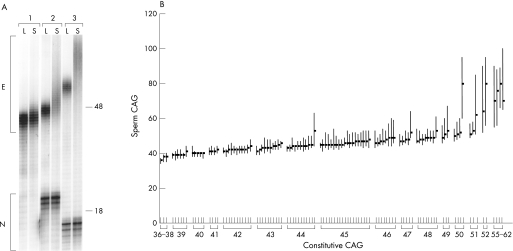
We evaluated the relationship of constitutive HD CAG repeat length on instability in sperm from 112 men (see supplementary data file; available at available at http://jmg.bmj.com/supplemental). Two typical HD heterozygotes and one HD homozygote (two mutant HD alleles) are shown in figure 1A. The normal (N) alleles of the former were identical in lymphoblastoid and sperm DNA, at 21 and 16 repeats, respectively, whereas the latter possessed no normal allele. In each case, the expanded (E) mutant alleles show greater length variation in sperm DNA than in lymphoblastoid DNA, with the extent of variation in sperm greater for the person with the larger constitutive CAG repeat.
Figure 1 The effect of constitutive HD CAG repeat size on instability in sperm. (A) Autoradiogram showing HD CAG repeats, amplified using PCR, from paired lymphoblastoid (L) and sperm (S) DNA samples from three subjects. PCR products from the expanded (E) and normal (N) HD alleles are shown (note: person 1 is homozygous for the HD mutation). The positions on the autoradiogram corresponding to 48 and 18 CAG repeats, as determined from cosmid clones of known CAG repeat size, are indicated. (B) The HD CAG repeat was amplified using PCR from sperm DNA of 112 subjects, and the modal CAG repeat size (black squares) and range of CAG repeat sizes in sperm (vertical black lines) were determined by densitometry of the autoradiograms. Mode and range were plotted versus the constitutive size of the expanded CAG repeat, as determined from lymphoblastoid DNA. For six people, the spread of the sperm PCR products was such that we were unable to determine the range of CAG sizes, although we were able to determine the modal repeat size. This figure therefore shows data plotted for 106 of the 112 subjects.
Figure 1B depicts the mode and range of CAG repeat lengths in sperm for 106 men, showing an increase in both of these measures with increasing constitutive CAG length We used the range of repeat lengths in sperm and the repeat‐length change in sperm, (ΔCAG), where ΔCAG = (modal sperm CAG length) − (modal lymphoblastoid (constitutive) CAG length), as measures of repeat instability. ΔCAG is, on average, zero for constitutive repeat lengths up to 42 CAGs, is weakly positive (0.4–2) for constitutive repeat lengths of 43–49 CAGs, and is strongly positive (4–20) for constitutive repeat lengths ⩾50 CAGs. Both ΔCAG and range and were strongly correlated with constitutive HD CAG repeat length (ΔCAG r = 0.60, p<0.001; range r = 0.72, p<0.001, both remaining significant after Bonferroni correction for multiple testing), showing that CAG length is a major determinant of male germline instability. However, as illustrated in figure 1B, people with the same constitutive repeat size appear to have different degrees of variation in sperm CAG repeat lengths, suggesting that factors other than the HD CAG repeat size influence instability.
Lack of effect of age on HD CAG repeat‐length variation in sperm
De novo mutations associated with advanced paternal age have been suggested to be due to increased frequency of mutations in sperm,14,15 thus we tested whether CAG repeat variation in sperm is influenced by male age at time of sampling.
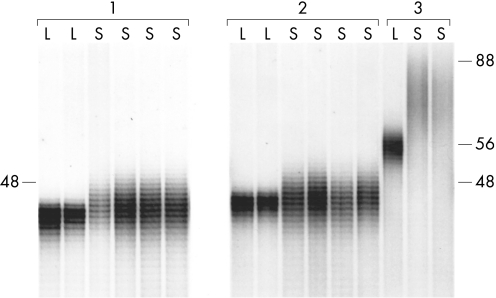
We assessed whether the degree of HD CAG repeat‐length variation changes over time in a single person. Figure 2 shows the HD CAG PCR products in the expanded allele range from three subjects heterozygous for HD, from whom multiple individual lymphoblastoid cell lines and sperm samples had been obtained. For subject 3, two sperm samples were collected within the same year. The spread of HD CAG repeat lengths in sperm DNA differed, as expected, between the three subjects, but was remarkably consistent over time for each subject, despite the samples being collected up to 2 years apart. Although these findings argue against a major effect of age on repeat‐length variation in sperm, modal analysis may not be sufficiently sensitive to detect more subtle effects within this limited time frame. Therefore, we performed a statistical analysis to determine whether instability in sperm was associated with age in a group of 112 HD male subjects, aged 14–70 years at the time of sperm collection. Not surprisingly, age was inversely correlated with repeat size (r = −0.44, p<0.001), indicating that people surviving to older ages had smaller HD repeats. We therefore controlled for constitutive HD CAG repeat size in our analysis. We found that age was not a significant predictor of ΔCAG (p = 0.28) or range of HD CAG repeats lengths in sperm (p = 0.06, Bonferroni corrected p = 0.18). These findings support the view that age does not play a major role in determining HD CAG repeat instability in the male germline.
Figure 2 HD CAG repeat variation in sperm does not vary over time. Autoradiogram showing expanded HD CAG repeats amplified by PCR from lymphoblastoid (L) and sperm (S) DNA samples from three subjects. The positions on the autoradiogram corresponding to 48 CAG repeats as determined from a cosmid clone of known CAG repeat size, and to 56 and 88 CAG repeats as determined from densitometric analysis of lymphoblastoid DNA and the first sperm sample of subject 3, are indicated. See supplementary data file (available at http://jmg.bmj.com/supplemental) for quantified data on subjects 1 (code 172), 2 (code 169) and 3 (code 51). For subject 1, two lymphoblastoid samples representing independent lines spanning 10 years and four sperm DNA samples collected over 2 years were analysed. Sperm sample 2 was collected 1 year after sperm sample 1; sperm samples 3 and 4 were collected 2 years after sperm sample 1. For subject 2, two lymphoblastoid samples spanning 9 years and four sperm DNA samples, collected over 2 years were analysed. Sperm samples 2 and 3 were collected one year after sperm sample 1; sperm sample 4 was collected two years after sperm sample 1.
Lack of effect of affected status on HD CAG repeat‐length variation in sperm
We also evaluated whether CAG repeat variation in sperm was influenced by the affected status of the males at the time of sperm collection. As expected from the relationship between HD CAG repeat length and onset of clinical symptoms, significant differences were found in the length of the constitutive CAG repeat (p<0.01) and in the age (p<0.001) of subjects who were affected versus those who were unaffected at the time of sperm collection. Therefore, we controlled for both constitutive CAG repeat size and age in order to determine the influence of affected status on sperm CAG instability. These analyses revealed no significant difference in ΔCAG (p = 0.17) or in the range (p = 0.23) of CAG repeat lengths in sperm between affected and unaffected subjects.
Transmission of the HD CAG repeat in the Venezuelan pedigree
Examination of transmission of the HD mutant CAG allele in parent–offspring pairs provides insight into the germline variation in parents, as revealed by the allele sizes present in the progeny. Table 1 summarises parent–offspring HD CAG repeat transmissions in the Venezuelan HD pedigree (see supplementary data file; available at http://jmg.bmj.com/supplemental). Male transmission differed markedly from female transmission for both mean repeat‐length change (p<0.001, t test) and for the proportion of expansions versus contractions (χ2 = 44.7, 2 d.f., p<0.001). These remained significant after Bonferroni correction for multiple comparisons. The mean repeat‐length change was +3.5 CAGs for male transmissions (range –6 to +41), which was similar to that seen in sperm (mean ΔCAG +2.4, over range –4 to +45).
Table 1 Transmission of the expanded HD CAG repeat in the Venezuelan pedigree.
| Total | Female | Male | |
|---|---|---|---|
| Transmissions, n | 495 | 311 | 184 |
| Mean CAG parent | 44.4 | 44.7 | 43.9 |
| Changed alleles, n (%) | 364 (74) | 214 (69) | 150 (82) |
| Contractions | 137 (28) | 107 (34.5) | 30 (16) |
| Expansions | 227 (46) | 107 (34.5) | 120 (65) |
| Repeat‐length change | |||
| Mean | +1.27 | −0.04 | +3.5 |
| Range | −7 to +41 | −5 to +10 | −7 to +41 |
A total of 206 HD carriers (83 male, 123 female) contributed to the total number of transmissions of the HD allele analysed. The size of the expanded HD CAG repeat in the parent and their offspring was used to determine the magnitude and direction of the repeat‐length change during transmission. There was no significant difference in transmission of the HD allele and the normal allele from male or female parents (data not shown) and the larger number of female compared with male transmissions reflects a sampling bias.
CAG repeat‐length changes are influenced by parental CAG repeat length but not by age or birth order
Repeat‐length variation in sperm was found to be dependent on parental CAG repeat length but not on the age of the transmitting male. We therefore examined the effect of these variables on inherited repeat‐length changes in the Venezuelan pedigree. Repeat‐length change was strongly influenced by parental CAG length in both maternal and paternal transmissions (both p<0.001; both remained significant after Bonferroni correction for multiple testing). The age of the transmitting women did not influence repeat‐length change (p = 0.61), whereas there was a weak, non‐significant effect of the age of the transmitting men on repeat‐length change (p = 0.05, Bonferroni‐corrected p = 0.26). To minimise possible confounding effects of interindividual variation we also assessed the role of parental age by determining whether birth order influenced repeat‐length change. Birth order was not found to predict repeat‐length change in either maternal (p = 0.93) or paternal (p = 0.23) transmissions.
CAG repeat‐length changes are influenced by the sex of the offspring
There is evidence in fragile X syndrome and in mouse models of HD and myotonic dystrophy that postzygotic mechanisms play a role in triplet repeat instability.16,17,18,19 In a mouse model of HD, a postzygotic mechanism has been proposed to explain the finding that expansions predominate in male offspring whereas contractions predominate in female offspring from the same parent.17,18
We therefore investigated whether instability of the HD CAG repeat in this Venezuelan pedigree is influenced by the sex of the offspring. To avoid masking an effect of offspring sex by the strong influence of the sex of the parent on repeat instability, we analysed maternal and paternal transmissions to sons and daughters separately (table 2). For maternal transmissions, we observed a significant difference between sons and daughters in the mean repeat‐size change (p<0.01, t test), and in the proportion of expansions and contractions (χ2 = 12, 2 d.f., p<0.01) (both remained significant after Bonferroni correction for multiple testing). However, in paternal transmissions, we did not find a significant difference between sons and daughters in either the mean repeat‐size change (p = 0.31, t test) or in the proportion of expansions and contractions (χ2 = 0.89, 2 d.f., p = 0.64). These findings provide evidence that progeny sex influences the direction of repeat‐size changes in maternal transmissions. The strong predominance of HD CAG repeat expansions in the male germline may mask the relatively mild offspring sex effect so that it is not readily apparent when men transmit the CAG repeat but is evident in maternal transmissions.
Table 2 Transmission of the expanded HD CAG repeat to female and male offspring in the Venezuelan pedigree.
| Mothers | Fathers | |||
|---|---|---|---|---|
| Daughters | Sons | Daughters | Sons | |
| Transmissions, n | 147 | 164 | 96 | 88 |
| Mean CAG parent | 44.8 | 44.5 | 44.3 | 43.6 |
| Changed alleles, n (%) | 99 (67) | 115 (70) | 79 (82) | 71 (81) |
| Contractions | 62 (42) | 45 (27) | 18 (19) | 12 (14) |
| Expansions | 37 (25) | 70 (43) | 61 (64) | 59 (67) |
| Repeat‐length change | ||||
| Mean | −0.4 | +0.28 | +3.0 | +4.0 |
| Range | −5 to +5 | −5 to +10 | −6 to +29 | −7 to +41 |
Transmissions from both mothers and fathers were analysed according to whether the HD CAG repeat was transmitted to daughters or sons. In total, 87 HD carrier mothers contributed to the transmissions to daughters and 93 to the transmissions to sons, and 65 HD carrier fathers contributed to the transmissions to daughters and 53 to the transmissions to sons. The size of the expanded HD CAG repeat in the parent and their offspring was used to determine the magnitude and direction of the repeat‐length change during transmission.
Instability is correlated between siblings
Differences in HD CAG repeat‐length variability in sperm DNA between subjects with similar constitutive repeat lengths (as shown in fig 1B), suggest that genetic factors other than CAG repeat length may play a role in repeat instability. To assess whether repeat instability tends to cluster in families, we evaluated whether the deviation from expected HD CAG repeat length, as predicted by the size of the repeat in the parent, was correlated between siblings in this Venezuelan pedigree. Multiple sibling pairs were derived from each sibship of >2 members. Analysis of 367 sibling pairs from maternal transmissions revealed a significant correlation (r = 0.28, p<0.001, significant after Bonferroni correction for multiple testing) between siblings in the deviation from expected HD CAG repeat length predicted by the size of repeat in the mother. Including the sex of the offspring in the computation of the expected repeat size did not change the sibling–sibling correlation appreciably. Analysis of 168 sibling pairs from paternal transmissions revealed a weak, borderline significant correlation (r = 0.16, p = 0.04) between siblings in the extent to which their HD CAG lengths deviated from the expected CAG length, which was not significant (p = 0.2) after Bonferroni correction for multiple comparisons. However, we did not observe any correlation (r = 0.08, p = 0.35) when the analysis of paternal siblings was repeated omitting the largest (>10 CAGs) repeat changes. These results imply that intergenerational repeat instability in the Venezuelan pedigree is, in part, heritable and that the large expansions in male transmissions contribute to the heritability.
Discussion
We investigated factors that influence the instability of the HD CAG repeat by analysing HD CAG repeat lengths in sperm from a large number of men carrying a wide range of constitutive HD repeat sizes, and by analysing the transmission of the HD CAG repeat in parent–offspring pairs from a large Venezuelan pedigree.
Consistent with previous observations,3,7,10,11 our analyses showed a strong positive correlation between the constitutive size of the HD CAG repeat and repeat‐length variation in sperm, implicating constitutive HD CAG repeat length as the main determinant of instability. This finding supports the hypothesis that instability is driven by the CAG repeat itself, as a result of a repeat length‐dependent propensity to form unusual secondary structures.20
In agreement with previous investigations of repeat instability in sperm of patients with HD and patients with myotonic dystrophy,11,21 we did not find a correlation between HD CAG repeat‐length variation in sperm and the age of the subjects at the time of sperm donation. This is supported by the absence of a significant effect of parental age or birth order on inherited repeat‐length changes in this Venezuelan pedigree. Analysis of spermatogenic cell populations in patients with HD has revealed that instability can occur before meiosis.22 If these CAG length alterations arose during the mitotic cell divisions of postpubertal spermatogonia, such a mechanism would be predicted to generate progressively increasing CAG length variation over time. Although a definitive test of an age effect would require examining sperm samples from the same subjects collected over many years, our present findings and those reported by Leeflang et al11 suggest that pre‐meiotic repeat instability may be more likely to arise between the time of primordial germ‐cell determination and postpubertal spermatogonial proliferation.
The absence of a positive correlation of HD CAG repeat‐length variation in sperm with age suggests further that instability in the male germline does not occur as a consequence of a progressively worsening disease. This is also indicated by the finding that there was no significant difference in repeat‐length variation in sperm from subjects who were clinically affected at the time of sperm donation compared with sperm from those who were presymptomatic. Thus, at least in the male germline, instability appears to be an inherent property of the expanded HD CAG repeat, not obviously affected by aging or degenerative processes. It would be of interest to determine whether HD CAG repeat length may also be a primary determinant of the dramatic somatic repeat expansions of the mutant HD CAG repeat recently observed in human HD brain,23 and whether these repeat expansions are associated with ongoing neurodegeneration. CAG repeat length was recently shown to be a major modifier of somatic repeat instability in buccal cells.24
Our analyses of repeat‐length changes in transmissions of the HD CAG repeat in the Venezuelan pedigree reveal an overall frequency of changes (74%) in this population, which fits well with the frequency of instability (70–83%) derived from transmission data in many other studies.4,7,10,25,26 Similar to other HD populations, this Venezuelan HD pedigree shows a tendency towards repeat expansion in paternal transmission, but no clear expansion trend in maternal transmission.4,7,10,25,26
We report for the first time that the sex of the offspring can influence the direction of inherited repeat‐size changes in maternal transmission, with male offspring displaying a tendency towards expansion, and female offspring displaying a tendency towards contraction. Although in a previous study, offspring sex was not found to influence the intergenerational repeat instability of the HD CAG repeat,27 the discrepancy between that study and our data may lie in the fact that we analysed transmissions from male and female HD carriers separately. Interestingly, the direction bias of repeat‐size changes we observed in male and female offspring parallels that seen in a transgenic HD mouse model.17,18
The lack of somatic instability in blood or in lymphoblastoid cell lines3,10 implies that differences in repeat‐size changes transmitted to male and female offspring do not reflect different degrees of somatic expansion between the sexes. Our data are consistent with a postzygotic component of repeat instability, influenced by X‐encoded or Y‐encoded factors, to generate the offspring sex‐specific differences observed. Concordance of HD CAG repeat sizes in monozygotic twins3 implies that a postzygotic event would have to occur very early (<5 days) after fertilisation.
Evidence from other triplet‐repeat disorders also points to postzygotic events that contribute to repeat instability. In fragile X syndrome, postzygotic contraction of the full CGG repeat mutation to a premutation size is thought to account for the presence of mosaic children carrying both full mutation and premutation sized alleles,16 although the role of offspring sex in this condition is unclear.28 In addition, in a transgenic mouse model of myotonic dystrophy, intergenerational CTG repeat instability was found to depend partly on the presence or absence of the mismatch repair gene Msh2 in the offspring, independent of the Msh2 genotype of the CTG repeat‐transmitting parent.19 Thus, events occurring after fertilisation may play a role in determining repeat size in HD as it does in other triplet‐repeat diseases.
Our analyses show that deviation from expected HD CAG repeat size is correlated between siblings in the Venezuelan pedigree, providing evidence that genetic factors influence repeat instability. Although there is a possibility that rare clonal selection may occur, our experience with genotyping of lymphoblastoid cell line DNAs and corresponding blood DNAs is that the modal CAG repeat length is the same in >99% of cases. Consequently, we do not believe that cell line‐mediated misassignments of the parental alleles have yielded systematic errors in the predicted‐observed intergenerational transmissions in our sibling–sibling analyses.
Given the present findings, genetic factors could influence either germline or postzygotic instability. As we did not find the sibling–sibling correlation to be influenced by the sex of the sibling, the genetic modifier effect that we observed is probably acting at the germline instability level. Genetic factors that influence repeat instability could potentially act either in cis or in trans. Although chromosomal context (cis‐acting sequences) can influence repeat stability,29 sequences closely linked to the HD gene are unlikely to be responsible for the correlation of repeat instability between siblings in the Venezuelan cohort, as the HD chromosomes derive from the same founder chromosome.30 Therefore, it is more likely that in these patients with HD the genetic factors that influence instability act in trans, as suggested in a previous study of a limited number of Venezuelan patients with HD.6 As indicated by studies in mouse models, such trans‐acting factors may be proteins involved in DNA metabolic pathways.19,31,32,33,34 Understanding the pathways that are responsible for repeat instability in patients with HD could ultimately provide the possibility of therapeutic strategies aimed at preventing CAG repeat expansion or inducing contraction.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
Acknowledgements
We thank Dr Walter Koroshetz for assistance with sample collection. This work was supported by the HD Center Without Walls NS16367, NINDS grant NS049206, the W M Keck foundation and an anonymous donor.
Abbreviations
HD - Huntington disease
APPENDIX
The US–Venezuela Collaborative Research Group are: Nancy S. Wexler, Judith Lorimer, Julie Porter, Fidel\a Gomez, Carol Moskowitz, Edith Shackell, Karen Marder, Graciela Penchaszadeh, Simone A Roberts, Kelly Posner Gerstenhaber, all from Columbia University, 1051 Riverside Drive, New York, NY 10032, USA; Javier Gayan, Denise Brocklebank, Stacey S Cherny, Lon R Cardon, all from Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK; Jacqueline Gray, Stephen R Dlouhy, Sandra Wiktorski, Marion E Hodes (deceased), P Michael Conneally, all from Indiana University, 975 West Walnut Street, Indianapolis, IN 46202, USA; Jack B Penney (deceased), James Gusella, Jang‐Ho Cha, Michael Irizarry, Diana Rosas, Steven Hersch, Zane Hollingsworth, Marcy MacDonald, Anne B Young, all from Massachusetts General Hospital, Boston, MA, USA; J Michael Andresen, David E Housman, all from Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA; Margot Mieja de Young, Ernesto Bonilla, Theresa Stillings, Americo Negrette (deceased), all from Universidad del Zulia, AP 1151, Maracaibo, Venezuela; S Robert Snodgrass, Mattel Children's Hospital at University of California, Los Angeles, CA 90095, USA; Maria Dolores Martinez‐Jaurrieta, Maria A Ramos‐Arroyo, both from Hospital Virgen del Camino, Irunlarrea, 4, 31008 Pamplona, Spain; Jacqueline Bickham, University of Texas, 1515 Holcombe Boulevard, Houston, TX 77030, USA; Juan Sanchez Ramos, University of South Florida, 12901 Bruce B Downs Boulevard, Tampa, FL 33612, USA; Frederick Marshall, Ira Shoulson, both from University of Rochester, 1351 Mount Hope Avenue, Rochester, NY 14620, USA; Gustavo J Rey, Miami Children's Hospital, 3100 SW 62nd Avenue, Miami, FL 33155, USA; Andrew Feigin, North Shore University Hospital, 444 Community Drive, Manhasset, NY 11030, USA; Norman Arnheim, all from University of Southern California, 835 West 37th Street, Los Angeles, CA 90089, USA; Amarilis Acevedo‐Cruz, Mount Sinai Medical Center, 4300 Alton Road, Miami Beach, FL 33140, USA; Leticia Acosta, Health and Human Services Agency, 4588 Market Street, San Diego, CA 92102, USA; Jose Alvir, all from New York University, 550 1st Avenue, New York, NY 10016, USA; Kenneth Fischbeck, National Institute of Neurological Disorders and Stroke, National Institutes of Health, 10 Center Drive, Bethesda, MD 20892, USA; Leslie M Thompson, University of California, 2121 Gillespie, Irvine, CA 92697, USA; Angela Young, Leon Dure, both from University of Alabama, Birmingham, AL 35294, USA; Christopher J O'Brien, Thomas Jefferson University, 233 South 10th Street, Philadelphia, PA 19107, USA; Jane Paulsen, University of Iowa, 200 Hawkins Drive, Iowa City, IA 52242, USA; Adam Brickman, Denise Krch, Shelley Peery, all from City University of New York, 365 5th Avenue, New York, NY 10016, USA; Penelope Hogarth, Oregon Health Sciences University, 3181 SW Sam Jackson Park Road, Portland, OR 97201, USA; Donald S Higgins Jr, Albany Medical College, 47 New Scotland Avenue, Albany, NY 12208, USA; and Bernhard Landwehrmeyer, University of Ulm, 89069 Ulm, Germany.
Footnotes
Competing interests: None declared.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
References
- 1.Huntington's disease collaborative research group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 199372971–983. [DOI] [PubMed] [Google Scholar]
- 2.McNeil S M, Novelletto A, Srinidhi J, Barnes G, Kornbluth I, Altherr M R, Wasmuth J J, Gusella J F, MacDonald M E, Myers R H. Reduced penetrance of the Huntington's disease mutation. Hum Mol Genet 19976775–779. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald M E, Barnes G, Srinidhi J, Duyao M P, Ambrose C M, Myers R H, Gray J, Conneally P M, Young A, Penney J, Shoulson I, Hollingsworth Z, Koroshetz W, Bird E, Vonsattel J P, Bonilla E, Moscowitz C, Penchaszadeh G, Brzustowicz L, Alvir J, Bickam Conde J, Cha J ‐ H, Dure L, Gomez F, Ramos‐Arroyo M, Sanchez‐Ramos J, Snodgrass S R, de Young M, Wexler N S, MacFarlane H, Anderson M A, Jenkins B, Gusella J F. Gametic but not somatic instability of CAG repeat length in Huntington's disease. J Med Genet 199330982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuhlke C, Riess O, Bockel B, Lange H, Thies U. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum Mol Genet 199322063–2067. [DOI] [PubMed] [Google Scholar]
- 5.Telenius H, Kremer B, Goldberg Y P, Theilmann J, Andrew S E, Zeisler J, Adam S, Greenberg C, Ives E J, Clarke L A, Hayden M R. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet 19946409–414. [DOI] [PubMed] [Google Scholar]
- 6.Leeflang E P, Zhang L, Tavare S, Hubert R, Srinidhi J, MacDonald M E, Myers R H, de Young M, Wexler N S, Gusella J F, Arnheim N. Single sperm analysis of the trinucleotide repeats in the Huntington's disease gene: quantification of the mutation frequency spectrum. Hum Mol Genet 199541519–1526. [DOI] [PubMed] [Google Scholar]
- 7.Norremolle A, Sorensen S A, Fenger K, Hasholt L. Correlation between magnitude of CAG repeat length alterations and length of the paternal repeat in paternally inherited Huntington's disease. Clin Genet 199547113–117. [DOI] [PubMed] [Google Scholar]
- 8.Telenius H, Almqvist E, Kremer B, Spence N, Squitieri F, Nichol K, Grandell U, Starr E, Benjamin C, Castaldo I. Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum Mol Genet 19954189–195. [DOI] [PubMed] [Google Scholar]
- 9.Andrew S E, Goldberg Y P, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman M A, Graham R K, Hayden M R. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet 19934398–403. [DOI] [PubMed] [Google Scholar]
- 10.Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, Gray J, Conneally P, Young A, Penney J, Hollingsworth Z, Shoulson I, Lazzarini A, Falek A, Koroshetz W, Sax D, Bird E, Vonsattel J P, Bonilla E, Alvir J, Bickham Conde J, Cha J & h y p h, Dure L, Gomez F, Ramos M, Sanchez‐Ramos J, Snodgrass S, de Young M, Wexler N, Moscowitz C, Penchaszadeh G, MacFarlane H, Anderson M, Jenkins B, Srinidhi J, Barnes G, Gusella J, MacDonald M. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet 19934387–392. [DOI] [PubMed] [Google Scholar]
- 11.Leeflang E P, Tavare S, Marjoram P, Neal C O, Srinidhi J, MacFarlane H, MacDonald M E, Gusella J F, de Young M, Wexler N S, Arnheim N. Analysis of germline mutation spectra at the Huntington's disease locus supports a mitotic mutation mechanism. Hum Mol Genet 19998173–183. [DOI] [PubMed] [Google Scholar]
- 12.Wexler N S, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts S A, Gayan J, Brocklebank D, Cherny S S, Cardon L R, Gray J, Dlouhy S R, Wiktorski S, Hodes M E, Conneally P M, Penney J B, Gusella J, Cha J H, Irizarry M, Rosas D, Hersch S, Hollingsworth Z, MacDonald M, Young A B, Andresen J M, Housman D E, De Young M M, Bonilla E, Stillings T, Negrette A, Snodgrass S R, Martinez‐Jaurrieta M D, Ramos‐Arroyo M A, Bickham J, Ramos J S, Marshall F, Shoulson I, Rey G J, Feigin A, Arnheim N, Acevedo‐Cruz A, Acosta L, Alvir J, Fischbeck K, Thompson L M, Young A, Dure L, O’Brien C J, Paulsen J, Brickman A, Krch D, Peery S, Hogarth P, Higgins D S, Jr, Landwehrmeyer B U. S.‐Venezuela Collaborative Research Project. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci U S A 20041013498–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SAS Institute I SAS/STAT users guide. Cary, NC: SAS Institute Inc, 1999
- 14.Wilkin D J, Szabo J K, Cameron R, Henderson S, Bellus G A, Mack M L, Kaitila I, Loughlin J, Munnich A, Sykes B, Bonaventure J, Francomano C A. Mutations in fibroblast growth‐factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am J Hum Genet 199863711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risch N, Reich E W, Wishnick M M, McCarthy J G. Spontaneous mutation and parental age in humans. Am J Hum Genet 198741218–248. [PMC free article] [PubMed] [Google Scholar]
- 16.Imbert G, Feng Y, Nelson D, Warren S, Mandel J. FMR1 and Mutations in Fragile X syndrome: Molecular Biology, Biochemistry, and Genetics. In: Wells R, Warren S, eds. Genetic instabilities and hereditary neurological diseases. San Diego: Academic Press, 1998;27–53,
- 17.Kovtun I V, Therneau T M, McMurray C T. Gender of the embryo contributes to CAG instability in transgenic mice containing a Huntington's disease gene. Hum Mol Genet 200092767–2775. [DOI] [PubMed] [Google Scholar]
- 18.Kovtun I V, Welch G, Guthrie H D, Hafner K L, McMurray C T. CAG repeat lengths in X‐ and Y‐bearing sperm indicate that gender bias during transmission of Huntington's disease gene is determined in the embryo. J Biol Chem 20042799389–9391. [DOI] [PubMed] [Google Scholar]
- 19.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair‐deficient mice. EMBO J 2003222264–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMurray C T. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci USA 1999961823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martorell L, Gamez J, Cayuela M L, Gould F K, McAbney J P, Ashizawa T, Monckton D G, Baiget M. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology 200462269–274. [DOI] [PubMed] [Google Scholar]
- 22.Yoon S R, Dubeau L, De Young M, Wexler N S, Arnheim N. Huntington diseaseexpansion mutations in humans can occur before meiosis is completed. Proc Natl Acad Sci USA 20031008834–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy L, Evans E, Chen C, Craven L, Detloff P, Ennis M, Shelbourne P. Dramatic tissue‐specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet 2003123359–3367. [DOI] [PubMed] [Google Scholar]
- 24.Veitch NJ Ennis M, McAbney J P, Shelbourne P F, Monckton D. Inherited CAG.CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair (Amst) . 2007;6789–796. [DOI] [PubMed]
- 25.Barron L H, Warner J P, Porteous M, Holloway S, Simpson S, Davidson R, Brock D J. A survey of the Huntington's disease associated trinucleotide repeat in the Scottish population. J Med Genet 1993301003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trottier Y, Biancalana V, Mandel J L. Instability of CAG repeats in Huntington's disease: relation to parental transmission and age of onset. J Med Genet 199431377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer B, Almqvist E, Theilmann J, Spence N, Telenius H, Goldberg Y P, Hayden M R. Sex‐dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am J Hum Genet 199557343–350. [PMC free article] [PubMed] [Google Scholar]
- 28.Nolin S L, Brown W T, Glicksman A, Houck G E, Jr, Gargano A D, Sullivan A, Biancalana V, Brondum‐Nielsen K, Hjalgrim H, Holinski‐Feder E, Kooy F, Longshore J, Macpherson J, Mandel J L, Matthijs G, Rousseau F, Steinbach P, Vaisanen M L, von Koskull H, Sherman S L. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genets 200372454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Libby R T, Monckton D G, Fu Y H, Martinez R A, McAbney J P, Lau R, Einum D D, Nichol K, Ware C B, Ptacek L J, Pearson C E, La Spada A R. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet 20031241–50. [DOI] [PubMed] [Google Scholar]
- 30.Wexler N S, Rose E A, Housman D E. Molecular approaches to hereditary diseases of the nervous system: Huntington's disease as a paradigm. Ann Rev Neurosci 199114503–529. [DOI] [PubMed] [Google Scholar]
- 31.Manley K, Shirley T L, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet 199923471–473. [DOI] [PubMed] [Google Scholar]
- 32.Van den Broek W J, Nelen M R, Wansink D G, Coerwinkel MM teRiele H, Gronen P J, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock in mice is differentially affected by Msh3 and Msh6 mismatch‐repair proteins. Hum Mol Genet 200211191–198. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler V C, Lebel L A, Vrbanac V, Teed A, Te Riele H, MacDonald M E. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet 200312273–281. [DOI] [PubMed] [Google Scholar]
- 34.Gomes‐Pereira M, Fortune M T, Ingram L, McAbney J P, Monckton D G. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet 2004131815–1825. [DOI] [PubMed] [Google Scholar]