Summary
Transcriptional profiling of antioxidant genes of M. tuberculosis was performed by real-time RT-PCR during mouse lung infection and during adaptation to gradual oxygen depletion in vitro. M. tuberculosis genes involved in major detoxification pathways of oxidative stress were not up-regulated during chronic mouse lung infection, which is established in response to expression of host adaptive immunity. This result suggests that a major function of bacterial antioxidant enzymes is to protect from oxidants generated during the early, acute phase of infection. In vivo transcription profiles of bacterial antioxidant enzymes differed from those seen under adaptation to low oxygen in vitro, indicating differences between growth arrest in vivo and that induced by hypoxia in vitro.
Keywords: Reactive oxygen species, Reactive nitrogen species, Mouse lung infection, Reverse transcription polymerase chain reaction, bacterial persistence, bacterial growth arrest
Introduction
The interaction between the intracellular pathogen Mycobacterium tuberculosis and mononuclear phagocytes typically leads to induction of host adaptive immune responses, which control infection without sterilizing the host 1, 2. Survival of M. tuberculosis to expression of adaptive immunity is presumably associated with entry into a “dormant” state, in which tubercle bacilli exhibit no or slow growth and low metabolic activity. As a result, a latent infection is established. When immunity fails to control infection, tubercle bacilli exit dormancy, resume growth and cause disease. Thus, adaptation to the conditions found inside the phagocytic cell is key to the survival and persistence of M. tuberculosis in vivo.
Characteristic of the intracellular environment of the activated macrophage is high-output generation of reactive nitrogen species (RNS) and reactive oxygen species (ROS) 3, 4. ROS are also produced during bacterial aerobic metabolism 5, 6. It is well established that M. tuberculosis has evolved multiple ways to detoxify RNS and ROS. First, superoxide can be directly detoxified by the sequential activity of superoxide dismutases (SODs) and catalase (KatG); peroxynitrite can be detoxified by KatG 7. Second, hydrogen peroxide and peroxynitrite can also be detoxified by thiol-specific redox systems, which also maintain the intracellular thiol-disulfide balance. These include alkyl hydroperoxide reductases (AhpC and AhpD) 8, thioredoxin and thioredoxin reductase (TrxB2) 9–11, and methionine and methionine sulfoxide reductase (MsrA) 12. Third, NO can be directly detoxified by hemoglobins. M. tuberculosis produces two small, truncated hemoglobins (trHbs), encoded by glnO and glbN 13, 14, and another hemoglobin (Hmp) 15 that is homologous to the flavohemoglobin of Escherichia coli. Although progress has been made toward characterizing the transcriptional response of M. tuberculosis to oxidative and nitrosative stress in culture and in infected cultured macrophages 16–18, more limited is our understanding of the antioxidant responses of tubercle bacilli at different stages of lung infection.
In this paper, we report results of transcriptional profiling of the nitrosative and oxidative stress responses of tubercle bacilli during murine infection. In the lung of mice infected by the respiratory route, tubercle bacilli multiply exponentially for ~ 20 days (acute infection) and then stop growing in response to expression of adaptive, Th1-mediated immunity. Stabilization of bacterial numbers characterizes a much longer, second phase of infection (chronic infection) 19. Since bacteriostasis is primarily induced by the generation of NO from activated macrophages (mice incapable of producing iNOS die rapidly from acute infection 20), this animal model is particularly appropriate to characterize the antioxidant response of M. tuberculosis during growth and persistence in vivo. For comparative purposes, we also characterized the bacterial antioxidant response of M. tuberculosis in cultures gradually starved for oxygen. This in vitro model, which was developed by L. Wayne 21, reflects some aspects of the in vivo situation, since it too causes bacterial growth arrest and induction of the “dormancy” regulon, a set of ~50 genes that is upregulated when tubercle bacilli stop (or slow down) growth in vivo and in vitro 16, 18, 22.
When quantitative RT-PCR was used to measure the abundance of M. tuberculosis transcripts involved in the detoxification of ROS and RNS during mouse lung infection, it was found that the transition from bacterial replication to growth arrest was characterized by decreased transcript levels for all genes tested. It was also found that some genes were selectively induced at specific stages of O2 starvation in vitro, thus providing the first example of key transcriptional differences between a mouse model and the Wayne model.
Materials and Methods
Mouse infection
C57BL/6 mice at the age of 8 to 10 weeks were infected with ~2 × 102 CFU of M. tuberculosis strain H37Rv via the respiratory route, as described previously 23. At selected times, lungs from three to four mice were harvested. Half of the lung was used to monitor bacterial growth by standard plating 24, and the other half was snap-frozen in liquid nitrogen for subsequent RNA extraction.
Bacterial culture under gradual oxygen depletion
M. tuberculosis H37Rv was grown in liquid culture at 37°C in Dubos Tween-albumin broth as described 21. Gradual oxygen depletion in the Wayne model was obtained by culturing bacteria in sealed tubes with a headspace ratio (HSR) of 0.5 with slow magnetic stirring 21. The density of the starting culture was 106 cells/ml. At selected times, cells from 2-ml cultures were harvested by rapid centrifugation and snap-frozen in liquid nitrogen for subsequent RNA extraction. CFU were enumerated by plating 10-fold serial dilutions of liquid cultures on Dubos oleic albumin agar.
Copy number measurement of bacterial transcripts
RNA extraction, reverse transcription (RT) and real time PCR were performed as per our published protocol 22 and at http://www.phri.org/research/res_pigennaro.asp. Briefly, infected mouse lungs or bacterial cultures were homogenized in guanidinium thiocyanate buffer in a Mini-Beadbeater (Biospec Products, Bartlesville, OK), followed by phenol extraction of total nucleic acids. Total RNAs were purified with TRI reagent (Molecular Research Center, Cincinnati, OH). RT was performed using ThermoScript™ Reverse Transcriptase (Invitrogen, Carlsbad, CA), and quantification of cDNA was carried out using real time PCR with molecular beacons 25. RT primers, PCR primers and molecular beacons are listed in Table 1. Copy numbers of M. tuberculosis 16S rRNA were used as normalization factor to enumerate bacterial transcripts per cell, since 16S rRNA copy numbers correlate well with CFU in culture (26 and Fig. 2) and during the course of lung infection 22.
Table 1.
Nucleotide sequences of RT primers, PCR primers, and molecular beacons for bacterial gene expression measurements.
| Gene | RT primers | PCR primers | Molecular beacons |
|---|---|---|---|
| katG (Rv1908) | tgacctcccacccgacttgtg | catgggtcccgttgcgagata cccggatctggctcttaaggc |
FAMagcgcgatccggtccctgcg gtcagcgcgctDabcyl |
| glbN (Rv1542c) | caggctgaagtggtgcatggtaa | caaacgtgagccgatcagcat cggcaagcacacgaacataga |
FAMccgcggcatgaggccatcga agtcgtcgcggDabcyl |
| ahpC (Rv2428) | tgttggggtcgacgataaaggtc | ggcgttcagcaagctcaatgac agcatcgggaagggtaacgtttt |
FAMccggcggcccagatcctggg ggtttcgcgccggDabcyl |
| trxB2 (Rv3913) | tcagcggcccgtgaagtgatac | ctggtcttcgagggcacgtct cccgcatctcatccatcaactc |
FAMacgcgcgacgtggagaacta cccgggagcgcgtDabcyl |
| msrA (Rv0137c) | tggggtagcgctgcaggtagt | cttccagatccacgacccgacaac gatccgcttttgctgctcatcgaa |
FAMgcgcgaacgaccggggga ccagctaccgcgcDabcyl |
| hmp (Rv3571) | gatccgattccagcgacttgaa | ctcgttgtgcagttcgccctac gagggttgtggggacgaagtt |
FAMcgggccaccgccgacgggt acgcctcggcccgDabcyl |
Nucleotide sequences were obtained from http://genolist.pasteur.fr/TubercuList/48. RT and PCR primers (direction: 5′ to 3′, one primer per line) were designed by using the software Oligo 6.6 (Molecular Biology Insights, Cascade, CO) and were purchased from Integrated DNA Technologies (Coralville, Iowa). Molecular beacons were synthesized by Biosearch Technologies (Novato, CA). The nucleotide sequences of primers and molecular beacons for M. tuberculosis 16S rRNA, sodA and sodC were published previously 22. FAM, iodoacetamide derivative of fluorescein (5-iodoactamidofluorescein); Dabcyl, 4-(4′-dimethylaminophenylazo)-benzoic acid) succinimidyl ester.
Figure 2. M. tuberculosis growth and changes of bacterial transcripts encoding antioxidant enzymes in liquid cultures during adaptation to gradual oxygen depletion.
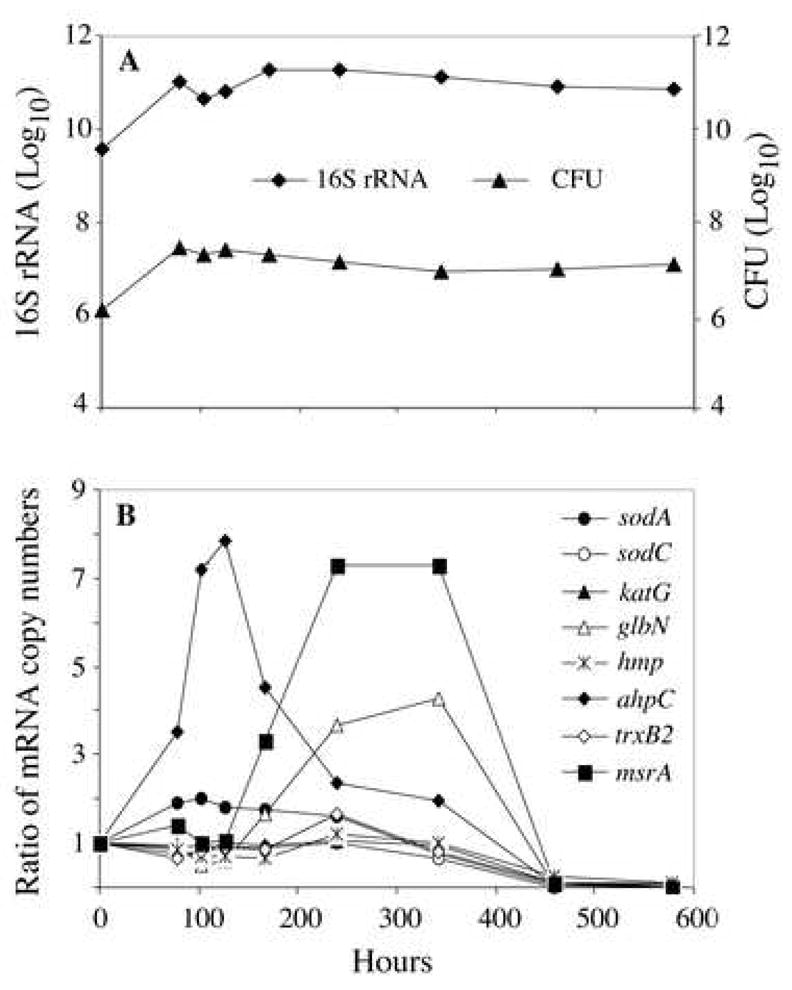
M. tuberculosis was cultured under conditions of gradual O2 depletion established by Wayne and Hayes 21. Bacterial RNA was extracted, and transcripts were enumerated by real-time RT-PCR with molecular beacons. Panel A: M. tuberculosis growth curve and 16S rRNA levels. Panel B: Changes of M. tuberculosis transcripts encoding antioxidant enzymes during gradual oxygen depletion. Levels of transcripts per cell were obtained by dividing mRNA copy number by the corresponding 16S rRNA copy number. Shown are ratios of normalized mRNA copy numbers determined at hour 78, 102, 126, 168, 240, 342, 460 and 578 relative to aerated mid-log cultures (hour 0). Replicate experiments produced similar results.
Results
Transcriptional profiles of M. tuberculosis genes encoding antioxidant enzymes in the mouse lung
To investigate the antioxidant response of M. tuberculosis during mouse lung infection by transcriptional profiling, total lung RNA was extracted at various times post-infection (the course of mouse lung infection is shown in Fig. 1A), and specific bacterial transcripts were enumerated by quantitative RT-PCR. As a reference gene we selected cydA, which encodes a subunit of cytochrome bd oxidase, an alternative terminal oxidase that is transiently upregulated at the transition from acute to chronic infection 27 (Fig. 1A). The first set of antioxidant genes probed included sodA, sodC and katG. These genes encode, respectively, the secreted, iron-dependent SodA 28, 29, the membrane bound, copper/zinc-dependent SodC 30–32 and catalase (KatG) 33. The two SODs and KatG sequentially convert superoxide into molecular oxygen and water 33. KatG also detoxifies peroxynitrite that arises from the reaction of nitric oxide (NO) and superoxide 7. Levels of sodA and sodC mRNAs were highest at day 12 post-infection and decreased starting at day 15, when the Th1 cytokine IFNγ is upregulated 22. At day 30 mRNA levels for sodA and sodC were 9- and 6-fold lower than at day 12 (Fig. 1B). The katG transcript levels were essentially stable during acute infection and showed a significant drop only at day 21. At day 30 katG mRNA was 2.5-fold lower than at day 12 and remained low thereafter (Fig. 1B).
Figure 1. M. tuberculosis growth and normalized copy number of bacterial transcripts encoding antioxidant enzymes in infected mouse lung.
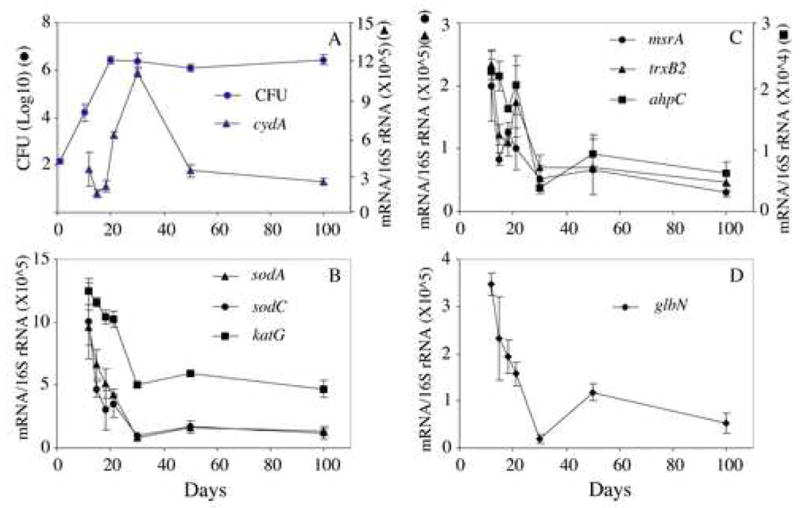
Panel A: M. tuberculosis growth and copy numbers of cytochrome bd oxidase (cydA) during infection. In lungs of C57BL/6 mice infected with M. tuberculosis H37Rv via the respiratory route, M. tuberculosis grew exponentially for about 20 days followed by chronic phase characterized with stabilization of bacterial counts (filled circles). Copy numbers of cydA transcripts were determined by real time RT-PCR and normalized against 16S rRNA (filled triangles, data from 27). Panels B–D: Copy numbers of bacterial transcripts encoding antioxidant enzymes. Normalized copy numbers of M. tuberculosis mRNAs against 16S rRNA were expressed as mean ± SD of data obtained from lungs of 3 or 4 mice per time point.
We also measured transcript levels for key enzymes in the thio-redox systems, which are involved in detoxification of H2O2 and peroxynitrite. The alkyl hydroperoxide reductase (AhpC and AhpD) system and the thioredoxin/thioredoxin reductase (TrxB2) system participate in detoxification of peroxide and peroxynitrite via oxidation-reduction of a conserved NH2-terminal cysteine 8–11. Similarly, the methionine and methionine sulfoxide reductase (MsrA) system scavenges ROS and RNS via cycles of oxidation and reduction of methionine residues in surface-exposed proteins 12. The ahpC transcript levels were essentially stable during acute infection and decreased after day 21 (Fig. 1C). At day 30 ahpC mRNA levels were 6-fold lower than at day 12. The transcript levels for trxB2 and msrA were highest at day 12 and decreased with bacterial growth arrest. By day 30 the transcript levels of these two transcripts were 3- to 4-fold lower than at day 12 and remained low thereafter (Fig. 1C).
From among enzymes that directly detoxify NO, we measured mRNA transcripts for the truncated hemoglobin TrHbN encoded by glbN. TrHbN exhibits an oxygen-dependent NO consumption activity 13, 14 and protects heterologous hosts from growth inhibition by NO 34. The copy number of the glbN transcript was highest at day 12 and decreased 18-fold by day 30 (Fig. 1D). At day 50 the glbN transcript levels showed a transient, 6-fold increase relative to day 30 and then decreased further by day 100 (Fig. 1E). Levels of the transcript encoding flavohemoglobin (hmp), were below detection (data not shown).
Transcriptional profiles of M. tuberculosis genes encoding antioxidant enzymes during gradual oxygen depletion in vitro
Gradual depletion of oxygen in vitro also causes bacterial growth arrest of M. tuberculosis, with similar transcriptional changes as seen in vivo 22, 27. In the Wayne model 21, tubercle bacilli stop growing at ~78 h, when the dissolved oxygen approaches 1% of saturation (the microaerophilic stage of non-replicating persistence, NRP-1) 21. Anaerobiosis (NRP-2) commences at ~200 hrs, when the dissolved oxygen content decreases below 0.06% saturation (Fig. 2A).
To compare the transcriptional profiles of genes encoding antioxidant enzymes in O2-starved cultures of M. tuberculosis with the in vivo data, M. tuberculosis transcripts characterized in the mouse lung were also enumerated in hypoxic cultures (Fig. 2B). We found that ahpC was induced during NRP-1 (up to 8-fold), while msrA and glbN were induced in NRP-2 (7.5-fold and 4-fold, respectively). All other transcripts showed no or modest (≤ 2-fold) increase. During the late stages of anaerobiosis, i.e., hour 460 through 578, all transcripts decreased drastically (Fig. 2B), when M. tuberculosis has very low metabolic activity but remains viable (26 and Fig. 2A).
Discussion
The antioxidant response of M. tuberculosis in vivo was characterized during the course of mouse respiratory infection by measuring levels of seven bacterial transcripts encoding enzymes involved in the oxidative and nitrosative stress responses. Transcript levels for sodA, sodC, glbN, trxB2, and msrA decreased starting at day 15, coincident with expression of host adaptive immunity 22, whereas mRNA levels for ahpC and katG began to decrease a few days later (day 21), when bacterial growth was halted. Thus the response of M. tuberculosis to host adaptive immunity in the mouse lung is characterized by the downregulation of antioxidant enzymes. These mouse data are consistent with earlier observations that the same bacterial transcripts were not induced either by IFNγ-activation of cultured macrophages infected with M. tuberculosis or by exposure of bacterial cultures to bacteriostatic concentrations of NO 16. Indeed, the data suggest the alternative possibility that the main function of the antioxidative pathways investigated in the present work is to detoxify harmful products produced by the bacterial aerobic metabolism of growing bacilli or by host defense mechanisms implemented during acute infection.
Several observations agree with the hypothesis proposed above. One is that a sodA mutant is attenuated for growth both in vitro and in mice 35. Another observation is that katG is essential for bacterial survival only between 2 and 4 weeks post-infection 36, that is, the same time interval in which respiratory-burst-deficient mice experience an increased load of wild-type M. tuberculosis in their lungs 37. Accordingly, the growth defect of an M. tuberculosis sodC mutant in murine macrophages is abolished when macrophages are derived from respiratory-burst-deficient mice but not from NOS2−/− mice 38. Further, none of M. tuberculosis mutants deleted for genes investigated in the present work shows decreased ability to persist during chronic infection of the mouse lung 39–42.
The downregulation of antioxidative pathways in growth-arrested tubercle bacilli in the mouse lung suggests that the bacterial defense against ROS and RNS produced by the host adaptive immunity may be carried out by enzymes other than those investigated in the present paper. One potential candidate is cytochrome bd oxidase, encoded by cydABDC gene cluster. This gene cluster is transiently upregulated when tubercle bacilli stop growing in the mouse lung (Fig. 1A), and it is required for the transition from acute to chronic mouse lung infection 27. Due to the high affinity of cytochrome bd oxidase for oxygen, this enzyme may act as oxygen scavenger and thus prevent cellular damage from potential excessive production of ROS, as postulated for other bacteria 43, 44.
Several considerations derive from the transcriptional profiles associated with gradual oxygen depletion in vitro. First, the upregulation of ahpC during NRP-1 may protect tubercle bacilli from oxidative stress and also prevent protein misfolding or unfolding 45 under microaerobic conditions. Second, the elevated levels of msrA during NRP-2 may help maintain cellular redox balance and deal with damage under anaerobic conditions. Third, the high expression of glbN during NRP-2 suggests the possibility that homodimeric hemoglobin, which displays extremely high oxygen binding affinity and cooperativity, facilitates oxygen diffusion to the terminal oxidase 13, 46. Since no upregulation of these genes was observed in the mouse lung, these pathways have presumably evolved in tubercle bacilli to adapt to hypoxic/anoxic conditions that are not found in the lung of infected mice. Transcriptome microarray analysis of M. tuberculosis in human lungs of tuberculosis patients also reveals no upregulation of M. tuberculosis antioxidant genes investigated in this study 47, suggesting that the physiological status of tubercle bacilli is similar in the murine model and in granulomas from human tuberculosis.
In conclusion, immunity-induced arrest of M. tuberculosis growth in the mouse lung is associated with an overall decrease of the levels of transcripts encoding antioxidant enzymes. These data imply that a major function of these genes is to defend tubercle bacilli from ROS and RNS produced by the host during acute infection and from ROS produced by the bacterial aerobic metabolism. The data add to existing evidence that the antioxidant response is not essential for M. tuberculosis persistence in vivo and lend further credence to the idea that the key survival strategy of this pathogen lies in its ability to enter a state of slow growth or no growth accompanied by decreased metabolic activity rather than to implement active defense mechanisms against the adaptive host immune response.
Acknowledgments
We thank Ron LaCourse (Trudeau Institute) for mouse infection and Karl Drlica for critical reading of this manuscript. L.S. is the recipient of a grant from the New York University Center For AIDS Research. The work was supported by grants from the NIH (AI-37844 and AI-059557) (R.J.N.), from the Medical Research Services of the U.S. Department of Veterans Affairs (C.D.S.), and from the NIH (AI-36989) and the Futura Foundation (M.L.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 2.North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 3.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol Oct. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266(11):6957–6965. [PubMed] [Google Scholar]
- 6.Huycke MM, Moore D, Joyce W, et al. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol. 2001;42(3):729–740. doi: 10.1046/j.1365-2958.2001.02638.x. [DOI] [PubMed] [Google Scholar]
- 7.Wengenack NL, Jensen MP, Rusnak F, Stern MK. Mycobacterium tuberculosis KatG is a peroxynitritase. Biochem Biophys Res Commun. 1999;256(3):485–487. doi: 10.1006/bbrc.1999.0358. [DOI] [PubMed] [Google Scholar]
- 8.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295(5557):1073–1077. doi: 10.1126/science.1067798. Epub 2002 Jan 1017. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger T, Budde H, Flohe L, et al. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;423(1):182–191. doi: 10.1016/j.abb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 10.Wieles B, Nagai S, Wiker HG, Harboe M, Ottenhoff TH. Identification and functional characterization of thioredoxin of Mycobacterium tuberculosis. Infect Immun. 1995;63(12):4946–4948. doi: 10.1128/iai.63.12.4946-4948.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Hillas PJ, Ortiz de Montellano PR. Reduction of peroxides and dinitrobenzenes by Mycobacterium tuberculosis thioredoxin and thioredoxin reductase. Arch Biochem Biophys. 1999;363(1):19–26. doi: 10.1006/abbi.1998.1056. [DOI] [PubMed] [Google Scholar]
- 12.Taylor AB, Benglis DM, Jr, Dhandayuthapani S, Hart PJ. Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine. J Bacteriol. 2003;185(14):4119–4126. doi: 10.1128/JB.185.14.4119-4126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couture M, Yeh SR, Wittenberg BA, et al. A cooperative oxygen-binding hemoglobin from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1999;96(20):11223–11228. doi: 10.1073/pnas.96.20.11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidon-Chanal A, Marti MA, Crespo A, et al. Ligand-induced dynamical regulation of NO conversion in Mycobacterium tuberculosis truncated hemoglobin-N. Proteins. 2006;10:10. doi: 10.1002/prot.21004. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Butcher PD, Mangan JA, Rajandream MA, Coates AR. Regulation of hmp gene transcription in Mycobacterium tuberculosis: effects of oxygen limitation and nitrosative and oxidative stress. J Bacteriol. 1999;181(11):3486–3493. doi: 10.1128/jb.181.11.3486-3493.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 18.Voskuil MI, Schnappinger D, Visconti KC, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orme IM. A mouse model of the recrudescence of latent tuberculosis in the elderly. Am Rev Respir Dis Mar. 1988;137(3):716–718. doi: 10.1164/ajrccm/137.3.716. [DOI] [PubMed] [Google Scholar]
- 20.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94(10):5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, Jung YJ, Tyagi S, Gennaro ML, North RJ. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc Natl Acad Sci U S A. 2003;100(1):241–246. doi: 10.1073/pnas.0136863100. Epub 2002 Dec 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung YJ, LaCourse R, Ryan L, North RJ. Virulent but not avirulent Mycobacterium tuberculosis can evade the growth inhibitory action of a T helper 1-dependent, nitric oxide Synthase 2-independent defense in mice. J Exp Med. 2002;196(7):991–998. doi: 10.1084/jem.20021186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn PL, North RJ. Virulence ranking of some Mycobacterium tuberculosis and Mycobacterium bovis strains according to their ability to multiply in the lungs, induce lung pathology, and cause mortality in mice. Infect Immun. 1995;63(9):3428–3437. doi: 10.1128/iai.63.9.3428-3437.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 26.Desjardin LE, Hayes LG, Sohaskey CD, Wayne LG, Eisenach KD. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J Bacteriol. 2001;183(18):5311–5316. doi: 10.1128/JB.183.18.5311-5316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Sohaskey CD, Kana BD, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A. 2005;102(43):15629–15634. doi: 10.1073/pnas.0507850102. Epub 12005 Oct 15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(2):453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 29.Tullius MV, Harth G, Horwitz MA. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect Immun. 2001;69(10):6348–6363. doi: 10.1128/IAI.69.10.6348-6363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spagnolo L, Toro I, D’Orazio M, et al. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J Biol Chem. 2004;279(32):33447–33455. doi: 10.1074/jbc.M404699200. Epub 32004 May 33423. [DOI] [PubMed] [Google Scholar]
- 31.D’Orazio M, Folcarelli S, Mariani F, Colizzi V, Rotilio G, Battistoni A. Lipid modification of the Cu, Zn superoxide dismutase from Mycobacterium tuberculosis. Biochem J. 2001;359(Pt 1):17–22. doi: 10.1042/0264-6021:3590017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu CH, Tsai-Wu JJ, Huang YT, Lin CY, Lioua GG, Lee FJ. Identification and subcellular localization of a novel Cu, Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 1998;439(1–2):192–196. doi: 10.1016/s0014-5793(98)01373-8. [DOI] [PubMed] [Google Scholar]
- 33.Heym B, Zhang Y, Poulet S, Young D, Cole ST. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol. 1993;175(13):4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathania R, Navani NK, Gardner AM, Gardner PR, Dikshit KL. Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli. Mol Microbiol. 2002;45(5):1303–1314. doi: 10.1046/j.1365-2958.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 35.Edwards KM, Cynamon MH, Voladri RK, et al. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am J Respir Crit Care Med. 2001;164(12):2213–2219. doi: 10.1164/ajrccm.164.12.2106093. [DOI] [PubMed] [Google Scholar]
- 36.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52(5):1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 37.Cooper AM, Segal BH, Frank AA, Holland SM, Orme IM. Transient loss of resistance to pulmonary tuberculosis in p47(phox−/−) mice. Infect Immun. 2000;68(3):1231–1234. doi: 10.1128/iai.68.3.1231-1234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. Cu, Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun. 2001;69(8):4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heym B, Stavropoulos E, Honore N, et al. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect Immun. 1997;65(4):1395–1401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Springer B, Master S, Sander P, et al. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect Immun. 2001;69(10):5967–5973. doi: 10.1128/IAI.69.10.5967-5973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dussurget O, Stewart G, Neyrolles O, Pescher P, Young D, Marchal G. Role of Mycobacterium tuberculosis copper-zinc superoxide dismutase. Infect Immun. 2001;69(1):529–533. doi: 10.1128/IAI.69.1.529-533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaushal D, Schroeder BG, Tyagi S, et al. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A. 2002;99(12):8330–8335. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duwat P, Sourice S, Cesselin B, et al. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J Bacteriol. 2001;183(15):4509–4516. doi: 10.1128/JB.183.15.4509-4516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das A, Silaghi-Dumitrescu R, Ljungdahl LG, Kurtz DM., Jr Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J Bacteriol. 2005;187(6):2020–2029. doi: 10.1128/JB.187.6.2020-2029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A. 2006;103(8):2552–2557. doi: 10.1073/pnas.0510770103. Epub 2006 Feb 2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh SR, Couture M, Ouellet Y, Guertin M, Rousseau DL. A cooperative oxygen binding hemoglobin from Mycobacterium tuberculosis. Stabilization of heme ligands by a distal tyrosine residue. J Biol Chem. 2000;275(3):1679–1684. doi: 10.1074/jbc.275.3.1679. [DOI] [PubMed] [Google Scholar]
- 47.Rachman H, Strong M, Ulrichs T, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun Feb. 2006;74(2):1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]


