Abstract
Pathogens, inflammatory signals, and stress cause acute transcriptional responses in cells. The induced expression of genes in response to these signals invariably involves transcription factors of the NF-κB and AP-1/ATF families. Activation of NF-κB factors is thought to be mediated primarily via IκB kinases (IKK), whereas that of AP-1/ATF can be mediated by stress-activated protein kinases (SAPKs; also named Jun kinases or JNKs). IKKα and IKKβ are two catalytic subunits of a core IKK complex that also contains the regulatory subunit NEMO (NF-κB essential modulator)/IKKγ. The latter protein is essential for activation of the IKKs, but its mechanism of action is not known. Here we describe the molecular cloning of CIKS (connection to IKK and SAPK/JNK), a previously unknown protein that directly interacts with NEMO/IKKγ in cells. When ectopically expressed, CIKS stimulates IKK and SAPK/JNK kinases and it transactivates an NF-κB-dependent reporter. Activation of NF-κB is prevented in the presence of kinase-deficient, interfering mutants of the IKKs. CIKS may help to connect upstream signaling events to IKK and SAPK/JNK modules. CIKS could coordinate the activation of two stress-induced signaling pathways, functions reminiscent of those noted for tumor necrosis factor receptor-associated factor adaptor proteins.
NF-κB is a ubiquitously expressed family of transcription factors controlling the expression of numerous genes involved in inflammatory and immune responses (1–3). NF-κB exists in a latent state in the cytoplasm, bound to inhibitors, collectively called IκBs (4). Various stimuli, including cytokines, pathogens, or pathogen-related factors, lead to proteasome-mediated degradation of inhibitory IκB proteins. This process is initiated by phosphorylation of IκB proteins on specific serine residues (S32 and S36 on IκBα), a step that marks these proteins for ubiquitination by the Skp1p-Cdc53p-F box protein (SCF) E3 ligase (5–15). The only mammalian kinases known to date to phosphorylate both critical serines on IκB proteins are IκB kinase α and β (IKKα and IKKβ) (16–20). They are composed of a catalytic kinase domain, a leucine zipper, and a helix–loop–helix (HLH) domain. The leucine zipper, is important for dimerization, whereas the HLH domain is required for full IKK activation, possibly through interactions with the kinase domain (21). IKKα and IKKβ knockout mice have been generated (22–27). Analysis of these mutant mice suggests that IKKβ is critical for activation of NF-κB by inflammatory cytokines, whereas IKKα is largely dispensable for this function, but is important during skin development, for example. It also has been reported, however, that if both IKKs are present, inflammatory signals may be transmitted in a hierarchical order, starting with activation of IKKα, and only then, IKKβ (28, 29).
IKKs are part of a large 900-kDa kinase complex that also includes the regulatory subunit NEMO (NF-κB essential modulator), also known as IKKγ or FIP3 (30–32). The essential role of NEMO/IKKγ in activating the NF-κB pathway is based, in part, on the inability of NEMO-deficient cell lines to respond to NF-κB activating signals, including lipopolysaccharide, IL-1, and Tax (31, 33). Antisense studies further demonstrate the essential role for NEMO/IKKγ in activation of the IKK complex, including the response to tumor necrosis factor (TNF) signaling (30). Finally, the phenotype of NEMO/IKKγ-deficient mice confirms the need for this protein in activation of NF-κB (34). NEMO/IKKγ contains a leucine zipper domain and several coiled–coil protein interaction domains. Despite the absolute requirement for NEMO/IKKγ in NF-κB activation, its mechanism of action is not understood. It has been speculated that NEMO/IKKγ may be required for the correct assembly of the IKKs and/or the recruitment to upstream signal transducing modules and/or depressing basal activity.
To better understand how the IKK complex is regulated and to find potential upstream connectors that link to NEMO/IKKγ, we used this essential subunit of the IKK complex as bait in yeast two-hybrid screens. Here we describe CIKS, a NEMO/IKKγassociated protein that connects to both the IKK and stress-activated protein kinase (SAPK)/Jun kinase (JNK) signaling complexes to mediate their activation.
Materials and Methods
Cell Culture and Biological Reagents.
Human embryonic kidney 293 and HeLa cells were maintained as described (35).
The CIKS polyclonal antibody was generated in rabbits and is directed against amino acids 562–574. Other antibodies were obtained commercially (Santa Cruz Biotechnology).
Mouse NEMO/IKKγ was cloned by PCR from a mouse liver cDNA library (CLONTECH). Truncation mutants of NEMO/IKKγ were generated by PCR. The human NF-κB inducing kinase (NIK) clone has been described (35). Full-length IKKα and IKKβ were PCR-amplified from a HeLa cell cDNA library. Full-length MEKK1 was a gift of M. Cobb, University of Texas Southwestern Medical Center, Dallas (36). cDNAs encoding NEMO/IKKγ, IKKα, IKKβ, NIK, and CIKS were cloned into pcDNA3.1-hemagglutinin (HA) or pcDNA3.1-FLAG (Invitrogen) for expression in mammalian cells. The 5′ end for construction of a full-length CIKS cDNA was obtained by PCR amplification with a 5′ primer that included the ATG as derived from the GenBank database (accession no. AI051544).
The kinase-deficient mutants of NIK (KK429–430AA), IKKα (K44A), and IKKβ (K44A) and of the catalytic domain of MEKK1 (K432A) were obtained by mutating the lysine residues in the ATP-binding domain to alanine. The kinase-deficient mutant of SEK1 (K390R) was a gift of J. Kyriakis, Massachusetts General Hospital, Charlestown.
Yeast Two-Hybrid Assays.
The cDNA encoding NEMO/IKKγ (amino acids 1–339) was cloned in-frame into the GAL4 DNA-binding vector pGBT9 (CLONTECH). This plasmid was used as bait in a two-hybrid screen of a human liver cDNA library in Saccharomyces cerevisiae Y190, according to the Matchmaker Two-Hybrid System Protocol (CLONTECH). Positive yeast clones were selected for their ability to grow in the absence of histidine. Yeast DNA was recovered, transformed into Escherichia coli HB101 carrying a leuB mutation, and sequenced.
In Vitro Translation and Glutathione S-Transferase (GST) Pull-Down Assays.
In vitro transcription and translation of 1 μg of CIKS* template (derived from the original isolate, lacking the first 87 aa) was performed with the TNT T7 Quick Master Mix kit (Promega) in the presence of [35S]methionine, according to the manufacturer's protocol.
Both the GST-NEMO/IKKγ fusion protein and GST alone were produced and purified as described (37). GST pull-down assays were performed by incubating an aliquot of GST-NEMO or GST, bound to glutathione-Sepharose beads, together with 5 μl of in vitro-translated HA-CIKS* in 200 μl of TWB buffer (20 mM Hepes, pH 7.9/60 mM NaCl/1 mM DTT/6 mM MgCl2/8.2% glycerol/0.1 mM EDTA) for 1 h at 4°C. Beads then were washed five times with 1 ml of NENTM buffer (250 mM NaCl/1 mM EDTA/20 mM Tris⋅HCl, pH 8/1.5% NP-40/0.5% dry milk), 1 mM, resuspended into migrating buffer and run on a SDS-polyacrylamide gel before autoradiography.
Cell Transfection, Immunoprecipitation, and Luciferase Assays.
HeLa cells (2 × 106) were transiently transfected as described (35). An aliquot (1/3) of the lysates was incubated for 2 h at 4°C with 2 μg of the indicated antibodies bound to agarose beads. The beads were washed extensively with lysis buffer and boiled in SDS sample buffer, and the supernatant was subjected to 10% SDS/PAGE.
For immunoprecipitation of endogenous NEMO/IKKγ, Jurkat cells (2.5 × 108) were stimulated with human TNF (2,000 units/ml) for the indicated period or left untreated. Cells were collected by centrifugation and lysed with 5 ml of 1% Triton X-100 lysis buffer. Nuclear and cellular debris was removed by centrifugation at 35,000 × g for 30 min at 4°C. Cell extracts were normalized for protein content, and 20 mg of extract was incubated for 8 h at 4°C with 2 μg of rabbit polyclonal anti-NEMO antibody (Santa Cruz Biotechnology). Immune complexes were collected by adding 30 μl of Protein A Sepharose, extensively washed with lysis buffer, and boiled in SDS sample buffer, and the resulting supernatants were subjected to 10% SDS/PAGE.
For luciferase assays, 293 or HeLa cells (4 × 105 cells per well) were seeded in 6-well (35 mm) plates. After 12 h, cells were transfected as described above with 0.5 μg of either the Ig-κB-luciferase or the CRE reporter gene plasmid (38) and with expression plasmids as indicated. The total amount of transfected DNA was kept constant by adding empty expression vector DNA as needed. Cell extracts were prepared 24 h after transfection, and reporter gene activity was determined by the luciferase assay system (Promega). A pRSV-β-galactosidase vector (0.2 μg) was used to normalize for transfection efficiencies. Cells were stimulated with human TNF (2,000 units/ml) or cAMP (1 mM) for 4 h before lysis.
Kinase Assay.
For SAPK/JNK activity, anti-HA immunoprecipitates were used in immune-complex kinase assays with GST–c-Jun as substrate as described (35).
For the IKK assay, transfected IKKα was immunoprecipitated by using anti-FLAG antibodies, and the kinase activity was assayed by using GST-IκBα (amino acids 1–54) as substrate.
Results
Identification of CIKS, a NEMO-IKKγ Interacting Protein.
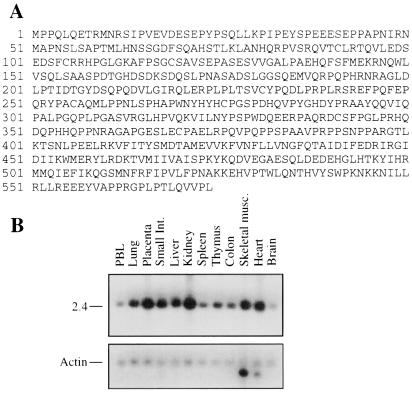
To gain insights into possible mechanisms of action of NEMO/IKKγ during activation of NF-κB, we searched by yeast two-hybrid analysis for proteins that physically interact with NEMO/IKKγ. As bait we used most of mouse NEMO/IKKγ (amino acids 1–339) fused to the DNA binding domain of the yeast GAL4 transcription factor. We screened a human liver cDNA library that expresses the encoded products as fusions with the GAL4 transactivation domain. Among 25 clones that were scored positive for interaction with the bait, three encoded portions of IKKα or IKKβ, proteins known to interact with NEMO/IKKγ. Two additional and identical cDNAs were homologous to uncharacterized expressed sequence tag sequences deposited in GenBank (the complete cDNA was obtained as outlined in Materials and Methods). The encoded protein of 574 aa was named CIKS (Fig. 1A). It has no obvious homology with known proteins in public databases. CIKS is ubiquitously expressed as a single, approximately 2.5-kB long mRNA transcript (Fig. 1B).
Figure 1.
(A) Predicted amino acid sequence of CIKS. (B) Detection of the ubiquitous 2.5-kB CIKS mRNA transcript on a Multiple Tissue Northern Blot (CLONTECH) (Upper). Actin mRNA is a control for RNA loading (Lower). The tissue source of the RNA is indicated.
To delineate the domain of NEMO/IKKγ responsible for its interaction with CIKS in yeast, we tested various truncation mutants of NEMO/IKKγ fused to GAL4BD for binding with the nearly full-length original CIKS clone. A C-terminal truncation mutant of NEMO/IKKγ lacking the last 100 aa (thus lacking the leucine zipper), and an N-terminal truncation mutant lacking the initial 50 aa were still able to interact with CIKS. However, no interaction could be detected between CIKS and an N-terminal deletion of NEMO/IKKγ lacking the first 100 aa (data not shown). This result suggests that the region between amino acids 50 and 100 of NEMO/IKKγ is necessary for interaction with CIKS. This region of NEMO/IKKγ also is required for binding to the IKKs (A.L. and U.S., unpublished data; ref. 39).
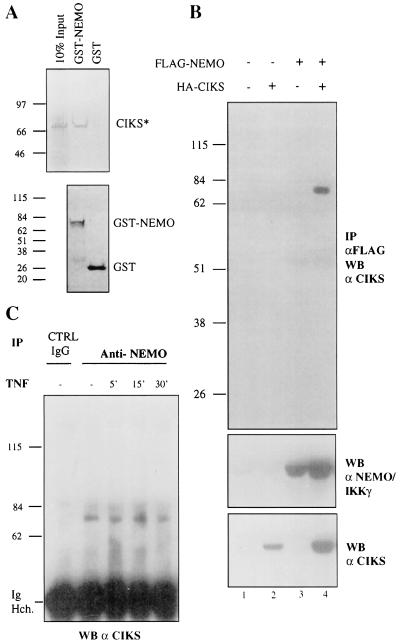
Several additional experiments were performed to confirm the interaction of NEMO/IKKγ and CIKS. First, a recombinant GST-mouse NEMO/IKKγ fusion could be shown to bind in vitro translated 35S-labeled CIKS (Fig. 2A; CIKS* refers to the protein generated from the original clone isolate). Second, epitope-tagged, exogenously expressed versions of NEMO/IKKγ and full-length CIKS could be coimmunoprecipitated in HeLa cell extracts. Immunoprecipitates of FLAG-NEMO/IKKγ contained HA-CIKS, as determined with anti-CIKS antibodies (Fig. 2B Top, lane 4; similar results were obtained with anti-HA antibodies, not shown). Of note, the amount of CIKS present in whole-cell lysates was consistently higher whenever NEMO/IKKγ was cotransfected (compare lanes 2 and 4, Fig. 2B Bottom), which may be caused by stabilization of CIKS by association with NEMO/IKKγ. Finally, and importantly, we were able to demonstrate an interaction of endogenous NEMO/IKKγ with endogenous CIKS. Untransfected Jurkat T cells, either stimulated with TNF or left untreated, were lysed and the extracts were immunoprecipitated with anti-NEMO/IKKγ antibodies (or control anti-CD4 antibodies). The immunoprecipitates were subjected to a Western analysis with anti-CIKS antibodies (Fig. 2C). A positive signal corresponding in size to CIKS was detected in anti-NEMO/IKKγ immunoprecipitates, but not in the control. The association of CIKS and NEMO/IKKγ did not appear to be modulated by stimulation of cells with TNF for up to 30 min. The demonstrated interaction of the endogenous proteins (as opposed to overexpressed, transfected proteins) strongly suggests a physiologic role. Taken together, the results identify CIKS as a previously unknown NEMO/IKKγ-interacting protein.
Figure 2.
Physical interaction between CIKS and NEMO/IKKγ. (A) GST pull-down assay: GST- NEMO/IKKγ or GST (negative control) (Upper, second and third lanes, respectively) was incubated with in vitro-translated CIKS* (amino acids 88–574) (Upper, first lane). (Lower) Aliquots of GST or GST-NEMO/IKKγ stained with Coomassie blue are shown. Molecular weight markers (in kDa) are indicated. (B) Anti-Flag immunoprecipitates (IP) of HeLa cell extracts transfected with FLAG-NEMO/IKKγ and CIKS were Western-blotted with anti-CIKS antibodies to detect coimmunoprecipitated CIKS (Top, lane 4). (Middle and Bottom) Western blots with anti-Flag (anti-NEMO) and anti-CIKS antibodies on whole-cell extracts are shown, respectively. (C) Coimmunoprecipitation of endogenous CIKS with endogenous NEMO/IKKγ. Immunoprecipitates of Jurkat cells with anti-NEMO antibodies were subjected to Western blot (WB) analysis with anti-CIKS antibodies. Jurkat cells were treated with TNF for the times indicated or left untreated. Extracts also were immunoprecipitated with an unrelated antibody (CTRL) raised against CD4. Ig Hch, Ig heavy chain.
CIKS Interacts with Other Components of the IKK-Complex.
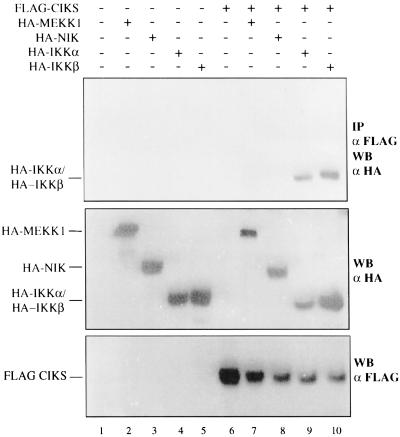
Next we investigated whether CIKS could interact with other components of the IKK complex or with known activators of the IKKs, NIK and MEKK1. HeLa cells were transfected with FLAG-tagged CIKS together with HA-tagged NIK, MEKK1, IKKα, or IKKβ (Fig. 3). Neither NIK nor MEKK1 were coimmunoprecipitated with CIKS (Fig. 3, lanes 7 and 8). However, both IKKα and IKKβ were coimmunoprecipitated, even in the absence of overexpressed NEMO/IKKγ (Fig. 3, lanes 9 and 10). This result raised the question whether endogenous NEMO/IKKγ mediated the detected interaction between CIKS and IKKα and IKKβ, or if these proteins also might be able to interact directly. This question was investigated further in yeast two-hybrid tests. NEMO/IKKγ, NIK, IKKα, and IKKβ coding sequences were cloned in-frame with the GAL4-DNA binding domain (in vector pGBT9), and CIKS was cloned in-frame with the GAL4 activation domain (in vector pGAD24). No interaction was detected in yeast between CIKS and NIK, as expected from the HeLa transfection experiments, whereas CIKS clearly interacted not only with NEMO/IKKγ, but also with IKKα and IKKβ (data not shown). Taken together these results demonstrate that CIKS can interact directly with NEMO/IKKγ and, based on the yeast two-hybrid data at least, it appears to also interact with the IKKs. The interaction of CIKS with components of the IKK complex in mammalian cells was further confirmed by detection of endogenous IκB-kinase activity in immunoprecipitates of epitope-tagged, transfected CIKS (data not shown).
Figure 3.
CIKS interacts with IKKα and IKKβ, but not with NIK or MEKK1 in transfected cells. HeLa cells (2 × 106) were cotransfected with an expression vector encoding FLAG-tagged CIKS (5 μg) together with expression vectors encoding HA-MEKK1 (3 μg), HA-NIK (3 μg), HA-IKKα (5 μg), or HA-IKKβ (3 μg). Anti-FLAG immunoprecipitates (IP) from cell lysates were analyzed by Western blotting (WB) with anti-HA antibodies to detect coprecipitated proteins (Top). Western blots with the anti-HA antibody (Middle) or anti-FLAG antibody (Bottom) were performed to check for expression of the constructs.
CIKS Activates IKK/NF-κB and SAPK.
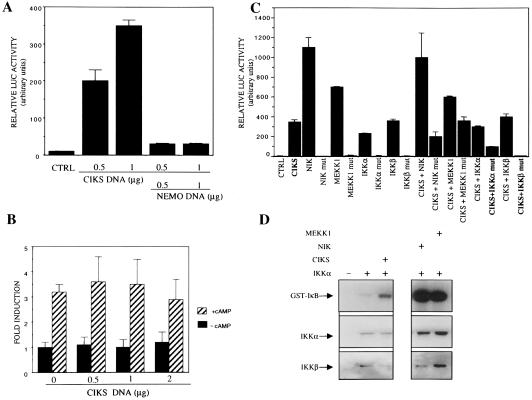
The strong interaction of CIKS with the IKK complex prompted us to investigate the effect of overexpression of CIKS on activation of NF-κB. HeLa cells were transfected with an Ig-κB luciferase reporter plasmid in the presence of increasing amounts of CIKS. CIKS activated the κB-dependent reporter in a dose-dependent manner (Fig. 4A). Moreover, the CIKS-mediated activation was blocked by cotransfected NEMO/IKKγ, most likely through binding and titration of CIKS away from functional signaling complexes. To evaluate the specificity of CIKS in reporter activation, we analyzed the effect of overexpressed CIKS on the activity of an unrelated reporter. HeLa cells were transfected with a CRE-dependent luciferase reporter in the presence of increasing amounts of CIKS, and cells were either stimulated with cAMP or left untreated. Neither basal nor cAMP-induced CRE activity was affected by CIKS, indicating that CIKS did not nonspecifically lead to transactivation of reporter plasmids (Fig. 4B).
Figure 4.
CIKS stimulates IKK and NF-κB activity. (A) Relative luciferase activity observed in HeLa cells transfected in triplicate with 0.5 μg of Ig-kB luciferase reporter plasmid, with or without CIKS and NEMO/IKKγ expression vectors, as indicated. Values shown (in arbitrary units) represent the means (± S.D) of at least three independent experiments, normalized for β-galactosidase activity of a cotransfected Rous sarcoma virus-β-galactosidase plasmid. (B) HeLa cells (4 × 105) were transfected in duplicate with 0.5 μg of CRE-luciferase reporter plasmid together with increasing amounts of CIKS expression vector, as shown. Twenty four hours after transfection, cells were either stimulated for 5 h with cAMP (1 mM final concentration) or left untreated and then harvested. Measurements were normalized for β-galactosidase activity and data (± SD) are shown as fold induction. (C) Analysis of the effects of wild-type or kinase-deficient mutants (mut) of NIK, MEKK1, IKKα, or IKKβ on CIKS-dependent NF-κB activation. HeLa cells (4 × 105) were transfected in triplicate with 0.5 μg of Ig-κB luciferase reporter plasmid, together with CIKS (0.5 μg) and NIK (0.2 μg), MEKK1 (0.4 μg), IKKα (0.4 μg), IKKβ (0.4 μg), NIK mut (0.2 μg), MEKK1 mut (0.2 μg), IKKα mut (0.4 μg), or IKKβ mut (0.4 μg). Data from a representative experiment, normalized for β-galactosidase activity, are shown as relative luciferase activity (± SD). Similar results were obtained in three independent experiments. (D) HeLa cells (3 × 106) were left untransfected, or transfected with an expression vector encoding Flag-IKKα (2 μg), alone or together with expression vectors encoding HA-CIKS (4 μg), HA-NIK (3 μg), or HA-MEKK1 (5 μg), as indicated. Twenty four hours after transfection cells were harvested and IKKα was immunoprecipitated with anti-FLAG. IKK kinase activity was assayed as described in Materials and Methods with GST-IkBα (amino acids 1–54) as substrate. Also shown are an immunoblot for transfected IKKα present in the various extracts (anti-FLAG) and an immunoblot for endogenous, coprecipitated IKKβ present in the in vitro kinase assays.
To further explore mechanisms involved in the CIKS-mediated activation of the κB-dependent reporter, we evaluated the effects of cotransfected wild-type or kinase-deficient mutants of NIK, MEKK1, IKKα, or IKKβ (Fig. 4C). Cotransfection of the wild-type kinases produced no apparent synergistic effects with CIKS. Cotransfection of kinase-deficient mutants of MEKK1 and NIK did not inhibit or only partially inhibited CIKS-mediated activation, respectively, whereas kinase-deficient IKKα or IKKβ mutants clearly did inhibit activation. This latter finding suggests that CIKS activated NF-κB in an IKK-dependent mechanism.
To more directly demonstrate IKK-dependent activation of NF-κB by CIKS, we determined the kinase activity of immunoprecipitates of Flag-tagged, transfected IKKα in the presence of cotransfected CIKS. HeLa cells were transfected with an IKKα expression vector in the presence of CIKS, NIK, MEKK1, or empty expression vectors. IKKα was immunoprecipitated with anti-Flag antibodies, and associated kinase activity was assayed by using GST-IκBα as a substrate (Fig. 4D). As expected, NIK and MEKK1 activated IKKα-associated kinase activity, but so did CIKS, further supporting the conclusion that CIKS induced κB-dependent reporter activity via IKKs (Fig. 4D Middle shows the relative amounts of transfected IKKα present in cell extracts). The anti-Flag immunoprecipitates also contained endogenous IKKβ (Fig. 4D Bottom), suggesting that transfected IKKα was at last partly assembled into complexes with IKKβ. Similar results were obtained when these assays were performed by transfection of 293 cells (data not shown).
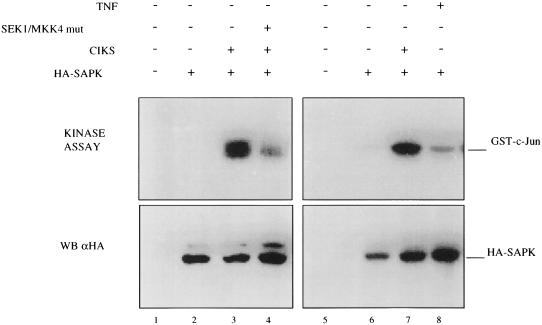
TNF, IL-1, and Toll receptors activate both IKK and SAPK/JNK and for each of these receptors the initial signal events that target these kinases are the same, although eventually the paths bifurcate. We investigated whether overexpressed CIKS also might activate SAPK/JNK. HeLa cells were transiently cotransfected with CIKS and HA-tagged SAPK/JNK expression vectors. Twenty four hours after transfection, cells were harvested and the activity of the transfected SAPK/JNK was assayed in an immune-complex kinase assay. Overexpressed CIKS potently activated SAPK/JNK (Fig. 5, lane 3), and this activation was partially blocked by a kinase-deficient mutant form of SEK (K390R), a direct upstream activator of SAPK/JNK. In contrast, CIKS failed to activate the p38 or extracellular signal-regulated kinase/mitogen-activated protein kinase in similar assays (data not shown). Therefore, CIKS activates both IKK and SAPK/JNK. The dual effect suggests a role for CIKS similar to that of TNF receptor-associated factor (TRAF) adaptors (40–45), but it does not imply any role for CIKS in TRAF-dependent pathways.
Figure 5.
Transfected CIKS activates SAPK/JNK activity. (Upper) HeLa cells (2 × 106) were cotransfected with an expression vector encoding HA-SAPK/JNK (2 μg) and an expression vector encoding CIKS (4 μg) together with, or without, an expression vector for a kinase-deficient mutant of SEK/MKK4 (2 μg). Twenty four hours after transfection, cells were stimulated with TNF (2,000 units/ml) for 15 min, or left untreated, and then harvested. SAPK/JNK activity was assayed as described in Materials and Methods with GST–c-JUN (amino acids 1–79) as substrate. (Lower) An HA immunoblot is shown to document the relative amounts of HA-SAPK/JNK present in the cell extracts.
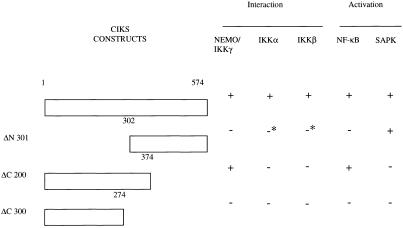
To explore whether distinct domains of CIKS may be involved in its activation of NF-κB and SAPK/JNK, we tested the ability of several CIKS truncation mutants to interact with IKK components and to activate IKK or SAPK/JNK upon overexpression (results summarized in Fig. 6). A CIKS mutant lacking the N-terminal 301 aa retains significant, albeit reduced, SAPK/JNK-inducing activity, whereas it lacks the ability to activate κB reporters. This mutant also is impaired in its ability to interact with IKKα or IKKβ, and it completely fails to interact with NEMO/IKKγ. By contrast, a CIKS mutant lacking the C-terminal 200 aa no longer activates SAPK/JNK, but retains significant, albeit reduced, ability to activate κB reporters. Consistent with its ability to activate NF-κB, this latter mutant also still interacts with NEMO/IKKγ. Finally, a C-terminal truncation of 300 aa cannot activate or interact in any of these assays. We infer that sequences between amino acids 302 and 574 are necessary and sufficient for SAPK/JNK activation, and that sequences located in the first 374 aa are necessary and sufficient for κB reporter activation. The data also suggest that NF-κB activation by CIKS correlates primarily with its ability to interact with NEMO/IKKγ.
Figure 6.
Distinct domains of CIKS required for activation of NF-κB and SAPK/JNK. CIKS mutants ΔN 301, ΔC 200, and ΔC 300 are truncations lacking the amino-terminal 301 aa, the carboxyl-terminal 200 aa, or the carboxyl-terminal 300 aa, respectively. The ability of these deletion mutants to interact with NEMO/IKKγ, IKKα, and IKKβ was assayed in coimmunoprecipitation experiments of transfected HeLa cells, as described for Fig. 3. The ability of these deletion mutants to activate NF-κB and SAPK/JNK was assayed with luciferase assays (as described for Fig. 4) and by immunecomplex kinase assay (as described in Fig. 5), respectively. +, positive assay; −, negative assay; *, a marginal effect in at least some assays.
Discussion
We have cloned and characterized CIKS, a previously undescribed protein that interacts with the IKK complex, and, when overexpressed, activates both IKK and SAPK/JNK activities. CIKS bears no obvious homologies with known proteins in public databases. The interaction of CIKS and NEMO/IKKγ, the regulatory component of the IKK complex, could be demonstrated in vitro and by coimmunoprecipitations from cells, including untransfected cells. Initial data suggest that CIKS also may interact with the two other components of the IKK core complex, the catalytic subunits IKKα and IKKβ. Activation of a κB reporter by overexpressed CIKS is blocked by kinase-deficient, interfering mutants of IKKα or IKKβ. Finally, CIKS stimulates the activity of the IKKs. The sum of the data implies CIKS can connect to both the IKK and SAPK/JNK modules, presumably mediating a signal response. This expands the network of proteins involved in NF-κB and SAPK/JNK activation. The apparent complexity may reflect the fact that many diverse signals affect these two pathways.
An important outstanding question is what signals CIKS may help to transmit. To date we have been unable to block activation of NF-κB in the presence of various exogenously introduced CIKS truncations (A.L., A.C., and U.S., unpublished observations). It is possible that we have not used the appropriate cell, signal, or mutant, assuming transfected CIKS mutants could be made to dominantly interfere with endogenous signaling pathways. The generation of mice deficient in CIKS should aid identification of pathways that require this protein.
Could CIKS play a role in more thoroughly described signaling pathways, such as those initiated by the TNF, IL-1, or Toll receptors? We have not observed an interaction between TRAF2 or TRAF6 and CIKS (A.L., A.C., and U.S., unpublished observations). Further, given that the NEMO/IKKγ-IKK complex appears to be directly recruited to the TNF receptor via interaction of NEMO with the receptor interacting protein (RIP)/TRAF2 complex (46, 47), a role for CIKS in this particular activation of NF-κB seems less likely, but cannot be ruled out. Of note in this regard, the association of CIKS with NEMO appears not to be modulated during a short-term stimulation with TNF. Much less is known about how the SAPK/JNK pathway is activated by the inflammatory receptors, although recent evidence suggests a role for MEKK1 in this process (48). Concerning a possible involvement of CIKS in the IL-1 or Toll pathways, this possibility also would seem less likely if it turns out that the IRAK/TRAF6 complex can directly associate with IKK and SAPK/JNK modules. IRAK/TRAF6 is essential for the IL-1 receptor and Toll signaling pathways and may function analogous to RIP/TRAF2 in the TNF pathway (49, 50). Aside from TNF and IL-1/Toll pathways, however, there are many other, much less well-understood signaling pathways leading to IKK and SAPK/JNK and these pathways are likely to require their own connectors, possibly CIKS, to communicate with these kinases.
CIKS may connect upstream signaling paths to only a subset of the IKK or SAPK modules. If so, this would explain why CIKS has not previously been identified as a copurifying component of the IKK complex, even though the association of endogenous CIKS and NEMO already can be detected in unstimulated cells. It is conceivable that CIKS helps to preassemble a fraction of IKK complexes with signal-responsive proteins to facilitate signal transmission. Given the fairly wide pattern of expression of CIKS, one may speculate that CIKS either functions in diverse pathways, or, alternatively, in a ubiquitous pathway.
CIKS is able to independently associate with NEMO as well as with the IKKα and IKKβ subunits, at least based on yeast two-hybrid analyses. Overexpression of CIKS deletions indicates a close correlation between NF-κB induction and NEMO-binding activities. Together, the data suggest a functionally important interaction between NEMO and CIKS that may be further augmented by binding of CIKS to the catalytic subunits of IKK. It remains to be determined, however, whether CIKS can simultaneously contact all three subunits of the IKK complex. As for the mechanism by which CIKS might activate the IKK complex, a potentially relevant finding might be the observation that transfected CIKS self-associates (data not shown).
The activities of CIKS loosely resemble those of TRAF proteins downstream of TNF, IL-1, and Toll receptor pathways, because overexpression of CIKS or of TRAF2, TRAF5, or TRAF6 activates SAPK/JNK and IKK (40–45). Activation of these two stress-inducible pathways (SAPK/JNK and IKK) may be frequently linked. Signaling through a common mediator like CIKS could allow for coordinated responses downstream of certain receptors and may further provide cells the opportunity to regulate, at a single point, the relative amount of signal the two pathways receive. Once SAPK/JNK- and IKK-derived signals have contributed to activation of both AP-1 and NF-κB transcription factors, these factors can converge during transcriptional activation of at least some target genes. It has been shown that these transcription factors also can physically associate with each other, leading to synergistic activation (51). Therefore, the AP-1 and NF-κB stress pathways can have multiple points of convergence, and CIKS may represent such a point.
Acknowledgments
We are grateful to Anthony Fauci for his support and encouragement, Keith Brown and Jürgen Müller for helpful discussions and plasmids, and Keith Brown for reading of the manuscript. We thank Drs. X. Li and G.R. Stark for exchanging information on their independent cloning and characterization of a protein identical to CIKS. A.C. also serves as a Research Assistant of the University of Liege, Belgium and is supported in part by postdoctoral grants from the NATO and the Fulbright Commission.
Abbreviations
- IKK
IκB kinase
- SAPK
stress-activated protein kinase
- JNK
Jun kinase
- TNF
tumor necrosis factor
- TRAF
TNF receptor-associated factor
- CIKS
connection to IKK and SAPK/JNK
- NEMO
NF-κB essential modulator
- NIK
NF-κB inducing kinase
- HA
hemagglutinin
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF272151).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190245697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190245697
References
- 1.Siebenlist U, Franzoso G, Brown K. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S J. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 4.Verma I M, Stevenson J K, Schwarz E M, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 5.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 6.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;5:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 8.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteside S T, Ernst M K, LeBail O, Laurent-Winter C, Rice N, Israel A. Mol Cell Biol. 1995;15:5339–5345. doi: 10.1128/mcb.15.10.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldi L, Brown K, Franzoso G, Siebenlist U. J Biol Chem. 1996;271:376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 11.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, et al. Proc Natl Acad Sci USA. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Nature (London) 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 14.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer E, Jiang J, Chen Z J. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 17.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 18.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 19.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 20.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 21.Delhase M, Hayakawa Y, Chen Y, Karin M. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Lu Q, Hwang J Y, Buscher D, Lee K F, Izpisua-Belmonte J C, Verma I M. Genes Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Van Antwerp D, Mercurio F, Lee K F, Verma I M. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 25.Li Z W, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, Fuentes M E, Yamaguchi K, Durnin M H, Dalrymple S A, Hardy K L, Goeddel D V. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 28.O'Mahony A, Lin X, Geleziunas R, Greene W C. Mol Cell Biol. 2000;20:1170–1178. doi: 10.1128/mcb.20.4.1170-1178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto Y, Yin M J, Gaynor R B. Mol Cell Biol. 2000;20:3655–3666. doi: 10.1128/mcb.20.10.3655-3666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothwarf D M, Zandi E, Natoli G, Karin M. Nature (London) 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 31.Yamaoka S, Courtois G, Bessia C, Whiteside S, T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, Wallach D, Horwitz M S. Proc Natl Acad Sci USA. 1999;96:1042–1047. doi: 10.1073/pnas.96.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harhaj E W, Good L F, Xiao G, Uhlik M, Cvijic M E, Rivera-Walsh I, Sun S-C. Oncogene. 2000;19:1448–1456. doi: 10.1038/sj.onc.1203445. [DOI] [PubMed] [Google Scholar]
- 34.Rudolph D, Yeh W C, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia A J, Mak T W. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- 35.Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U. J Biol Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- 36.Xu S, Robbins D J, Christerson B L, English J M, Vanderbilt C A, Cobb H M. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chariot A, Princen F, Gielen J, Merville M P, Franzoso G, Brown K, Siebenlist U, Bours V. J Biol Chem. 1999;274:5318–5325. doi: 10.1074/jbc.274.9.5318. [DOI] [PubMed] [Google Scholar]
- 38.Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg P H, Journot L. Nature (London) 1993;365:170–175. doi: 10.1038/365170a0. [DOI] [PubMed] [Google Scholar]
- 39.Harhaj E W, Sun S-C. J Biol Chem. 1999;274:22911–22914. doi: 10.1074/jbc.274.33.22911. [DOI] [PubMed] [Google Scholar]
- 40.Ishida T K, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, Yamamoto T, Inoue J. Proc Natl Acad Sci USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano H, Oshima H, Chung W, Williams-Abbott L, Ware C F, Yagita H, Okumura K. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 42.Reinhard C, Shamoon B, Shyamala V, Williams L T. EMBO J. 1997;16:1080–1092. doi: 10.1093/emboj/16.5.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baud V, Liu Z G, Bennett B, Suzuki N, Xia Y, Karin M. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wajant H, Grell M, Scheurich P. Cytokine Growth Factor Rev. 1999;10:15–26. doi: 10.1016/s1359-6101(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 46.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. Immunity. 2000;12:419–429. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S Q, Kovalenko A, Cantarella G, Wallach D. Immunity. 2000;12:301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, Makris C, Su B, Li E, Yang J, Nemerow G R, Karin M. Proc Natl Acad Sci USA. 2000;97:5243–5248. doi: 10.1073/pnas.97.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wesche H, Henzel W J, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 50.Lomaga M A, Yeh W C, Sarosi I, Duncan G S, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]