Abstract
Motivation: The relationship between nucleosome positioning and gene regulation is fundamental yet complex. Previous studies on genomic nucleosome positions have revealed a correlation between nucleosome occupancy on promoters and gene expression levels. Many of these studies focused on individual nucleosomes, especially those proximal to transcription start sites. To study the collective effect of multiple nucleosomes on the gene expression, we developed a mathematical approach based on autocorrelation to relate genomic nucleosome organization to gene regulation.
Results: We found that nucleosome organization in gene promoters can be well described by autocorrelation transformation. Some promoters show obvious periods in their nucleosome organization, while others have no clear periodicity. The genes with periodic nucleosome organization in promoters tend to be lower expressed than the genes without periodic nucleosome organization. These suggest that regular organization of nucleosomes plays a critical role in gene regulation. To quantitatively associate nucleosome organization and gene expression, we predicted gene expression solely based on nucleosome status and found that nucleosome status accounts for ∼25% of the observed gene expression variability. Furthermore, we explored the underlying forces that maintain the periodicity in nucleosome organization, namely intrinsic (i.e. DNA sequence) and extrinsic forces (i.e. chromatin remodeling factors). We found that the extrinsic factors play a critical role in maintaining the periodic nucleosome organization.
Contact: jiang.qian@jhmi.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Eukaryotic DNA is condensed into a compact structure through the aid of histone and associated proteins. The specialized complex of DNA and proteins is known as chromatin and its fundamental packing unit is the nucleosome (Kornberg, 1974; Kornberg and Lorch, 1999). Nucleosomes are composed of a stretch of DNA of ∼147 bp that is sharply bent and wrapped almost twice around a histone core complex (Richmond and Davey, 2003). The location and binding of nucleosomes have important consequences on gene regulation, replication and recombination (Groth et al., 2007; Lee et al., 2007).
The development of high-throughput technologies such as DNA microarrays (Lee et al., 2007; Yuan et al., 2005) and next generation sequencing (Albert et al., 2007; Barski et al., 2007) has transformed the determination of nucleosome location on DNA to the genomic scale, providing an opportunity to explore global relationships between nucleosome organization and biological function. Early studies demonstrated that nucleosomes are depleted in active regulatory regions (Bernstein et al., 2004; Lee et al., 2004; Yuan et al., 2005), while more recent higher resolution studies of nucleosome positioning have revealed that nucleosome-free regions are located upstream to transcription start sites (TSSs), often flanked by two well-positioned nucleosomes centered at about -200 bp and 100 bp (Ioshikhes et al., 2006; Lee et al., 2007; Segal et al., 2006).
While these studies added significantly to our understanding of the relationship between nucleosome positioning and gene regulation, the analyses often emphasized individual nucleosome positioning, especially nucleosomes close to gene TSSs. Although some studies considered the effect of multiple nucleosomes, they often simply focused on nucleosome density in a region and did not consider their structural arrangement. Here, we argue that multiple nucleosomes work in concert to participate in regulating gene expression. To study the overall organization of nucleosome positions in a genomic region, we used autocorrelation transformation of original nucleosome occupancy signal to describe the collective behavior of multiple nucleosomes. Autocorrelation is a function of correlation between a profile and shifted versions of itself (See Section 2). The method can reveal signal periodicity that may not be evident by examining the intensities alone and manifest the general periods of multiple nucleosomes within whole spatial ranges of interest. By autocorrelation transformation of the occupancy of multiple nucleosomes, we were able to describe nucleosome organization better than by just using intensity profiles or the density of the nucleosomes alone. Periodic nucleosome positioning has recently been discovered in genome, especially in transcribed regions (Mavrich et al., 2008; Shivaswamy et al., 2008; Yuan et al., 2005). However, there is no systematic study on nucleosome periodicity in promoters, where the periodicity is much subtler and difficult to be detected. More importantly, we related the nucleosome organization in promoters to gene regulation. This analysis provides an added level of information to help understand the role of nucleosome organization in biological function.
Furthermore, we attempted to understand the underlying forces that determine and/or maintain the periodicity of nucleosome organization. Previous studies have attempted to predict nucleosome occupancy based solely on DNA sequence features (Kaplan et al., 2009; Peckham et al., 2007; Segal et al., 2006; Yuan and Liu, 2008). The relatively high success rate of these sequence-based algorithms in predicting in vivo nucleosome positions indicates that DNA sequence plays an important role in maintaining the nucleosome organization. On the other hand, experimental evidence indicates that chromatin-remodeling complexes can alter the nucleosome positions in vivo and in vitro (Whitehouse et al., 2007; Yang et al., 2006). These trans-factors can act as antagonistic forces to reposition nucleosomes in vivo. In other words, nucleosome organization in vivo can be dynamically modified in response to environmental conditions with the aid of these remodeling complexes. In essence, it is the interplay of the intrinsic DNA sequence and extrinsic factors that maintain, modify and position nucleosomes on DNA. In this article, we also examined the relative contribution from these intrinsic and extrinsic forces that determine the structures of nucleosome organization.
2 MATERIALS AND METHODS
2.1 Datasets
In this article, we studied nucleosome positioning and its relation to gene expression, occurrence of transcriptional factors (TFs), histone occupancy and modification. The experimental data were collected from following sources:
Nucleosome occupancy intensity, DNA sequence and gene expression levels were downloaded from the website of T. Hughes's lab (Lee et al., 2007).
The gene expression under different physiological conditions were obtained from the paper by Beer and Tavazoie (2004). We performed quantile normalization before further analysis.
The functional TFs binding sites were achieved from file http://fraenkel.mit.edu/improved_map/p001_c1.gff (MacIsaac et al., 2006).
The data of histone occupancy (Bernstein et al., 2004; Lee et al., 2004; Zhang et al., 2005) and histone modification (Bernstein et al., 2002; Kurdistani et al., 2004; Millar et al., 2006; Rao et al., 2005; Xu et al., 2005) were downloaded from the database ChromatinDB (www.bioinformatics2.wsu.edu/ChromatinDB/).
In vitro dataset was obtained from GEO web site with accession number GSE13622 (Kaplan et al., 2009).
2.2 Autocorrelation of nucleosome occupancy
Autocorrelation is widely used in signal processing to find hidden periodic patterns in either time domain or space domain. Here, autocorrelation coefficient is defined as
| (1) |
where L is shifted distance, x is chromosomal coordinate and I(x) is the nucleosome occupancy intensity obtained from microarray at position x. The region of interest is [x1, x2]. We normalized R(L) by R(0). Therefore, R(L) measures the cross-correlation of the nucleosome intensities on two segments with pair starting points apart from a distance of L. If the profile is periodic and the shift distance is close to the periodicity, we will observe a relatively large value for R(L). And vice verse, the high-value peak R(L) at a specific L indicates that the signal exhibits a period of L in that all nucleosomes of interest almost occupy their neighboring nucleosomes' positions after shifting a distance L. For each gene, we applied the autocorrelation transformation to three regions: (i) minus signal −1000 bp relative to the TSSs to 1000 bp of transcribed regions; (ii) only the negative signal −1000 bp relative to the TSSs, which we called promoter regions; (iii) only the downstream 1000 bp relative to the TSSs, which we called transcribed regions.
2.3 Gene Ontology analysis
Gene Ontology (GO) analysis was performed on four gene groups, respectively. We used hypergeometric distribution to calculate P-value of each GO term for the gene groups. The final P-values were modified by false discovery rate correction for multiple testing (Benjamini and Hochberg, 1995; Shaffer, 1995).
3 RESULTS
3.1 Autocorrelation of nucleosome occupancy
High-resolution nucleosome occupancy was inferred by the intensity of hybridization of nucleosome-enriched DNA on a microarray (Lee et al., 2007; Yuan et al., 2005). We employed autocorrelation to analyze this nucleosome occupancy data. To demonstrate the difference between nucleosome intensity and its autocorrelation pattern, we show several examples in Figure 1. While all six genes share similar regular nucleosome organization in their transcribed regions, they exhibit distinct patterns of nucleosome organization in their promoter regions. From Figure 1a, it can be seen that the top three genes have similar period in autocorrelation profiles. Despite not having obvious relationships in their intensity profiles. In contrast, the autocorrelations of the other three genes in Figure 1b do not demonstrate periodicity in nucleosome organization in their promoters.
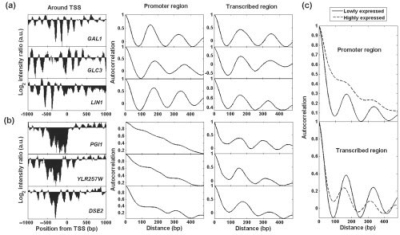
Fig. 1.
Nucleosome occupancy intensities, autocorrelation patterns and gene expression levels. (a) and (b) are six gene examples. From left to right are intensity ratio signal, autocorrelation in promoter and transcribed region. In (a) are three lowly expressed genes: GAL1 (0.04), GLC3 (0.32) and LIN1 (0.08). The number in the bracelet following gene name represents gene expression level. Three highly expressed genes are shown in (b): PGI1 (4.03), YLR257W (3.75) and DSE2 (4.13). (c) Average autocorrelation profiles for top 250 highly expressed (dashed lines), bottom 250 lowly expressed genes (solid lines) in promoter and transcribed region, respectively.
A potential biological significance of nucleosomal periodicity is that it tends to be correlated with gene expression activity. The three genes shown in Figure 1 that have periodicity in both their promoter and transcribed regions (GAL1, GLC3 and LIN1) have low expression (<0.4—normalized intensity from microarray analysis), whereas the three genes with periodicity only in the transcribed region (name genes) show high expression (>3.70, indicating an almost 10-fold expression difference). This finding suggests that periodicity in the promoter region, upstream to the TSS, may be important in gene regulation. To test the significance of this observation, we expanded the analysis. The average autocorrelation was determined for the top 250 highly expressed genes and the bottom 250 low-expressed genes. The first group showed no periodic behavior in the promoter region, while the second group demonstrated clear periodicity of about 164–168 bp (Fig. 1c). The periodicity for low-expressed genes in their transcribed regions is also more apparent than that for high-expressed genes.
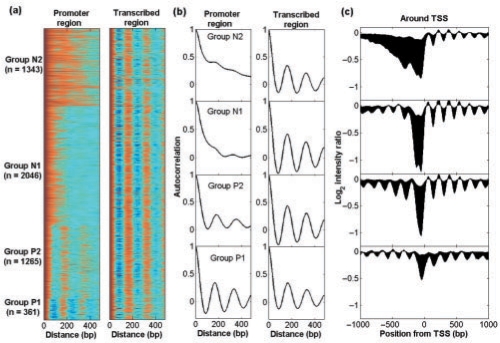
We next performed the autocorrelation analysis on the nucleosome intensity profiles for all 5015 yeast genes (Lee et al., 2007). The autocorrelation patterns of transcribed region look similar, showing strong periodicity in nucleosome organization. In contrast, their promoters exhibit high diversity. When all the gene promoters are compared with each other based on upstream nucleosome autocorrelation profiles using the k-mean clustering algorithm, four groups of genes were obtained, each with distinct nucleosome organization patterns in their promoters (Fig. 2). (We tried k = 5, 6 and 7 for which results were quite similar, but with small difference in details.) Both Groups P1 and P2 show periodicity, whereas Groups N1 and N2 have little or no periodicity. For the periodic genes, those in Group P1 appear more tightly compacted, with a nucleosome period of 164–168 bp (the smallest distance observed for all nucleosomes in all gene regions); on the other hand, genes in P2 exhibit a looser organization with a period of 176–180 bp. While genes in the N1 and N2 groups are similar in that they show minimal periodicity; they differ in that N1 genes demonstrate a sharp drop in autocorrelation and N2 genes reveal a more gradual decay in autocorrelation profiles (Fig. 2). This suggests that N1 genes have a higher frequency fluctuation in their occupancy profiles.
Fig. 2.
Genes clustered by autocorrelation profiles in promoter region. The ordering of genes is based on k-means (k = 4) clustering of promoter autocorrelation. (a) Gene nucleosome autocorrelation profile in promoter and transcribed region, respectively. Color magenta represents high autocorrelation coefficient whereas blue is low one. (b) Average autocorrelation for each gene group in promoter and transcribed region, respectively. (c) Average original intensity profiles within ±1 kb around TSSs for each gene group.
To gain biological insight of the gene groups by nucleosome organization, we studied biological functions significantly enriched in each group. The four groups of genes generally have distinct functions. The functional difference is especially clear between the gene groups with and without nucleosome periodicity (Supplementary Table S1). For example, Group P1 is enriched for ‘signalosome’, and Group P2 is enriched for ‘DNA repair’ and ‘DNA metabolic process’. In contrast, Groups N1 and N2 are enriched for ‘RNA helicase activity’ and ‘nuclear nucleosome’, respectively.
It is also interesting to note that the genes from same group tend to be close to each other on chromosomes, suggesting that the genes are partially organized on chromosomes with similar nucleosome organization (Supplementary Fig. S2).
3.2 Comparison of clustering methods
Previous studies have also clustered genes based on their nucleosome occupancy profiles (Lee et al., 2007). We compared our gene clustering in autocorrelation space with that in nucleosome intensity space. The clustering results are obviously different to each other (Fig. 2c). When we compared the average nucleosome occupancy intensity profiles among the four groups, the average intensity profiles of the four groups show less significant difference than the average autocorrelation profiles do. For example, although the groups of P2 and N1 had clear difference in periodicity behavior in autocorrelation space, their average original intensities did not exhibit much differently.
We evaluated the gene clustering approaches according to the level of co-expression within each groups. We calculated the correlation coefficient of gene expression profiles across multiple experimental conditions (Beer and Tavazoie, 2004) and defined co-expressed genes as gene pairs whose correlation coefficient is greater than 0.9. We found 1122 co-expressed gene pairs for the entire yeast genome. Compared with cluster analysis based on nucleosome intensity profiles, the autocorrelation result correlates better with gene expression than do original intensity profiles (Ioshikhes et al., 2006; Lee et al., 2007), indicating that autocorrelation transformation is an efficient and sensitive method to detect the nucleosome organization that is associated with gene regulation (see Supplementary Fig. S3 for a detailed analysis).
3.3 Relating nucleosome organization and gene regulation
To further understand the influence of periodicity on gene regulation, we examined the general characteristics for the four gene groups defined by autocorrelation profiles. We first calculated the gene expression level for the four groups and found that the genes from the different groups tend to express at different levels (Fig. 3a). As we would predict, the gene expression level is the lowest for genes with tightly periodic nucleosome organization, namely groups P1 and P2. In contrast, genes with no periodic nucleosome organization, namely groups N1 and N2, had higher expression. Group N2 for which nucleosomes are depleted close to TSSs in the promoters has the highest expression level. Besides the gene expression, we also checked the variation of gene expression across 250 cellular states for each gene (Beer and Tavazoie, 2004). We found that the genes from Group N2 had the greatest variation in expression (Fig. 3b), perhaps due to the fact that Group N2 genes have sufficient space for nucleosome repositioning, which could in turn lead to variation of gene expression level.
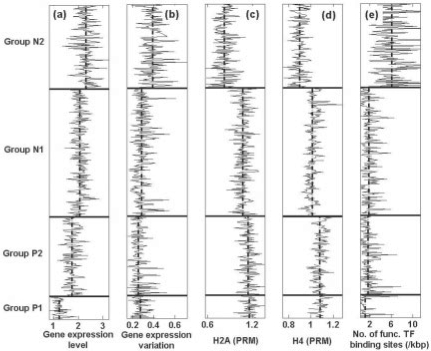
Fig. 3.
Relation between nucleosome organization (gene group categorized) and (a) gene expression, (b) gene expression variation across different states, (c) histone H2A density in promoter region, (d) histone H4 density in promoter region and (e) number of functional TF binding sites. The vertical dash-dotted lines are average values for each gene group to exhibit the difference.
We next examined whether the gene expression differences among the four groups could be simply due to histone density difference in the promoters, since it is known that depletion of nucleosome in a gene's promoter region leads to increased expression (Bernstein et al., 2004; Lee et al., 2004). Consistent with this hypothesis, we found that Group N2 both has the lowest histone density and the highest overall gene expression level. However, Groups P1 and P2 have almost the same histone density values while their expression levels are significantly different (Fig. 3a, c and d). These results indicate that nucleosome organization is a better predictor of gene expression than simple histone density, and suggest that the pattern of nucleosome positioning may provide an additional level of regulation on top of that provided by nucleosome density.
A possible link between patterns of nucleosome positioning and gene expression is the occurrence of functional TF binding sites within gene promoters. To test this possible association, the number of functional TF binding sites in the promoter regions obtained from ChIP-chip experiments (MacIsaac et al., 2006) was analyzed (Fig. 3e). The genes from Group P1 have the lowest number of functional sites and Group N2 has the highest number of such sites, suggesting that high regularity of nucleosome organization inhibits TF binding. Interestingly, we also observed that functional TF binding sites in N2 groups have the greatest variability (Fig. 3e).
3.4 Component of gene expression variation attributable to chromatin structure
We have demonstrated that the periodicity in nucleosome organization can be related to the gene expression level. Here, we attempted to quantitatively relate the chromatin structure to gene expression. Gene expression, at least in theory, can be determined from a combination of two general categories of data. One is genetic sequence data, notably the configuration of TF binding sites in the promoter; another is epigenetic non-sequence-based information, such as chromatin structure. Efforts have been made to predict gene expression directly from promoter sequences using TF binding site configuration as the independent variable (Beer and Tavazoie, 2004; Bussemaker et al., 2001; Conlon et al., 2003; Smith et al., 2006). However, there have been few studies that examine gene expression control as a function of chromatin structure. Here, we explored how much regulatory information is contained in chromatin structure. In other words, to what extent can we predict gene expression is solely based on chromatin structural information without any input of TF binding site data.
We used histone configuration data from 42 experiments to predict gene expression levels (as discussed in Section 2). The variables analyzed include nucleosome organization groups (i.e. P1, P2, N1 and N2), histone occupancies and modifications in promoter and transcribed regions (O'Connor and Wyrick, 2007). In principle, we performed categorical regression based on the four gene groups defined by clustering in autocorrelation space. The optimal histone occupancies and modifications were selected through sequential forward floating selection (Pudil et al., 1994) in regression. The final model obtained an R2-value of ∼0.25, indicating that chromatin structure alone can account for 25% of the observed variation in gene expression. This is a similar level to the prediction based on TF binding sites (Bussemaker et al., 2001; Conlon et al., 2003) and histone acetylation with or without considering motif scores and nucleosome density (Yuan et al., 2006).
3.5 Intrinsic and extrinsic effect on nucleosome organization
Having demonstrated the importance of the regularity of nucleosome organization on gene regulation, we turned to investigating the underlying forces that cause the regularity of nucleosome organization. As previously mentioned, there are two general contributions to nucleosome organization: the intrinsic properties of DNA sequences (cis-regulatory elements) and the extrinsic chromatin remodeling factors (trans-regulatory factors). While it is likely that both determine nucleosome organization, it is important to quantify the contribution of each of them. One approach is based on direct comparison of in vivo and in vitro nucleosome occupancy. If the in vitro and in vivo nucleosome occupancy correlates well in a particular genomic region, the nucleosome organization in that region is mainly determined by the DNA sequence. On the other hand, more limited correlation between in vitro and in vivo nucleosome occupancy patterns could reflect the importance of chromatin effects. The difference between the in vitro and in vivo can thus be viewed as an estimate of the effect of extrinsic forces such as chromatin-remodeling factors.
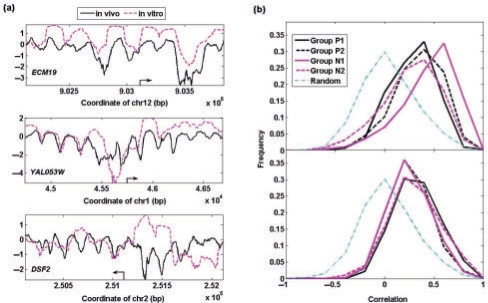
By comparing the in vitro and in vivo nucleosome occupancy intensities, only a small fraction of genes keep the same nucleosome organization in promoters in vivo as in vitro (e.g. gene ECM19 in Fig. 4a). While some genes partially keep the same nucleosome organization (e.g. gene YAL053W in Fig. 4a), others are forced to occupy ‘unfavorable’ DNA segments from their preferred positions in vitro (e.g. gene DSF2 in Fig. 4a). To quantify the difference between the in vitro and in vivo nucleosome occupancy, we calculated the correlation of nucleosome intensity in vitro and in vivo. As noted above, higher correlation represents higher contribution from DNA sequence; and lower correlation indicates higher contribution from extrinsic factors. First, we observed that all four groups show better agreement in the in vitro and in vivo nucleosome occupancy than seen in a random simulation (Fig. 4b), in which the in vitro and in vivo occupancy profiles were permutated among the genes. Second, different gene groups demonstrate relatively different levels of agreement between the in vitro and in vivo nucleosome occupancy. The genes from Group P1 with strongest periodicity in nucleosome organization have the least correlation between the in vivo and in vitro nucleosome organization, suggesting that extrinsic factors contribute significantly to maintaining the periodicity in the nucleosome occupancy. As a comparison, the transcribed regions from all four groups are not significantly different from each other and lie between the two extremes.
Fig. 4.
(a) Comparison of the in vivo nucleosome intensity with the in vitro nucleosome occupancy data for three genes as examples; (b) distribution of correlation between nucleosome intensity in vitro and in vivo for the same gene in terms of different gene groups. As comparison, the dash-dotted lines in the plots are correlation between the in vivo nucleosome occupancy of one gene and in vitro data of another randomly chosen gene.
Another independent approach to evaluate the intrinsic and extrinsic effect on nucleosome organization is to examine how nucleosome organization changes after removing a chromatin remodeling factor. We compared the nucleosome organization in autocorrelation space between wild-type and mutant yeast with a defect in the chromatin-remodeling factor Isw2 (Whitehouse et al., 2007). The majority of promoters did not show dramatic changes in nucleosome organization in the mutant. We chose two groups of genes with the most significant changes (correlation R < 0.8) of nucleosome organization either in promoters or transcribed regions (Supplementary Fig. S4). These are the genes with strong periodicity in nucleosome organization in wild type as seen in their autocorrelation profiles. However, the periodicity becomes weaker in mutant with defect Isw2, suggesting that Isw2 can enhance the periodic structure in nucleosome organization. This is consistent with previous findings (Fyodorov et al., 2004; Ito et al., 1997; Tsukiyama et al., 1999; Yang et al., 2006) that some extrinsic factors affect the periodicity in nucleosome organization.
3.6 Nucleosome organization in humans
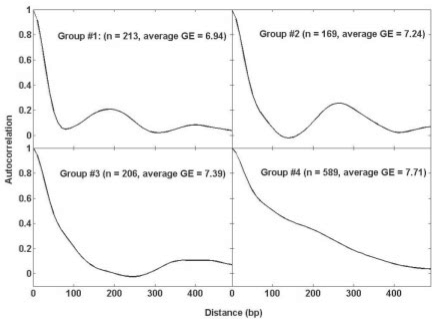
Having examined the nucleosome organization in yeast, we then performed the analysis of nucleosome organization in humans. We obtained the nucleosome occupancy in human cell line A375 (Ozsolak et al., 2007). We clustered the genes based on their autocorrelation profiles and identified four groups of genes (Fig. 5). Two groups shows clear periodicity in nucleosome organization (with period of ∼190 bp and 260 bp, respectively), while the other two do not have clear periodicity. Interestingly, the nucleosome organization also correlates with gene expression. The genes with periodicity tend to have lower gene expression, indicating that the observation we made in yeast could be generalized to the higher eukaryotic genomes.
Fig. 5.
Nucleosome organization in humans. Average autocorrelation profiles are shown for four gene groups with corresponding number of genes and average gene expression level.
4 DISCUSSION
In order to take into consideration the collective effect of nucleosomes on gene expression, we developed a computational method to analyze nucleosome organization in autocorrelation space. When clustered according to their nucleosome organization in promoter regions, four distinct groups of genes emerged. Gene groups with different nucleosome organization have distinct properties such as gene expression and TF occupancy. Periodicity in nucleosome organization at the upstream regions indicates heterochromatin status and low gene expression. Genes without periodic nucleosome organization had higher levels of gene expression and demonstrated increased expression variation. We also explored the possible underlying forces that maintain the periodicity of the nucleosome organization and found that extrinsic nucleosome remodeling complexes play a critical role in maintaining the regularity of nucleosome organization.
4.1 Advantage of autocorrelation
Autocorrelation analysis can exhibit the hidden periodicity of signals in the range of interest. We have demonstrated that it can efficiently capture the subtle signatures in nucleosome occupancy profiles and found that the organizational structure is strongly correlated with gene expression. Another advantage of autocorrelation analysis is that we can avoid the prediction of nucleosome positions using computational methods such as hidden Markov model (HMM), where inaccuracy in the nucleosome position prediction can introduce additional ambiguity in the further analysis. Furthermore, traditional gene clustering methods often align the nucleosome profiles respect to one location such as TSSs. Some genes may have similar nucleosome organization, but with certain position shift relative to each other. If we align the nucleosomes for these genes and obtain an average nucleosome profile for the genes, nucleosome signals will be cancelled out due to the position shifting among these genes. In contrast, the autocorrelation transformation is position independent. Similar nucleosome profiles must yield similar autocorrelation profiles regardless their relative positions. This is one of the reasons why our clustering of genes is better correlated with co-expression.
4.2 High-order structure of nucleosomes
We found that the genes with regular nucleosome organization tend to be low expressed. We speculate that the regularity of nucleosomes might be related to their specific high-order structure. The regular nucleosomes could facilitate the formation of high-order structures. Researchers have found an optimal linker DNA length for specific high-order nucleosome organization, e.g. the 30 nm fiber (McGhee et al., 1983; Widom and Klug, 1985). Therefore, the genes in Groups P1 and P2 might form high-order structures and prevent the access of other TFs. However, the structure of 30 nm fiber in Saccharomyces cerevisiae is still in debate. We need further evidence to confirm the link between the nucleosome periodicity and high-order structure.
4.3 Predicting gene expression from chromatin structure
From this study, we found that the regulatory contribution solely from chromatin structure, without any input from TFs, accounted for 25% of variation in gene expression. This finding suggests that epigenetic structure contains no less gene regulatory information than genetic coding does. However, we would like to point out that the information contained in chromatin structures is not totally independent to that in genetic coding. Since the nucleosome positioning is partially determined by DNA sequences, the DNA sequences encode simultaneously both the information for nucleosome positioning and TF regulation. This perhaps can explain the success of sequence-only predictions and the added power of nucleosome organization (Yuan et al., 2006).
4.4 Dynamic nucleosome organization
Nucleosome positioning is dynamically regulated in response to various cellular environments and stimuli. Under different physiological conditions, nucleosome positions have to be re-arranged. The intrinsic DNA sequence preference for nucleosome formation and position is a static property, one that cannot guide the modulation of nucleosome positions. On the other hand, extrinsic chromatin modifiers could influence nucleosome rearrangement when needed. In this article, we also explored the possible underlying forces that maintain the periodicity of nucleosome organization and found that extrinsic nucleosome remodeling complexes play a critical role in maintaining the regularity of nucleosome organization. Recently, it has been found that intrinsic DNA sequence preferences of nucleosomes play a central role in nucleosome organization (Kaplan et al., 2009). The work by Field et al. (2009) also suggests that the gene expression divergence between species can be attributed to the difference of nucleosome organization that is largely encoded by DNA sequences. These apparent contradictions might be explained by the fact that previous studies analyzed the overall trend of nucleosome organization, while we compared the relative difference of underlying forces that shape the nucleosome organization among four groups. We observed that correlation level between in vitro and in vivo nucleosome organization is different among the gene groups. The observation is to some extent supported by the statement by Kaplan et al. (2009) that ‘the correlation between the maps is not uniform across the genome’. We found that the groups with periodic nucleosome organization tend to be more controlled by extrinsic factors than other groups. It suggests that the periodicity of nucleosome organization might be a dynamic property that is maintained and created by chromatin remodeling complexes.
ACKNOWLEDGEMENTS
We thank Dr Xin Chen, Dr Xueping Yu and Dr Jef Boeke for helpful discussions.
Funding: National Institutes of Health (Grant EY017589); Funds supporting the Guerrieri Center for Genetic Engineering and Molecular Ophthalmology; generous gift from Mr and Mrs Robert and Clarice Smith.
Conflict of Interest: none declared.
REFERENCES
- Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, et al. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, et al. Regulatory element detection using correlation with expression. Nat. Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- Conlon EM, et al. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, et al. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat. Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, et al. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, et al. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, et al. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Ito T, et al. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, et al. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Lee CK, et al. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, et al. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Millar CB, et al. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TR, Wyrick JJ. ChromatinDB: a database of genome-wide histone modification patterns for Saccharomyces cerevisiae. Bioinformatics. 2007;23:1828–1830. doi: 10.1093/bioinformatics/btm236. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, et al. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Peckham HE, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudil P, et al. Floating search methods in feature selection. Pattern Recognit. Lett. 1994;15:1119–1125. [Google Scholar]
- Rao B, et al. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JP. Multiple hypothesis testing. Ann. Rev. Psychol. 1995;46:561–584. [Google Scholar]
- Shivaswamy S, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, et al. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc. Natl Acad. Sci. USA. 2006;103:6275–6280. doi: 10.1073/pnas.0508169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, et al. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, et al. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- Xu F, et al. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Yang JG, et al. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Yuan GC, et al. Statistical assessment of the global regulatory role of histone acetylation in Saccharomyces cerevisiae. Genome Biol. 2006;7:R70. doi: 10.1186/gb-2006-7-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]