Abstract
Objective
Free-text clinical reports serve as an important part of patient care management and clinical documentation of patient disease and treatment status. Free-text notes are commonplace in medical practice, but remain an under-used source of information for clinical and epidemiological research, as well as personalized medicine. The authors explore the challenges associated with automatically extracting information from clinical reports using their submission to the Integrating Informatics with Biology and the Bedside (i2b2) 2008 Natural Language Processing Obesity Challenge Task.
Design
A text mining system for classifying patient comorbidity status, based on the information contained in clinical reports. The approach of the authors incorporates a variety of automated techniques, including hot-spot filtering, negated concept identification, zero-vector filtering, weighting by inverse class-frequency, and error-correcting of output codes with linear support vector machines.
Measurements
Performance was evaluated in terms of the macroaveraged F1 measure.
Results
The automated system performed well against manual expert rule-based systems, finishing fifth in the Challenge's intuitive task, and 13th in the textual task.
Conclusions
The system demonstrates that effective comorbidity status classification by an automated system is possible.
Introduction and Background
The application of Natural Language Processing (NLP) techniques within clinical medicine is of growing interest to both physicians and machine learning theorists. These methods can potentially be used to reduce the amount of time and money spent carrying out repetitive text-related tasks, and to increase biomedical knowledge by recognizing complex patterns across large datasets that are impossible for humans to recognize.
The overall goal of the Integrating Informatics with Biology and the Bedside (i2b2) 2008 NLP Obesity Challenge Task was to assess classification algorithms for determining patient disease status with respect to Obesity and 15 of its comorbidities, based on the information extracted from medical discharge summaries. Two human clinical experts annotated each discharge summary as either Positive, Negative, Questionable, or Unknown, for each comorbidity (listed in ▶), using either “textual” or “intuitive” decision criteria. In the textual task annotators used only the information explicitly stated in a record, whereas in the intuitive task they used their best clinical judgment, informed by the content of the record. Because of this, the “Unknown” class label was excluded from the intuitive task. In the textual task, disagreements between the annotators were resolved by a third obesity expert, whereas in the intuitive task annotator disagreement led to the record being excluded from that particular disease's dataset.
Table 1.
Table 1 Macro-averaged F1 textual and intuitive scores for our best-performing submission in the 2008 i2b2 Obesity Challenge Task. In addition to presenting scores for each comorbidity within each task, our system scores are included, which was the basis for ranking team submissions by the i2b2 Challenge organizers
| Macro-Averaged F1 |
||
|---|---|---|
| Textual | Intuitive | |
| System | 0.598 | 0.634 |
| Asthma | 0.483 | 0.970 |
| CAD | 0.600 | 0.630 |
| CHF | 0.710 | 0.612 |
| Depression | 0.962 | 0.935 |
| Diabetes | 0.641 | 0.915 |
| Gallstones | 0.632 | 0.961 |
| GERD | 0.486 | 0.579 |
| Gout | 0.959 | 0.981 |
| Hypercholesterolemia | 0.484 | 0.912 |
| Hypertension | 0.827 | 0.899 |
| Hypertriglyceridemia | 0.727 | 0.876 |
| OA | 0.945 | 0.631 |
| Obesity | 0.489 | 0.973 |
| OSA | 0.654 | 0.653 |
| PVD | 0.955 | 0.623 |
| VI | 0.675 | 0.725 |
CAD = coronary artery disease; CHF = congestive heart failure; GERD = gastroesophageal reflux disease; OA = osteo arthritis; OSA = obstructive sleep apnea; PVD = peripheral vascular disease; VI = venous insufficiency.
Methods
General System Description
Our approach to the 2008 i2b2 Obesity Challenge Task incorporates a variety of automated techniques, and consists of five steps: preprocessing, tokenization, vectorization, filtering, and classification. We used 5 × 2-way cross-validation on the training data to select and tune the best-performing procedure for each step, including the best in our official task submission. Our best-performing submission used automated hot-spot passage isolation (AutoHP), whitespace, and punctuation-based tokenization, binary feature vectorization, either with or without negation term detection (NegEx), empty feature vector filtering (ZeroVF), and classification using Error-Correcting Output Codes (ECOC) with inverse class frequency-weighted linear support vector machine (SVM) classifiers.
System Components
Preprocessing: Automated Isolation of Hotspot Passages (AutoHP) and Negation Detection
Our preprocessing procedure identified and extracted passages of text that were likely to contribute the most relevant information to the classification task. We have previously shown that manual identification and isolation of such passages, or hot-spots, in patient discharge summaries can greatly improve the accuracy of medical text classification. 1 We hypothesized that a similar approach would be effective here, and that automating our previously described technique would lead to improved performance.
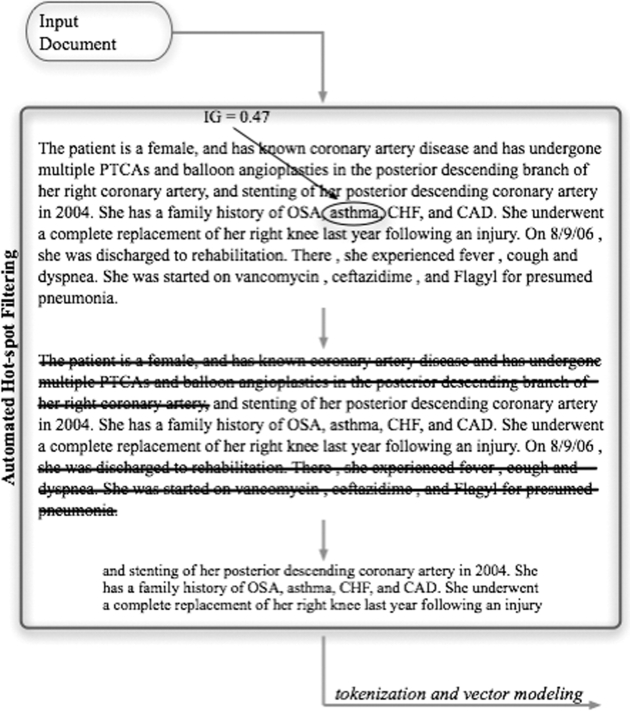
The AutoHP technique takes a set of features from a text collection as input, orders them based on their information gain (IG), and identifies those meeting a specified cut-off value. These features are located in the original document, and all text within 100 characters on either side of the feature is kept as the sample text, tokenized, and modeled as a binary feature vector the text outside the window is discarded (see ▶, for an example).
Figure 1.
Diagrammatic example of our automated hot-spot filtering procedure. In this example, the information gain associated with the word asthma identifies it as a hot-spot feature, so a 100-character window around it is extracted as the hot-spot passage and passed on to the tokenization and vector modeling steps.
This hotspot passage, along with all the others that were identified for the comorbidity, are tokenized and modeled as a binary feature vector; all features not found in these hotspots would be included in this model. For the i2b2 Obesity challenge, the optimal cut-off setting was determined by cross-validation on the training data. We found that the best-performing IG cut-off value was marginally different for each comorbidity (data not shown), but that, overall, the range of optimal values was fairly narrow (μtextual = 0.10, σtextual = 0.06; μintuitive = 0.08, σintuitive = 0.05).
Previous work has shown that clinical narratives contain useful information within their negated and pseudonegated terms. 2 We hypothesized that the accuracy of our classifier would be improved by taking such information into account. In our implementation of Chapman et al.'s NegEx negation identification procedure (AutoHP+ NegEx), any individual negated hot-spot features (identified in the previous step) were identified within the hotspot passages using the NegEx regular expressions; these terms were marked as negated features before the passage was passed to the tokenization and vectorization procedures, and the original nonnegated feature was discarded. An important difference between our use of NegEx and Chapman's original implementation, is that Chapman et al. use an NLP-based named entity recognition engine, (e.g., MetaMap 3 ) to identify negated concepts. Our implementation was simpler, only examining the source text for negations of hotspot terms. Since the hotspot terms were highly associated with the disease status, we hypothesized that these would be the most important concepts with which to recognize instances of negation.
Simple Tokenization and Binary Vectorization
All passages isolated by the AutoHP and AutoHP+ NegEx techniques were tokenized into individual features using the same simple algorithm, in which individual words were tokenized into features based on space and punctuation separation.
The tokenized set of features was next modeled as a binary vector, in which each position corresponded to a feature retained after preprocessing the training set. Text samples were assigned binary values at each position of the vector, indicating the presence or absence of the corresponding feature (or negated feature, where relevant).
Zero-vector Filtering (ZeroVF)
Pre-processing sometimes resulted in a zero-valued feature vector, especially for those comorbidities where the optimal AutoHP IG threshold was moderately high (e.g., Gout and Asthma). We hypothesized that these samples would not contribute any useful information to the classification algorithm, thus, we automatically assigned such samples to the most common class label in the training set. The results of cross-validation experiments showed that the ZeroVF procedure was almost universally helpful across comorbidities (data not shown); it was therefore included in all subsequent cross-validation experiments.
Classification: Error-correcting Output Codes and SVM
In both the textual and intuitive problems, samples were classified using the ECOC technique, an approach that has proven effective for multiple classification problems. 4,5 Briefly, the ECOC approach formulates a multiple classification problem as a set of several binary classification decisions between all possible subsets of the original classes. When classifying new documents, each of the subclassifiers makes a prediction, and the document is classified according to the class with the most similar result vector, based on the sum of the fractional bitwise differences (L1-distance) between them. Where an error made by a lone classifier would result in misclassification, with ECOC, an error made by any one subclassifier is less likely to affect the final classification. For a fuller treatment of how ECOC is used in multiway text classification, see Cohen 1 or Dietterich and Bakiri. 6
While any classification algorithm could be used to carry out the binary subclassification problems with ECOC, we and other authors 1,7,8 have obtained good results with the libSVM implementation of Vapnik's linear SVM technique. 9 One advantage of libSVM, is that it provides a parameter for specifying class-specific weights. We used this parameter to adjust the cost of misclassifying a sample according to the prior probabilities of the class:
where N is the total number of samples, and N class is the number of samples in a particular class.
i2b2 Evaluation
The i2b2 Obesity Challenge Task was evaluated using the Macro F1 measure, with precision and recall weighted equally (β = 1). The Micro F1 measure was also computed and used as a secondary performance measure. The textual and intuitive tasks were evaluated separately by the task organizers, with the overall performance of systems across comorbidities used to assign a score to each submission.
Follow-up Experiments
We conducted several post-hoc experiments to understand how the comorbidity-specific characteristics of the training and test collections affected performance. In particular, we wished to understand the relative contribution of AutoHP and AutoHP+ NegEx to the performance of our system, and the characteristics of the data that contributed.
Characteristics of the Document Collections
We were interested in the role that sample size played in the less-than-ideal performance of our system on certain comorbidities. To address this, we compared the performance of our submitted system on the combined training and test collections, using 2-, 4-, and 8-way cross-validation, stratified by class count. In contrast with standard cross-validation, here the classifier was trained on the smaller partition (one-half, one-quarter, or one-eighth of the combined dataset), and evaluated the performance on the remaining partition. This allowed us to estimate performance over a range of training data sizes, and quantify the contribution of small sample sizes to the performance of the submitted system.
Relative Contribution of the Pre-Processing Procedures
To examine the efficacy of AutoHP and AutoHP+ NegEx, we compared the performance of each to a baseline system not using preprocessing, and using a linear SVM classifier without preprocessing or ECOC. We also wished to understand why, for certain comorbidities, the addition of NegEx to the preprocessing stage failed to improve performance over and above AutoHP alone. Examining the negated terms extracted by the algorithm revealed that, although it often correctly identified negated terms, sometimes these negations were incorrect. For example, both “… she does not have a history of asthma or copd”, and “… no beta blockers, given history of asthma” were identified as containing a negated form of asthma, even though this is true only in the former case. The negative class was often very rare, so the NegEx-induced error rate was high enough to compromise its overall effectiveness as input to the classifier. We hypothesized that NegEx's contribution could be improved by following it with a classifier trained to distinguish NegEx negations associated with the negative class from those associated with the other classes.
As a step in this direction, we developed an SVM classifier to distinguish “true negations” (those associated with the negative class) from “false negations” (those not associated with the negative class). Using the combined training and testing document collections and replacing all the hotspot terms with $TERM$ as a placeholder, we located occurrences of NegEx-matched comorbidity-related phrases in the full texts, assigning “true negation” to the phrases from the negative class, and “false negation” to all others. We tested our classifier individually on each comorbidity after training it on the negation terms found in the combined data collections from the other comorbidities, and compared its performance to that of the NegEx algorithm alone, as implemented in our i2b2 submission.
Results and Discussion
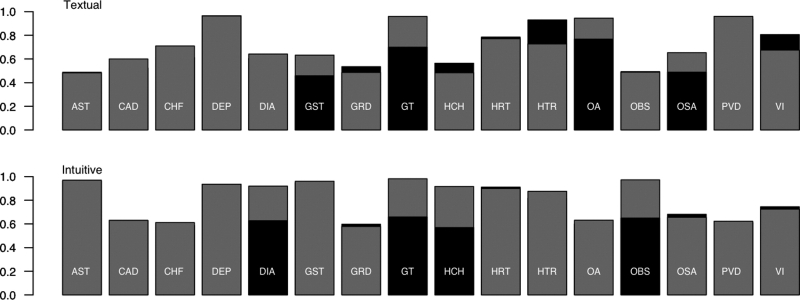
Our cross-validation results are depicted in ▶ (black); the performance of our top submission is shown in gray. We found that the performance of our best method on cross-validation studies was positively correlated with that on the test collection (▶, and the gray bars in ▶; textual: correlation: 0.762, t(14) = 4.407 [p < 0.05]; intuitive: correlation: 0.559, t(14) = 2.523 [p < 0.05]). This system scored 13th out of all runs submitted for the textual task, and fifth out of those submitted for the intuitive task.
Figure 2.
Macro-averaged F1 scores across comorbidities for cross-validation studies on the training document collection (black), and training on the training collection, and testing on the test collection (gray), for both the textual (top) and intuitive (bottom) tasks. Bars for which only one color is visible indicate that the difference between training and testing performance was not significant. Abbreviations: AST—Asthma, CAD—Coronary Artery Disease, CHF—Congestive Heart Failure, DEP—Depression, DIA—Diabetes, GST—Gallstones, GRD—Gastroesophogeal Reflux Disease, GT—Gout, HCH—Hypercholesterolemia, HRT—Hypertension, HTR—Hypertriglyceridemia, OA—Osteoarthritis, OBS—Obesity, OSA—Obstructive Sleep Apnea, PVD—Post-viral Depression, VI—Venous Insufficiency.
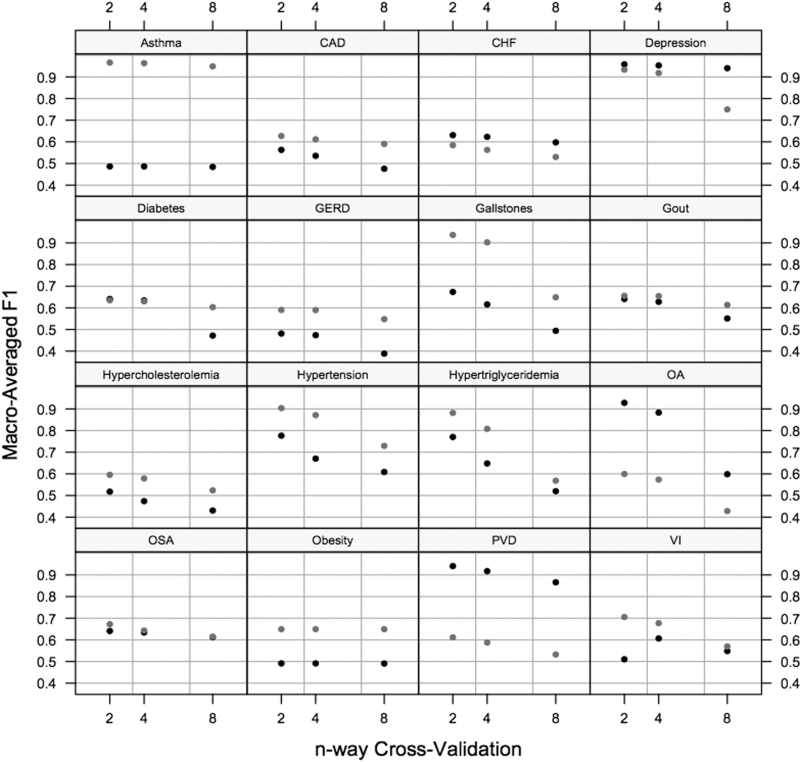
Our textual submission suffered on those comorbidities having one or two rare disease classes. We found that no matter what adjustments were made to our system, instances of these classes tended to be mislabeled as the most prevalent class in the training collection, especially in the textual task. In contrast, our performance on the intuitive task for many of these comorbidities was dramatically better (e.g., Asthma or Hypercholesterolemia). Since these comorbidities did not have as many rare classes in the intuitive task, this supports the hypothesis that the problem in the textual task can be attributed to misclassification of rare classes. Such misclassifications are common to scalable machine learning-based approaches applied to highly skewed data. 10 Where possible, the best solution to this is obtaining more examples of the rare classes—an approach that worked for us in this instance. We combined the training and testing collections into a single dataset, and performed 2-, 4-, and 8-way cross-validation using the smaller partition for training, and the larger for testing using our submitted system. Therefore, the larger the number of cross-ways corresponds to having less training data per iteration. ▶ depicts our results for both the textual (black) and intuitive (gray) classification tasks. For many comorbidities—hypercholesterolemia included—performance improved with the size of the training set. In these situations, one could reasonably expect that additional training data (especially for the rare classes) would improve performance. The data support this conclusion on 12 of the 16 textual and intuitive tasks.
Figure 3.
Macro-averaged F1 scores by comorbidity for 2-, 4-, and 8-way cross-validation using the combined training and testing document collections in both the textual (black) and intuitive (gray) tasks. For most comorbidities, performance decreased with smaller datasets, for a few it remained invariant.
There were, however, also situations where performance did not significantly vary with the size of the data (e.g., Asthma or Obesity for the textual). In these cases, additional data would not likely improve performance. For intuitive Asthma and textual Depression, the performance was already very high. For Asthma, on the textual task, and Obesity, on both tasks, it would be necessary to improve the classification algorithm or the feature set itself.
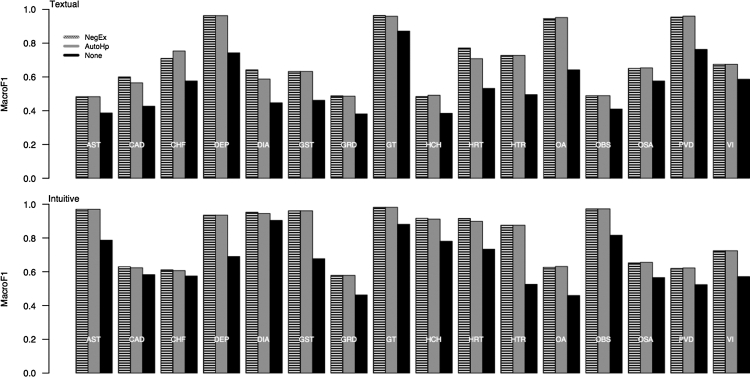
Post-hoc experiments indicated that AutoHP provided the most significant contribution to our system's performance. ▶ compares the AutoHP (light gray) and AutoHP+ NegEx (dark gray) preprocessing procedures against that of a system using no preprocessing procedure (black), for both tasks. For some comorbidities, AutoHP provided as much as a 0.30 performance increase over baseline (e.g., OA in the textual task, or Gallstones in the intuitive task). It is likely that, in these situations, only a small textual region of the discharge summary is important for classification and that including more text will mislead the classifier with irrelevant features.
Figure 4.
Macro-averaged F1 for the AutoHP (light gray), AutoHP+ NegEx (dark gray), and None (black) preprocessing procedures across comorbidities for the textual (top) and intuitive (bottom) classification tasks. The addition of NegEx only provided small improvement in performance over and above that provided by AutoHP for a few topics, which showed consistent improvements over the system having no pre-processing procedure. See ▶ for abbreviation definitions.
Although NegEx never significantly decreased performance, the addition of NegEx to AutoHP only improved the CAD, Diabetes, and Hypertension comorbidities in the textual task. To address why this might have happened, we examined the comorbidity-related terms negated by NegEx, and the classes with which they were most often associated. Quite frequently, negated features were found in multiple classes for a single comorbidity, decreasing their predictive power for binary classification. In its ideal form, a negation-detection procedure should distinguish between negations that are associated with the negative class, and those which are not (false negations).
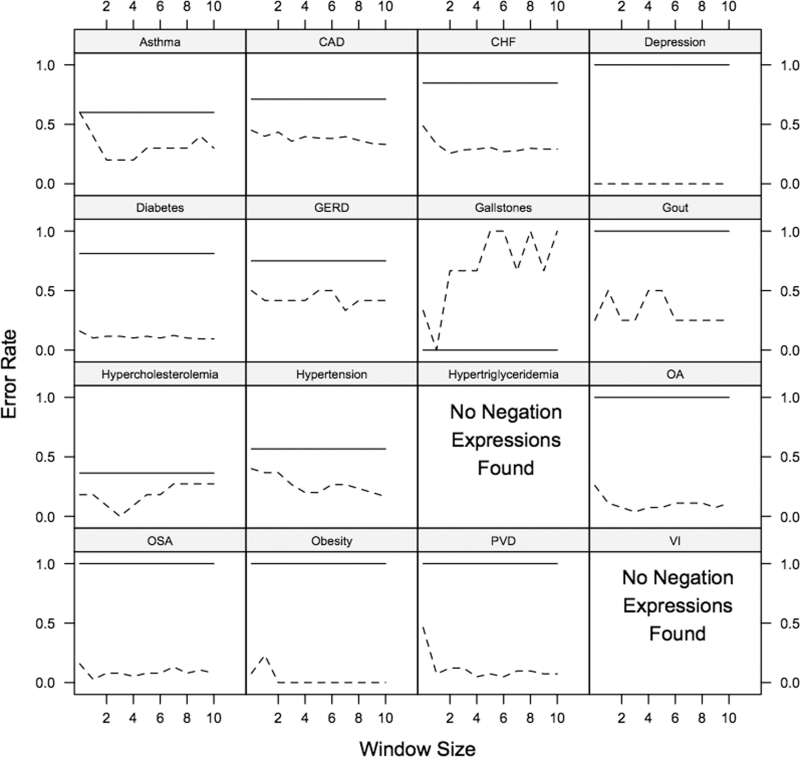
To see whether we could extend the NegEx procedure to avoid false negations, we trained an SVM classifier to use the features surrounding a negated hot-spot feature to distinguish false negations from those associated with the negative class. We compared the performance of this negation system to that of our standard NegEx procedure by examining their respective error rates (▶). The SVM+ NegEx's improved accuracy in all but one comorbidity, achieving up to 100% separation of true and false negations (e.g., Depression, Obesity, OSA). In future work, we will further develop this idea and examine how automated negation detection can be incorporated into a clinical narrative text classification system.
Figure 5.
Error rate for the plain NegEx (solid line) regular expressions and Enhanced using Support Vector Machine (SVM) (dashed line) procedures across comorbidities and varying window sizes during 2-way cross-validation on the combined training and testing documents collections for the textual task. For all but one comorbidity, the Automated Negation Finder tended to extract fewer falsely negated terms (negated terms not actually associated with the negative class). For the Hypertriglyceridemia and Venous Insufficiency comorbidities, no NegEx features were found.
Conclusions
We have demonstrated the effectiveness of several automated techniques for multiple-classification with clinical narrative text. All our techniques are generalizable and fully scalable for classification problems including many more diseases, especially if a large amount of data is available. No aspect of our system requires a-priori knowledge of a disease, expert medical knowledge, or manual examination of the patient records beyond the training labels themselves. Future work will focus on automated negation detection, and explore methods for efficiently using additional training data.
References
- 1.Cohen A. Five-way smoking status classification using text hot-spot identification and error-correcting output codes J Am Med Inform Assoc 2008;15:32-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries J Biomed Inform 2001;34:301-310. [DOI] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: The MetaMap Program Proc AMIA Symp. 2001. pp. 17-21. [PMC free article] [PubMed]
- 4.Dietterich TG. Ensemble methods in machine learning Lecture Notes in Computer Science 2000(1857):1-15.
- 5.Ghani R. Using error-correcting output codes for text classificationIn: Langely P, editor. Proceedings of the 17th International Conference on Maching Learning (ICML)-2000, San Francisco. United States: Morgan Kaufmann Publishers; 2000. pp. 303-310.
- 6.Dietterich TG, Bakiri G. Solving Multiclass Learning Problems via Error-Correcting Output Codes J Artif Intell Res 1995;2.
- 7.Cohen AM. An effective general purpose approach for automated biomedical document classification AnnuSymp Proc. 2006. pp. 161-165. [PMC free article] [PubMed]
- 8.Cohen AM, Yan J, Hersh WR. A comparison of techniques for classification and ad hoc retrieval of biomedical documentsProc of the Fourteenth Annu Text Retrieval Conference. 2005. Gaithersburg, MD.
- 9.Vapnik VN. The Nature of Statistical Learning Theory2nd edn. New York: Springer; 2000.
- 10.Chawla NV, Japkowicz N, Kotcz A. Editorial: Special issue on learning from imbalanced datasets ACM SIGKDD Explor Newsl 2004;6(1):1-6. [Google Scholar]