Abstract
The ATPase retinoid acid-inducible gene (RIG)-I senses viral RNA in the cytoplasm of infected cells and subsequently activates cellular antiviral defense mechanisms. RIG-I recognizes molecular structures that discriminate viral from host RNA. Here, we show that RIG-I ligands require base-paired structures in conjunction with a free 5′-triphosphate to trigger antiviral signaling. Hitherto unavailable chemically synthesized 5′-triphosphate RNA ligands do not trigger RIG-I-dependent IFN production in cells, and they are unable to trigger the ATPase activity of RIG-I without a base-paired stretch. Consistently, immunostimulatory RNA from cells infected with a virus recognized by RIG-I is sensitive to double-strand, but not single-strand, specific RNases. In vitro, base-paired stretches and the 5′-triphosphate bind to distinct sites of RIG-I and synergize to trigger the induction of signaling competent RIG-I multimers. Strengthening our model of a bipartite molecular pattern for RIG-I activation, we show that the activity of supposedly “single-stranded” 5′-triphosphate RNAs generated by in vitro transcription depends on extended and base-paired by-products inadvertently, but commonly, produced by this method. Together, our findings accurately define a minimal molecular pattern sufficient to activate RIG-I that can be found in viral genomes or transcripts.
Keywords: immunostimulatory RNA, melanoma differentiation-associated protein 5, retinoid acid-inducible gene-I-like helicases, virus infection, interferon production
Viral infections are sensed by pattern-recognition receptors (PRRs) of the innate immune system that recognize pathogen-associated molecular patterns (PAMPs), and trigger antiviral gene programs, including the production of IFN type-I (1). Viral RNA serves as a PAMP and can be recognized by toll-like receptor (TLR)-3, TLR-7/8, double-stranded (ds)RNA-activated protein kinase (PKR), and the retinoid acid-inducible gene (RIG)-I-like helicase (RLH) family members RIG-I, melanoma differentiation-associated protein 5 (MDA-5), and laboratory of genetics and physiology (Lgp)2 (2–4). There is evidence that RIG-I signals on infection by many RNA viruses, including important human pathogens (5, 6). After ligand-mediated activation critically involving ATPase activity and the C-terminal regulatory domain (RD) RIG-I binds via its amino-terminal caspase-activation and recruitment domain (CARD) to the adaptor protein Cardif (MAVS, VISA, Ips-1) that then triggers the ΝFκΒ and IRF signaling pathways (7). The exact nature of the PAMP that allows RIG-I to discriminate viral from host RNA in the cytosol is highly controversial. Kim et al. (8) have shown that RNA produced by in vitro transcription (IVT) bearing a 5′-triphosphate end is able to trigger IFN production in cells. Thereafter, our laboratory and others have reported that an essential feature of the viral RNA ligand of RIG-I is a free 5′-triphosphate that is absent from host cytoplasmic RNA due to eukaryotic RNA metabolism (9, 10). Using 5′-triphosphate RNAs produced by IVT, these studies concluded that both single-stranded (ss) and dsRNAs activate RIG-I as long as they carry the 5′-triphosphate (8–10). The RD of RIG-I has subsequently been characterized as the structural entity that binds 5′-triphosphate and, thus, aids in defining ligand specificity (11, 12). However, the concept that the 5′-triphosphate modification in cytosolic RNA represents the complete PAMP recognized by RIG-I was challenged recently by several prominent studies, suggesting that (i) 3′-monophosphate RNAs, as produced by RNase L, might be RIG-I ligands (13); (ii) a 5′-triphosphate end is dispensable if the RNA ligand is double stranded and carries either 5′-monophosphates or is long enough (12, 14); and (iii) RIG-I ligands require uridine- or adenosine-rich sequences (15). These reports raise the question whether the 5′-modification with (tri)phosphate is sufficient, merely required, or in some cases dispensable for physiological RIG-I ligands. For this report, we have investigated the structural requirements to activate RIG-I-mediated antiviral signaling using defined ligands including synthetic 5′-triphosphate RNA.
Results
Ligand-Induced ATPase Activity Is Triggered by a Feature Other than the 5′-Triphosphate Moiety.
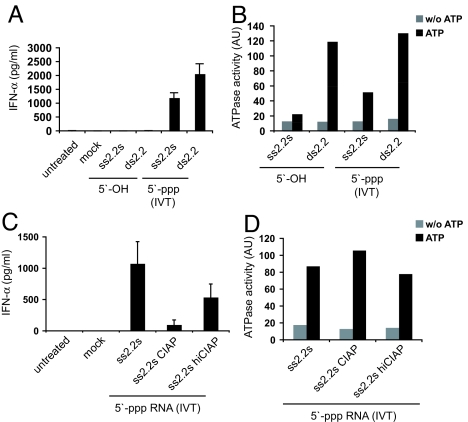
Previous studies have shown that short 5′-triphosphate RNAs of 19 to 21 bases generated by IVT are potent RIG-I ligands that induce IFN in human monocytes independently of TLRs (8–10). To identify a minimal pattern sufficient to trigger RIG-I signaling, we analyzed different versions of a 19-mer model-RNA named 2.2 (Table S1). The chemically synthesized 5′-OH version of ss2.2 sense (s) RNA, its complementary antisense (as) strand, and their annealed base-paired version (ds2.2) failed to induce IFN-α when transfected into human monocytes (Fig. 1A). However, as expected from previous studies, IVT (5′-triphosphate) 2.2 RNA induced strong IFN production either as ssRNA (ss2.2s) or as dsRNA (ds2.2) (Fig. 1A). Of note, in our hands, chemically synthesized ds or ss 3′- or 5′-monophosphorylated RNAs did not show significant immunostimulatory activity when we transfected them into human monocytes using the 2.2 sequence or a 25-mer sequence published to be active in mouse cells by Takahasi et al. (Fig. S1A) (12). However, they were active when transfected into human PBMCs containing plasmacytoid dendritic cells, indicating that these RNAs can be recognized by a different PRR like TLR7 (Fig. S1B) (16, 17). When we examined IVT ss2.2s and ds2.2 RNAs, they both induced ATPase activity of purified RIG-I in vitro, whereas ss 5′-OH-RNA did not (Fig. 1B). Double-stranded 5′-OH-RNA, even though unable to induce IFN, clearly triggered ATPase activation (Fig. 1B). To check whether the 5′-triphosphate modification in IVT RNAs is sufficient to trigger ATPase activity of RIG-I, we dephosphorylated IVT ss2.2s RNA with calf-intestine alkaline phosphatase (CIAP), and examined its immunostimulatory activity and ability to induce ATPase activity in RIG-I. Surprisingly, even though dephosphorylated ss2.2s RNA largely lost its immunostimulatory activity, indicating that its 5′-triphosphate modification has been quite efficiently removed (Fig. 1C), it retained its full ability to induce RIG-I ATPase activity (Fig. 1D). Together, these results confirmed the importance of the 5′-triphosphate modification for activation of RIG-I, and suggested that IVT ss2.2s RNA has a feature other than the 5′-ppp modification that triggers the ATPase activity of RIG-I and is missing in the synthetic 5′-OH-version of ss2.2s RNA.
Fig. 1.
In vitro transcribed ssRNA has a feature other than the 5′-triphosphate necessary for RIG-I activation. (A) Primary human monocytes were stimulated with ss- and dsRNAs that were chemically synthesized (5′-OH) or generated by IVT (5′-ppp). After 36 h, IFN-α was quantified in the supernatant by ELISA. (B) The same set of RNA ligands was assayed for their ability to induce ATPase activity in recombinant purified full-length RIG-I protein. Each condition was done in the presence and absence of ATP. (C) Untreated or CIAP-treated 5′-ppp-ssRNA (IVT) was tested for immunostimulatory capacity on monocytes as in A. RNA treatment with heat-inactivated (hi)CIAP was used as a control. (D) Fluorescence signal of the ATPase assay was measured after incubation of full-length RIG-I protein with RNA oligonucleotides from C.
RNAs Generated by IVT with T7 Polymerase Contain Unexpected Hairpin-Forming By-Products That Activate RIG-I.
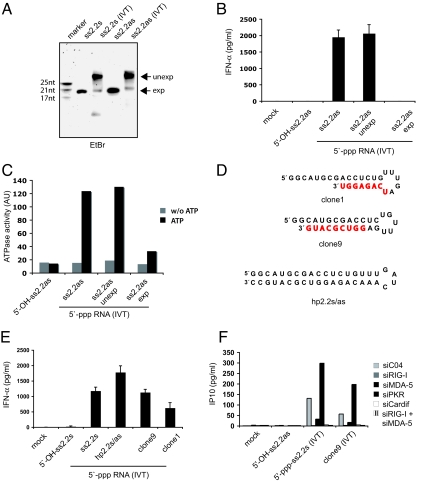
These results prompted us to examine the RNA products of our IVT reactions in more detail. Denaturing polyacrylamide gels showed that synthetic ss2.2s and ss2.2as RNAs could be observed at sizes according to their expected length (Fig. 2A). However, the RNA preparations produced by IVT contained pronounced amounts of products significantly longer than expected in addition to varying amounts of RNA of the expected size (Fig. 2A). When we reisolated IVT ss2.2as RNA from bands of the expected size and transfected it into primary human monocytes, no IFN production was seen (Fig. 2B). However, when the same amount of RNA isolated from a band of the unexpected size was tested, it induced amounts of IFN comparable with those induced by unpurified IVT ss2.2as RNA. Consistent with these results, only reisolates from the unexpected band were able to induce RIG-I ATPase activity (Fig. 2C). Comparable results were observed with ss2.2s RNA. Together, these results showed that the immunostimulatory activity of RNA produced by IVT resulted exclusively from unexpected, potentially base-paired RNA species.
Fig. 2.
5′-triphosphate RNAs generated by IVT contain base-paired by-products that activate RIG-I. (A) Chemically synthesized 5′-OH-ssRNAs and 5′-ppp-ss2.2 RNAs generated by IVT were analyzed by denaturing gel electrophoresis and ethidium bromide (EtdBr) staining. Products of the expected (lower arrow in all samples), and unexpected (upper arrow in lanes containing 5′-ppp-RNA generated by IVT) size were reisolated for further analysis. (B) Reisolated IVT products were compared with 5′-OH-ss2.2as RNA and unpurified 5′-ppp-ss2.2as (IVT) RNA, for their ability to induce IFN-α production in human monocytes. (C) Stimulation of ATPase activity of recombinant, full-length RIG-I protein with reisolated and control RNAs. (D) Clones 1 and 9 are 2 RNA-sequences identified by 5′-ppp-ss2.2 RNA (IVT) small RNA cloning and sequencing. Nucleotides not encoded by the DNA template are indicated in red. Also, the sequence of a designed hairpin RNA (hp2.2s/as) based on the 2.2 model sequence is displayed. Secondary structures (minimum free energy) of clone 1, clone 9, and hp2.2s/as are as predicted by the Mfold software (21). (E) RNAs of clone 1, clone 9, and hp2.2s/as were generated by IVT and transfected into human monocytes. Production of IFN-α was measured by ELISA after 36 h. (F) The 1205Lu human melanoma cells were treated with the indicated siRNAs for 48 h and subsequently transfected with the indicated RNAs; 12 h after stimulation, supernatants were subjected to IP10 analysis by ELISA.
It has been described that phage RNA polymerases such as T7, which is commonly used to produce IVT RNAs, possess an RNA-dependent RNA polymerase activity in addition to being a DNA-dependent RNA polymerase (18, 19). Therefore, RNA with self-complementarity at the 3′-end generated from a DNA-template can serve as a self-primed RNA-template that is elongated in a second step by the RNA-dependent-RNA polymerase activity of T7, potentially supporting the formation of nontemplated self-complementary products. Similar phenomena have been described for viral polymerases during viral replication as so called “copy-back” mechanisms (20). Due to the low degree of self-complementarity required, this phenomenon was found to be very common in IVT reactions (19). Indeed, using a previously analyzed large panel of permutated DNA-templates categorized into 3 groups according to their yield of expected and extended RNA products (type A, only the expected RNA; type B1.1, up to 2% extended products; type B2.1, up to 90% extended products; see ref. 19), we found that only in vitro transcribed RNAs that form high amounts of extended products were potent IFN inducers (Fig. S2A). Similarly, 100-mer poly(A) RNA generated by IVT as described by Saito et al. (15) was inactive in our hands if devoid of extended by-products (Fig. S2 B and C).
To prove that the unexpected bands derived from the 2.2 sequence do contain self-complementary structures, we sequenced clones of cDNA libraries constructed from RNA isolated from the expected and unexpected bands of IVT ss2.2s and ss2.2as. Indeed, whereas sequencing of RNAs from the expected band only identified sequences encoded by the respective DNA-template, the clones isolated from the unexpected bands contained sequences with 3′-extensions complementary to the original DNA-templated sequence; these extensions facilitate stable hairpin formation according to the prediction by the Mfold webserver (21). Two examples, clones 1 and 9, are shown in Fig. 2D. When we transcribed these sequences in vitro (for purity, see Fig. S3), they were able to induce significant amounts of IFN-α in human monocytes (Fig. 2E). The levels were comparable with those induced by ss2.2s IVT RNA or an optimized IVT hairpin artificially designed by fusion of the sense and antisense strand of the 2.2 sequence (hp2.2s/as). Interestingly, the amount of IFN induced by these RNAs seemed to correlate with the size of the stem structure. To investigate whether RIG-I mediates the recognition of these 5′-triphosphate hairpin RNAs, we silenced potential PRRs involved in 1205Lu melanoma cells using siRNA. Indeed, silencing of RIG-I, of its signaling adapter Cardif, and of both RIG-I and MDA-5 abolished IP10 production after stimulation with IVT 2.2 RNAs and clone 1 highlighting the requirement of RIG-I for recognition (Fig. 2F). These data were confirmed by experiments using mouse dendritic cells lacking Cardif and using HEK 293 cells overexpressing RIG-I (Fig. S4 A and B). Silencing MDA-5 alone led to a moderate decrease in IP10 production for these RNAs, suggesting that MDA-5 is not essential, but might be involved in a full signaling response. Depletion of PKR did not decrease IP10 production in any of the conditions tested. Together, these data show that small RNAs that carry both a base-paired hairpin structure and a 5′-triphosphate group in its proximity are potent RIG-I ligands. We propose that many observations that have been made with respect to RIG-I ligands using enzymatically synthesized RNAs might at least in part be due to unexpected RNAs that contain base-paired secondary structures in addition to the 5′-triphosphate end.
RNAs Require a Free 5′-Triphosphate and Base-Paired Stretches to Activate RIG-I.
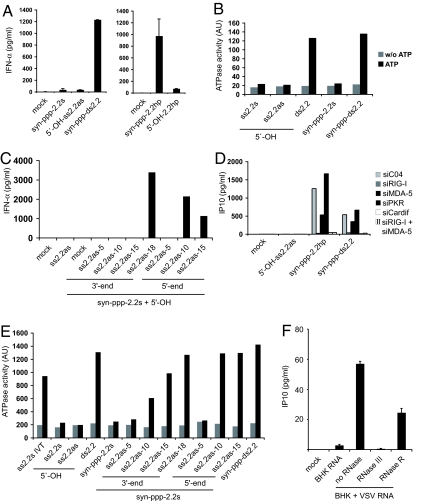
To investigate RIG-I activation in a system where the effect of base-pairing and 5′-triphosphate can be clearly separated, we resorted to defined, chemically synthesized 5′-triphosphate RNA (syn-ppp-ss2.2s). Of note, the production of 5′-triphosphate RNAs by nonenzymatic chemistry is a challenge, and these reagents were not available so far. We consistently found that syn-ppp-ss2.2s RNA, which like the expected IVT product of 2.2s cannot form stable secondary structures, was unable to induce IFN in cells (Fig. 3A Left). However, annealing of a complementary 5′-OH strand (5′-OH-ss2.2as) before transfection rescued IFN production, indicating that a base-paired structure was necessary and, together with a 5′-triphosphate, sufficient to trigger RIG-I (Fig. 3A Left). Thus, formation of a loop structure was dispensable in this system. Still, a chemically synthesized 5′-triphosphate hairpin RNA (syn-ppp-2.2hp) designed to incorporate base-paired secondary structure and 5′-triphosphate into one molecule lead to high-level IFN induction in human monocytes (Fig. 3A Right). Importantly, the corresponding 2.2 hairpin RNA lacking a 5′-triphosphate did not induce IFN (Fig. 3A Right). Signaling of chemically synthesized 5′-triphosphate RNAs showed the same RIG-I-dependence seen before with ligands generated by IVT (Fig. 3D; Fig. S4 A and B). Assaying RIG-I ATPase activity of the chemically produced ligands showed that only base-paired, but not unstructured 5′-triphosphate RNA, can induce ATP hydrolysis (Fig. 3B). This difference in activity was not due to a higher stability of base-paired RNA, because neither ssRNA nor dsRNA showed degradation under assay conditions (Fig. S4C). These results support our hypothesis and provide proof that the 5′-triphosphate end is not sufficient to mediate RNA-induced RIG-I activation and cannot trigger ATPase activity on its own.
Fig. 3.
A small base-paired stretch and the 5′-triphosphate are necessary and sufficient to activate RIG-I. (A) Chemically synthesized 5′-triphosphate 2.2 RNA (syn-ppp-2.2s) either alone or after hybridization (syn-ppp-2.2ds) to 5′-OH-ss2.2as RNA (Left) and chemically synthesized 2.2 hairpin RNA with (syn-ppp-2.2hp) or without (5′-OH-2.2hp) a 5′-triphosphate (Right) was transfected into human monocytes. IFN-α secretion was analyzed after 36 h. (B) Activation of ATP hydrolysis by full-length RIG-I protein was studied with chemically synthesized RNAs bearing either a 5′-OH group or a 5′-ppp group. (C) syn-ppp-2.2s RNA was hybridized to complementary OH-RNA of variable length. The length gradually increased from either the 3′- or the 5′-end. IFN-α production was assessed in human monocytes as in A. (D) 1205Lu human melanoma cells were stimulated with chemically synthesized RNAs after treatment with the indicated siRNAs for 48 h. IP10 levels were measured after 12 h by ELISA. (E) RNA oligonucleotides used in C were examined for their induction of RIG-I ATPase activity in full-length protein. (F) 1205Lu cells stimulated with total RNA isolated from BHK cells infected or not with VSV for 16 h at a multiplicity of infection of 0.2. RNA isolates from VSV-infected BHK cells were treated with equal activities of the RNases III or RNase R before transfection. IP10 levels were determined by ELISA 14 h after stimulation.
To define the structural requirements for RIG-I activation in more detail, we hybridized successively longer 5′-OH-RNAs complementary to the 3′- or 5′-end of syn-ppp-ss2.2s RNA, and tested their ability to induce IFN production in monocytes and ATPase activity of recombinant RIG-I. When oligonucleotides of 5-, 10-, or 15-nts length were hybridized to syn-ppp-ss2.2s RNA from the 3′-end, no IFN production was observed (Fig. 3C); only fully complementary RNA (Fig. 3A) or an 18-mer hybridized to the 3′-end of syn-ppp-ss2.2s RNA could rescue IFN production (Fig. 3C). However, when 5-, 10-, or 15-nts complementary RNA were hybridized to the 5′-end of syn-ppp-ss2.2s RNA, 10 nts were sufficient to rescue IFN induction, suggesting that the extent of base-pairing as well as its relative position to the 5′-triphosphate-end are important determinants of immunostimulatory activity (Fig. 3C). Assaying the same set of RNAs for ATPase activity showed that ATP-hydrolysis depended on a base-paired stretch of >5 nts, but was independent of its relative position to or the presence of the 5′-triphosphate (Fig. 3E). Interestingly, complementary strands producing a short 3′-overhang at the 5′-triphosphate-bearing end of the oligonucleotide supported RIG-I ATPase activity, but did not induce IFN. The short overhang in direct proximity to the 5′-triphosphate group seemed to interfere with its correct recognition, highlighting the importance of a free 5′-triphosphate for signaling activity (Fig. S5 A and B). Together, these results show that a 5′-triphosphate modification on RNA is not sufficient to activate RIG-I signaling. They support a model in which a minimal pattern that can be recognized by RIG-I is a rather small stretch of base-paired RNA in addition to a 5′-triphosphate group. Of note, this finding does not seem to be an effect that is observed exclusively with short RNAs below 20 bases. A 70-mer RNA designed not to form secondary structures (Table S1) is unable to induce IFN as an in vitro transcript. However, in line with our previous data, cytokine production and ATPase activity can be rescued by addition of a complementary strand (Fig. S6). Stressing the importance and requirement of the 5′-triphosphate modification for optimal activation of RIG-I, two chemically produced base-paired 70- and 40-nt RNAs lacking a 5′-triphosphate did not lead to detectable cytokine production in human 1205Lu cells (Fig. S6).
To investigate the importance of base-pairing in the setting of viral infection, we isolated total RNA from BHK cells infected with vesicular stomatitis virus (VSV), a virus known to be recognized by RIG-I (22). When we transfected 1205Lu cells with the RNA isolated from uninfected cells, no cytokine production was observed, because cellular RNA is not recognized by RIG-I. However, when we transfected RNA from virus-infected cells, IP10 production triggered by viral RNA ligands in the preparation could be observed. In line with our model, we found that the ds specific RNase III abolished signaling by these ligands, whereas the ss specific RNase R only had a minor effect on the activity of the RNA preparation (Fig. 3F). Specificity of the RNases was confirmed using synthetic ss or ds oligonucleotides (Fig. S7). Together, these results indicate that base-pairing is an important additional feature of the RIG-I ligands that generate the signal after viral infection.
Both Features of the PAMP Contribute to RIG-I Dimerization.
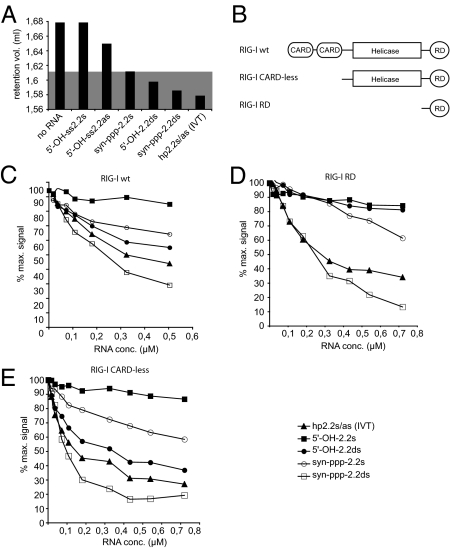
Next, we wanted to examine the molecular basis of RIG-I activation by ligands carrying the bipartite PAMP. Dimerization of RIG-I molecules has been proposed to have a crucial role in promoting RIG-I signaling (11). Thus, we sought to examine the association state of RIG-I molecules in the presence of defined, synthetic RNA ligands in vitro. Recombinant purified RIG-I protein formed complexes with chemically synthesized RNAs when they were base-paired or carried a 5′-triphosphate group as assayed by native PAGE analysis (Fig. S8A). For a more detailed analysis, we performed gel filtration analysis of purified full-length RIG-I incubated with different synthetic RNAs. As expected, RIG-I incubated with ssRNA carrying a 5′-OH group (5′-OH-ss2.2s RNA) showed no change in the elution volume compared with RIG-I protein analyzed in the absence of RNA (Fig. 4A; Fig. S8B). When we incubated RIG-I with synthetic ss 5′-triphosphate RNA or a base-paired 5′-OH RNA, both induced an elution volume shift of RIG-I indicative of dimerization (Fig. 4A; Fig. S8B). Interestingly, when we incubated RIG-I protein with ligands that were base-paired and carried a 5′-triphosphate (synthetic or IVT), the shift in elution volume was more pronounced. Together, these results demonstrate that both features of the PAMP, base-pairing and 5′-triphosphate, independently contribute to the structural changes in RIG-I leading to its higher order assembly in vitro. They also suggest that the bipartite PAMP leads to a structural change different from that induced by the single features that possibly reflects the conformation of the signaling competent complex in vivo.
Fig. 4.
Double-stranded 5′-triphosphate RNAs have multiple binding sites on RIG-I and induce dimerization of RIG-I. (A) Different RNAs were examined for their ability to induce homodimerization of recombinant full-length RIG-I protein. The protein-RNA mixture was loaded onto a gel filtration column, and the absorbance at 280 nm was determined. RIG-I elution volume peaks of the different mixtures are represented as columns. The gray area indicates the range of values interpreted as dimeric RIG-I-RNA complexes. (B) Schematic representation of the recombinant RIG-I proteins used in C, D, and E. (C) Recombinant RIG-I full-length protein prebound to fluorescently labeled 5′-ppp-hp2.2s/as RNA (IVT) was incubated with increasing amounts of unlabeled competitor RNAs. Fluorescence anisotropy was determined multiple times after each addition and equilibration. Binding and competition of the unlabeled RNAs is seen as a progressive loss of anisotropy signal. Background values were subtracted, and the anisotropy value of labeled RNA bound to RIG-I in the absence of a competitor was set as 100%. (D) The RNAs used in C were assayed by using recombinant RIG-I RD domain. (E) The RNAs used in C were assayed by using recombinant RIG-I protein lacking both CARD domains.
RIG-I Binding to Base-Paired 5′-Triphosphate RNA Is Mediated by Distinct Binding Sites.
The structure of the RIG-I carboxyl-terminal RD (RIG-I RD or CTD) has been solved recently by X-ray crystallography and NMR spectroscopy, and a binding groove for the 5′-triphosphate and dsRNA structures was identified in the RIG-I RD domain (11, 12). To gain further insight into how the structural features of the bipartite RIG-I PAMP, base-paired stretch and 5′-triphosphate end, are recognized by RIG-I, we used a competitive binding assay based on fluorescence anisotropy. Different purified RIG-I proteins (Fig. 4B) prebound to fluorescently labeled IVT hp2.2s/as RNA were incubated with increasing amounts of unlabeled synthetic ligands, and the loss of the aniso-tropy signal was measured. As expected, 5′-OH ssRNA could not compete for binding with IVT hp2.2s-as RNA on full-length RIG-I (Fig. 4C). However, when we added RNAs bearing a base-paired stretch or a 5′-triphosphate end or both (5′-OH-ds2.2s, syn-ppp-ss2.2s or syn-ppp-ds2.2), a pronounced loss of fluorescence anisotropy could be observed, indicating that these RNAs compete with IVT hp2.2s-as RNA for binding to RIG-I (Fig. 4C). Consistent with our model, loss of anisotropy was more pronounced with a ligand that contained both features of the PAMP (syn-ppp-ds2.2 and IVT hp2.2s-as RNA). When we competed from recombinant purified RIG-I RD protein in a very similar experiment, we found that, as with the full-length protein, a synthetic ligand that carried a base-paired stretch and a 5′-triphosphate was a superior competitor over a molecule bearing a 5′-triphosphate only, indicating that there is a contribution of base-paired stretches to binding the RD domain (Fig. 4D). However, the observation that a base-paired molecule that lacked a 5′-triphosphate could not compete showed that this contribution is small and the binding to the RD-domain is mainly determined by the 5′-triphosphate (Fig. 4D). Interestingly, when we used full-length RIG-I or protein that lacked the CARDs, but contains the helicase and RD domains, base-paired RNA lacking the 5′-triphosphate component was a better competitor than 5′-triphosphate RNA missing a base-paired stretch (Fig. 4E). Comparing the competition patterns of these 3 constructs provides molecular evidence that the helicase domain interacts strongly with the base-paired part of the molecule, and that this interaction constitutes a significant part of the overall binding of RNA ligands to RIG-I. Together, our observations are in line with a model in which the 5′-triphosphate end serves as an initial selective binding motif that associates with the RD domain, and formation of a stable and productive signaling complex occurs in a subsequent step, involving interaction of the base-paired part of the molecule with the helicase domain.
Discussion
We have studied the ligand requirements of RIG-I-mediated antiviral signaling using defined model ligands, including hitherto unavailable synthetic 5′-triphosphate RNA and RNA from virus-infected cells. We conclude that only the combination of a ds stretch of RNA, together with a 5′-triphosphate end, defines a sufficient PAMP recognized by RIG-I. Of note, the minimal extent of base-pairing that we find necessary for RIG-I activation is small (in the range of 5 to 10 bases). The observation that viral infections recognized by RIG-I are specifically characterized by the lack of high amounts of long (>20 bp) dsRNA in infected cells hints that our short model ligands might indeed be in the size range of natural viral ligands of RIG-I (10, 23). In vitro transcribed RNAs have been widely used to probe both the ligand requirements for RIG-I signaling in cells and the mechanisms of RIG-I activation in vitro. Our results clearly show that IVT products must be used with caution when conclusions about ligand requirements for RIG-I signaling are drawn (24). In our study, the functional analysis of IVT products revealed that nontemplated hairpin RNAs generated through the RNA-dependent RNA polymerase activity of T7 were the immunostimulatory RNA species. Interestingly, the self-templated copy-back activity of T7 presumably responsible for those RNA-species has also been demonstrated for the viral RNA polymerases of hepatitis C virus (HCV) and Sendai virus, which are both well described activators of RIG-I (20, 25, 26). It has been observed that RLHs, in particular MDA-5, are able to recognize the dsRNA analog poly(I·C) in the cytoplasm (22, 27, 28). However, for true dsRNA made by chemical synthesis, data are scarce. A recent publication that advocates the dispensability of the 5′-triphosphate end for dsRNA-recognition by RIG-I and differential recognition of long dsRNAs by RIG-I or MDA-5 shows that a chemically synthesized dsRNA of 70 nt without 5′-triphosphate leads to IFN responses that are minimal (>100-fold less) compared to stimulation with a natural RIG-I-dependent ligand (14). Together with our results that showed no activation of RIG-I by chemically produced dsRNA of up to 70 bases in the absence of a 5′-triphosphate, these findings in our view argue against the possibility that RIG-I is activated purely by dsRNA in a physiologically relevant way. The footprint of the ATPase-domain of SF2-family helicases is ≈6 to 7 base pairs (29). How structurally similar helicases distinguish RNAs by length in a range several 100 times larger than their footprint is not clear. Also, a current study shows differential recognition of poly I:C by RIG-I and MDA-5 in a much lower size range (30). Few reports exist that show direct recognition of particular viral RNAs by RIG-I in the setting of infection with a living virus (5, 31). In cases where data of this kind is available, these RNAs (the leader RNAs of negative-stranded RNA viruses and the EBERs of EBV) are comparably small, and contain hairpin-like structures in addition to and in proximity of a 5′-triphosphate end. In line with those data, we found that RNA isolated from virus-infected cells remains immunostimulatory when treated with a ss specific RNase, but not when base-paired RNA is digested. Recently, it was suggested that viral RNA requires A- or U-rich stretches to activate RIG-I (15). The mechanism of sequence-specific ligand recognition has not been investigated in this context. We did not systematically test the influence of sequence content on RIG-I activation. However, the different short sequences we found to be active were neither A nor U rich (<50% A plus U). Therefore, the ligand features that have been identified by Saito et al. (15) may certainly be highly relevant in the context of HCV recognition, but do not seem to present a generalized structural motif that explains RIG-I ligand recognition. Our biochemical binding and competition data suggest that 5′-triphosphate end and base-paired stretch, the 2 parts of the PAMP, bind distinct sites on RIG-I. Even though the functional cellular assays clearly prove that only a bipartite ligand induces changes in RIG-I that lead to a signaling-competent complex, at the moment, we can only speculate on the nature of these changes. A parallel study showing that RIG-I translocates along dsRNA after binding the 5′-triphosphate provides a further important piece in the puzzle (32). In the future, the structural analysis of RIG-I protein with synthetic RNA ligands will provide us with more information on RIG-I activation mechanisms.
Materials and Methods
RNAs.
RNAs (unmodified, monophosphate, and siRNA) and DNAs were purchased from Metabion. Eurogentec kindly produced the chemically synthesized 5′-triphosphate RNA (for details, see SI Methods). In vitro transcribed RNAs were synthesized by using the Megashortscript kit (Ambion). DNA templates were generated as previously described (9). RNAs were extracted with phenol/chloroform, precipitated with ethanol, and passed through a Mini Quick Spin Column (Roche). The treatment of IVT RNA with CIAP was carried out as in Hornung et al. (9). Small interfering RNA was applied to 1205Lu cells at 30 nM with Lipofectamine RNAiMax (Invitrogen) as a transfection reagent. After 48 h, the culture medium was exchanged, and cells were stimulated. For a detailed list of all RNA oligonucleotides used for stimulation, see Table S1. For DNA templates and primers, see Table S2, and for the siRNAs, see Table S3.
Cell Stimulation and Cytokine Measurement.
Unless indicated otherwise, all primary cells and cell lines were stimulated at 200 ng/mL RNA using Lipofectamine 2000 according to the manufacturer's manual. The 1205LU cells were transfected with Lipofectamine RNAiMax. R848 was from 3M Pharmaceuticals. IFN-α was measured 36 h after stimulation in the supernatant of human monocytes, PBMCs, and murine dendritic cells using the IFN-α module set from Bender MedSystems and PBL, respectively. Human IP10 was analyzed 12 h after stimulation in the culture medium of 1205LU cells using the opteia set from BD. Induction of the IFN-ß promoter was detected with a reporter assay in HEK 293 cells as described in Rothenfusser et al. (3); 24 h after transfection, the cells were stimulated with the indicated RNA oligonucleotides for 12 h.
Recombinant Protein and ATPase Activity Assay.
The ATPase activity of recombinant, purified RIG-I-protein was measured using the ADP Quest HS Assay (DiscoveRx). Full-length RIG-I was expressed in insect cells and purified as described previously (11). The reaction mixture was prepared on ice in a total volume of 10 μL, containing 1 ng/μL purified RIG-I-protein, 1 ng/μL purified RNA oligonucleotide, and 100 μM ATP. Reactions were initiated by the addition of ATP and incubated for 2 h at 37 °C.
Isolation and Purification of RNA from Polyacrylamide Gels.
Stained RNA bands of interest were cut out on a UV table. The gel slices were fragmented, and the RNA was eluted by adding elution buffer (0.5 M ammonium acetate/1 mM EDTA/0.2% SDS) for 12 h at 37 °C. Subsequently, the eluted RNA was extracted and precipitated as described above.
Size-Exclusion Chromatography.
All experiments were carried out at room temperature, using a GE Ettan LC system equipped with a Superose 6 PC 3.2/30 (GE Healthcare) size-exclusion column. The size-exclusion column was equilibrated with buffer containing 30 mM Tris·HCl, pH 7.5/150 mM NaCl/2 mM DTT/10 μM ZnCl2. After sample injection, UV absorption at both 260 and 280 nm wavelength was recorded. The column was calibrated with Gel Filtration Standard (Bio-Rad) before use.
Fluorescence Anisotropy Measurement.
Fluorescence anisotropy experiments were performed with a FluoroMax-P fluorimeter (Horiba Jobin Yvon); 1 mL of buffer (30 mM Tris·HCl, pH 7.5/150 mM NaCl/5 mM 2-mercaptoethanol/10 μM ZnCl2) and indicated amounts of fluorescently labeled RNA (in vitro transcribed hp2.2s/as RNA with incorporated Alexa Fluor 488–5-UTP) were preequilibrated in a quartz cuvette at 12 °C. Recombinant protein and competitor RNAs were added in a stepwise manner and mixed by gentle pippetting. Preequilibration was used until anisotropy signals were stabilized. The anisotropy data were collected using an excitation wavelength of 495 nm and monitoring the emission at 516 nm. A maximum number of 10 repeats were performed until <2% deviation of the signal was reached.
Supplementary Material
Acknowledgments.
We thank M. Linke and M. Ruesenberg from Eurogentec for organizing the synthesis of 5′-ppp-RNAs, Gerhard Steger for insightful discussions on RNA structure, and Julia Vorac and Simone Willms for excellent technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grants DFG RO 2525/3-1 (to S.R.), DFG GK 1202 (to T.S., W.H, J.C.H., S.R., R.B., and S.E.), and LMUexcellent (Center for Integrated Protein Science Munich 114, research professorship), Sonderforschungsbereich-TR 36, and EN 169/7-2 (to S.E.). This work is part of the thesis of T.S., W.H., and J.C.H. at the Ludwig-Maximilian University Munich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900971106/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 5.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumpter R, Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: A priceless link to innate immunity. Trends Mol Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 9.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 10.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 11.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Hornung V, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 18.Cazenave C, Uhlenbeck OC. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc Natl Acad Sci USA. 1994;91:6972–6976. doi: 10.1073/pnas.91.15.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nacheva GA, Berzal-Herranz A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur J Biochem. 2003;270:1458–1465. doi: 10.1046/j.1432-1033.2003.03510.x. [DOI] [PubMed] [Google Scholar]
- 20.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 23.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlee M, et al. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 25.Leppert M, Kort L, Kolakofsky D. Further characterization of Sendai virus DI-RNAs: A model for their generation. Cell. 1977;12:539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhong W, et al. Nucleoside triphosphatase and RNA helicase activities associated with GB virus B nonstructural protein 3. Virology. 1999;261:216–226. doi: 10.1006/viro.1999.9871. [DOI] [PubMed] [Google Scholar]
- 27.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 29.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjith-Kumar CT, et al. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2008;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plumet S, et al. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.