Abstract
Proper organization of microtubule arrays is essential for intracellular trafficking and cell motility. It is generally assumed that most if not all microtubules in vertebrate somatic cells are formed by the centrosome. Here we demonstrate that a large number of microtubules in untreated human cells originate from the Golgi apparatus in a centrosome-independent manner. Both centrosomal and Golgi-emanating microtubules need γ-tubulin for nucleation. Additionally, formation of microtubules at the Golgi requires CLASPs, microtubule-binding proteins that selectively coat non-centrosomal microtubule seeds. We show that CLASPs are recruited to trans-Golgi network (TGN) at the Golgi periphery by the TGN protein GCC185. In sharp contrast to radial centrosomal arrays, microtubules nucleated at the peripheral Golgi compartment are preferentially oriented toward the leading edge in motile cells. We propose that Golgi–emanating microtubules contribute to the asymmetric microtubule networks in polarized cells and support diverse processes including post-Golgi transport to the cell front.
Introduction
Microtubules (MTs) serve as highways for intracellular transport arranging appropriate distribution of organelles and signals within a cell. Precise spatial and temporal regulations of MT distribution are essential for numerous cell functions.
In animal cells, centrosomes serve as the principal MT-organizing centers (MTOCs). Centrosomes organize symmetric MT arrays of uniform polarity, where MT minus ends are embedded in the centrosome while the highly dynamic plus ends extend toward the cell periphery. MT nucleation can also occur via centrosome-independent mechanisms. MT nucleation events were described at the cell periphery far from the centrosome (Yvon and Wadsworth, 1997), and cells lacking centrosomes form relatively normal MT arrays (Khodjakov et al., 2000). A number of MT-organizing structures have been identified in interphase cells. Among these are the nuclear envelope in myotubes (Bugnard et al., 2005), plasma membrane of polarized epithelia (Reilein and Nelson, 2005) and melanosomes in pigment cells (Malikov et al., 2004). However, these sites appear to be functional only in specialized cell types. The question of where non-centrosomal MTs are nucleated in non-differentiated cells remains open.
There have been reports that purified Golgi membranes support MT nucleation. In cell reforming MTs upon nocodazole washout, short MTs consistently associate with the Golgi (Chabin-Brion et al., 2001). This work suggested that the Golgi could serve as an MTOC. However, it remained ambiguous whether Golgi-associated MTs found in nocodazole washouts were in fact nucleated at the Golgi or if they were nucleated by the centrosome but consequently released and captured by the Golgi (Rios et al., 2004). This later scenario is probable as MT minus ends are known to have affinity for Golgi membranes (Rios et al., 2004).
Indeed, is very difficult to prove de novo MT nucleation at the Golgi. During interphase, the Golgi complex consists of membrane cisternae stacks with distinct polarity (Ladinsky et al., 2002) arranged in a complex “ribbon” situated very close to the centrosome. For this reason Golgi-associated MT arrays could be easily confused with those originating from the centrosome. We have overcome this difficulty by developing a technique that allows us to trace individual MTs back to their point of origin in live cells. This approach reveals that the Golgi nucleates MTs under physiological conditions. In sharp contrast to the centrosome, MT arrays organized by the Golgi are inherently asymmetric.
Our data demonstrate that MT nucleation at the Golgi requires the MT +TIP proteins CLASPs, which have been previously localized to the Golgi (Akhmanova et al., 2001). Here, we provide evidence that CLASPs associates specifically with the trans-Golgi network (TGN) protein GCC185. Thus, CLASPs concentrate only in the TGN leading to the asymmetry of the MT array nucleated at the Golgi.
Results
Identification of MT nucleation sites in interphase cells
MT nucleation at centrosomes was previously analyzed by tracking fluorescently labeled plus tip-binding protein (Piehl et al., 2004). We have adopted this approach to detect the origin of non-centrosomal MTs in retinal pigment epithelial cells (RPE1) cells (Fig.1 A-D) during interphase. MT tips were visualized by fluorescently labeled EB3 (Figs. 1, 5) or CLIP170 (Fig.S1). MTs that carried EB3 signal in the first frame of the video sequence had been nucleated before we initiated our observations, and thus their origin could not be determined (Fig.1 A). Such MT tracks (Fig.1 B, magenta) were excluded from further analysis. All MT tracks that were initiated during the recording were divided in two distinct groups. First, MTs that originated from a common perinuclear site (∼2μm in diameter) were regarded as centrosomal. These MTs consistently formed a radial symmetric array (Fig.1 B,D,F yellow). Parallel analysis of similarly obtained EB3 tracks in cells co-expressing GFP-centrin revealed that the centrosome was always in the middle of these radial arrays (not shown). The second group of MTs originated from a larger common area spatially separated from the centrosome (Fig.1 B, cyan). In addition to these two major groups a few MT tracks emerged close to the cell periphery. These tracks likely corresponded to MTs that were not truly nucleated during our observations but rather rescued as the result of dynamic instability. It is important to emphasize that while MT rescues are relatively frequent in the peripheral parts of cytoplasm, they are extremely rare in the center of the cell (Komarova et al., 2002), our unpublished data). Thus, most of those tracks that originate near the cell center represent outgrowth of newly nucleated MTs.
Fig. 1. Golgi complex is an additional MTOC.
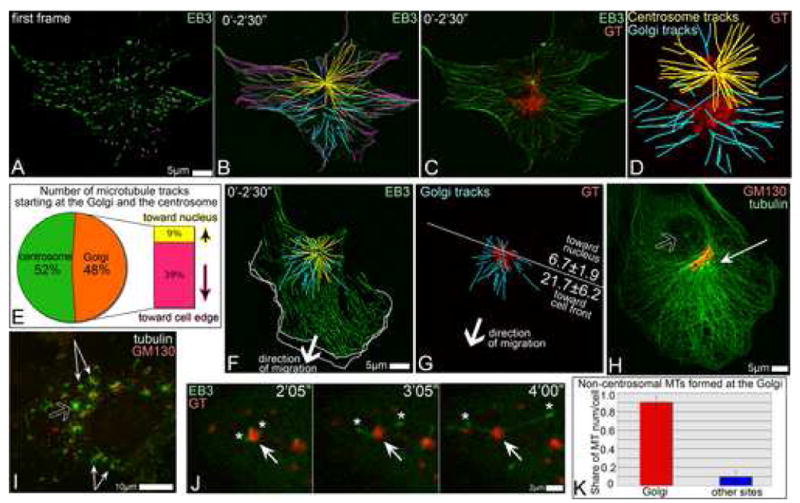
A-D. Detection of Golgi-originated MTs in time lapse recording of GFP-EB3 and mCherry-GT expressing RPE1 cell (5sec/frame). A. GFP-EB3 in the first frame of the video (green). Currently growing MTs are marked by magenta dots. B. Overlaid GFP-EB3 showing MT tracks within 2.5′. Magenta, tracks started at the frame one. Yellow, centrosomal tracks. Cyan, non-centrosomal tracks. C. Overlaid GFP-EB3 (green) and mCherry-GT (red) images within 2.5′. D. centrosomal (yellow) and non-centrosomal (cyan) MT tracks in the cell center and their relation to the Golgi position (GT, red). E. Percentage and directionality of Golgi-associated tracks (583 tracks in 10 cells, analyzed as above). F, G. Directionality of Golgi-associated tracks in a motile cell. Arrows indicate the direction of the cell re-location. F. Overlaid mCherry-EB3 tracks (2.5′, false-colored green). Centrosomal (yellow) and Golgi-associated (cyan) tracks in the cell center are shown. Outlines of the protruding cell front in the first and the last frame of the 5′ video are shown as white lines. G. Golgi (YFP-GT, false-colored red) and associated tracks (cyan). A line is drawn perpendicular to the direction of cell movement. Average number of MTs in 9 cells growing forwards or backwards are shown. H. Prominent MT array (thin arrow) in a polarized cell is rather associated with the Golgi (red) than with the centrosome (hollow arrow). Tubulin (green) and GM130 (red), immunostained. I, RPE1 cell fixed and stained 45″ after nocodazole washout. MTs (green) radiate from the Golgi mini-stacks (thin arrows) and the centrosome (hollow arrow). Tubulin, green. GM130, red. J. Live cell images of nocodazole washout. GFP-EB3-rich plus tips (green, asterisks) grow away from the mCherry-GT-marked Golgi stack (arrow, red). K. Number of non-centrosomal MTs nucleated at the Golgi (red) and elsewhere (blue) after nocodazole washout per cell, based on live recordings of mCherryEB3 and YFP-GT expressing cells (521 MTs in 8 cells).
Fig. 5. Golgi-originated MTs in steady state require CLASPs presence at the TGN.
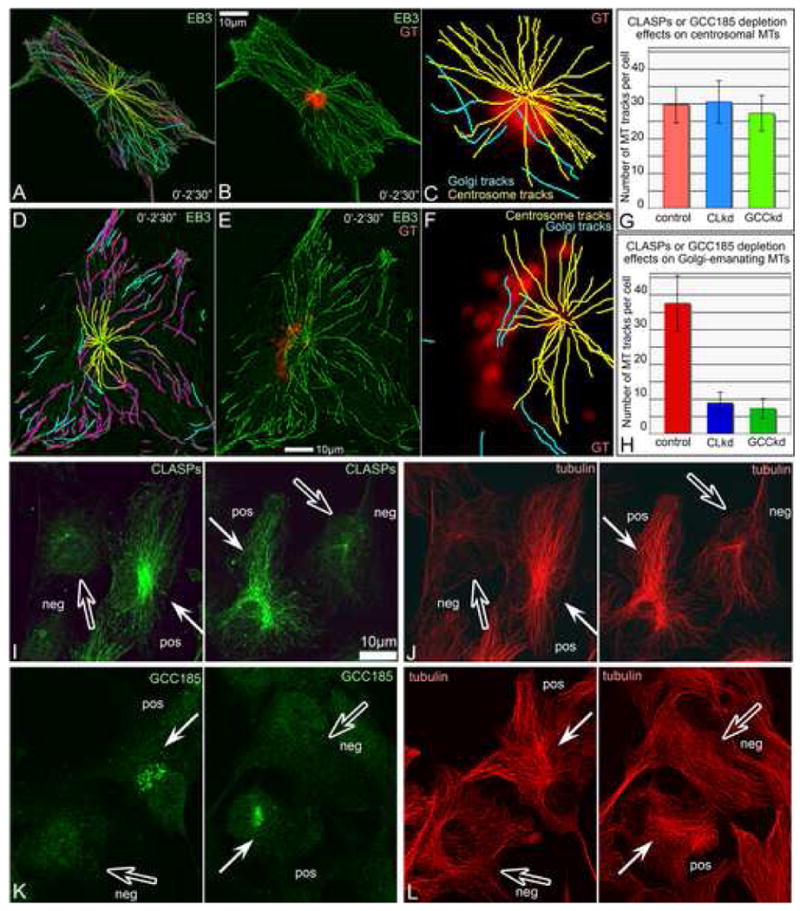
A-C. GFP-EB3 and mCherry-GT-expressing CLASP-depleted cell analyzed as in Fig. 1 AD. A, Overlaid GFP-EB3 showing MT tracks within 2.5′. Magenta, tracks started at the frame one. Yellow, centrosomal tracks. Cyan, non-centrosomal tracks. B, Overlaid GFP-EB3 (green) and mCherry-GT images within 2.5′. C, centrosomal (yellow) and few non-centrosomal (cyan) tracks in the cell center and their relation to the Golgi position. D-F. GFP-EB3 and mCherry-GT-expressing GCC185-depleted cell analyzed as in Fig. 1 A-D. D, Overlaid GFP-EB3 showing MT tracks within 2.5′. Magenta, tracks started at the frame one. Yellow, centrosomal tracks. Cyan, non-centrosomal tracks. E, Overlaid GFP-EB3 (green) and mCherry-GT images within 2.5′. C, centrosomal (yellow) and few non-centrosomal (cyan) tracks in the cell center and their relation to the Golgi position. G, No alteration of centrosomal MT tracks upon CLASP (CLkd, blue) or GCC185 (GCCkd, green) knockdown. H, Decrease in Golgi-associated MT track number upon CLASP (CLkd, blue) or GCC185 (GCCkd, green) knockdown. I-J, CLASPs (I) and MTs (J) in mixed culture of CLASP-positive (white arrows) and CLASP-depleted (hollow arrows) cells. K-L, GCC185 (K) and MTs (L) in mixed culture of control (white arrows) and GCC185-depleted (hollow arrows) cells. CLASP or GCC185-depleted cells lack Golgi-associated MT array. Immunostainings.
Non-centrosomal MTs are nucleated at the Golgi complex
The fact that most of the non-centrosomal MTs originated from a discrete site suggested that they are nucleated by a common mechanism. Overlaying the tracks of non-centrosomal MTs with the Golgi marker GT (membrane binding domain of galactosyl transferase) revealed that the non-centrosomal MTs grew from the Golgi (Fig.1 C,D). This was particularly obvious in cells where the centrosome and the Golgi were spatially separated. The number of MTs originating from the Golgi was slightly lower than those nucleated at the centrosome (22.5±3.2 versus 30.5±5.5 per cell in 2.5 minutes).
MT nucleation at the centrosome was resulted in symmetrical arrays, consistent with the previous finding (Salaycik et al., 2005). In contrast, MTs growing from the Golgi (Fig.1 E) were biased toward the cell edge (away from the nucleus). In motile cells (Fig.1 F,G), where the Golgi complex is oriented to the leading cell edge, Golgi-originated MTs were preferentially directed toward the cell front likely contributing to a directional MT array (Fig.1 H) that is considered important for motile cell polarization.
To further clarify the nature of MT nucleation sites in spread cells, we depolymerized MTs with nocodazole and then followed MT re-polymerization pattern. Tubulin stainings after short periods of re-growth revealed numerous non-centrosomal MTs along with the radial centrosomal array (Fig.1 I, Fig. S2). Golgi complex in the absence of MTs was dispersed to mini-stacks spread throughout the cytoplasm. The majority of non-centrosomal MTs was found attached to the mini-stacks consistent with previous reports (Chabin-Brion et al., 2001).
Time-lapse imaging of cells expressing 3GFP-EMTB or GFP-EB3 together with mCherry-GT revealed that non-centrosomal MTs were directly initiated at Golgi membranes (Fig.1 J, 4 G). Statistic analysis confirmed that the loci of first appearance of detectable EB3 dots co-localized with the Golgi stacks (Fig.1 K). MTs in these experiments were detected at the Golgi and the centrosome simultaneously. Recordings of EB3-marked MT plus ends revealed that the plus ends polymerized away from the Golgi stacks while the minus ends stayed attached to the Golgi (Fig.1 J). These results support the conclusion that the Golgi microenvironment is necessary for successful non-centrosomal MT formation.
Fig.4. siRNA CLASP knockdown suppresses MT formation at the Golgi upon nocodazole washout.
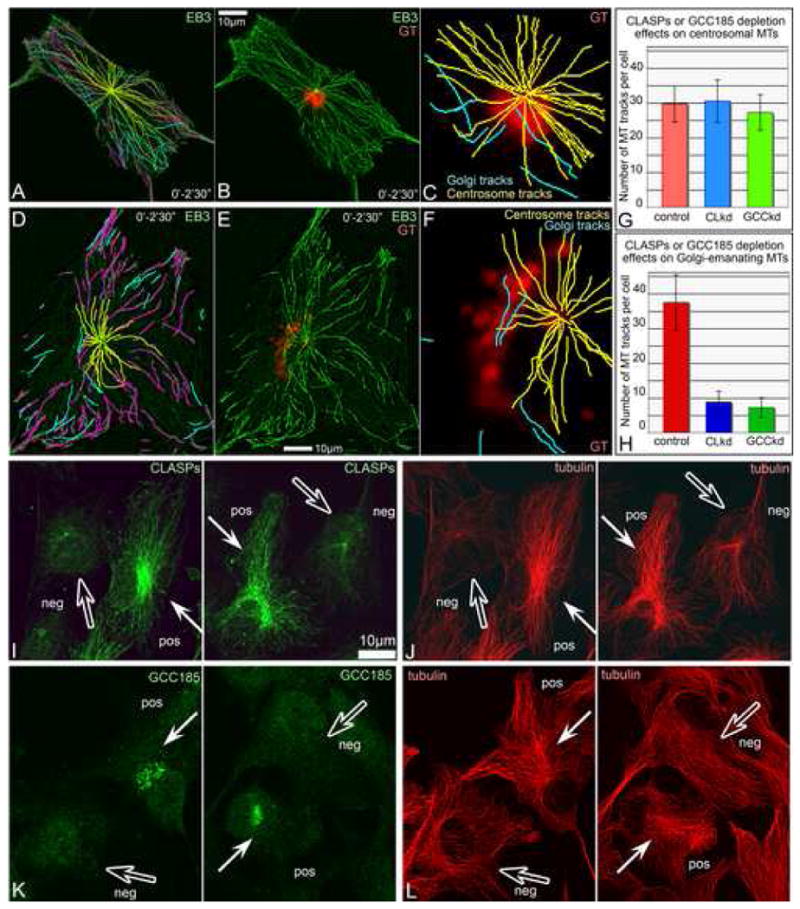
A, Western blotting illustrating ∼75% decrease of both CLASP1 and CLASP2 on the 3rd day after siRNA transfection as well as expression of non-silenceable GFP-CLASP2. Regions of interest are shown. B. Numbers of non-centrosomal MTs formed 45″ after nocodazole washout in CLASP-positive (red, 30 cells) and CLASP-depleted cells (blue, 30 cells), as well as upon rescue by non-silenceable GFP-CLASP2 (green, 16 cells). Based on tubulin immunostaining. C-F, Mixed culture of CLASP-positive (pos) and CLASP-depleted (neg) cells 45″ after nocodazole washout. Tubulin (red), GM130 (blue), CLASPs (green). Immunostaining. C, In CLASP positive cell, MTs radiate from Golgi stacks (arrows). D, In CLASP-depleted cell, Golgi stacks are not associated with rare MTs (hollow arrows). E, CLASPs and F, MTs and Golgi stacks at low magnification. Boxes enlarged in C and D. G, H. Video frames illustrating MT formation at the Golgi (arrows) in mRFP-EB3 (false-colored green) and YFP-GT (false-colored red) in nocodazole washout. Time after nocodazole removal in shown. Left, frames showing whole cell. Areas in boxes are enlarged to the right. G, Control cell. H, CLASP-depleted cell.
MT nucleation at the Golgi is centrosome-independent but requires γ-tubulin
The fact that MTs appear simultaneously in different parts of the cytoplasm during recovery from nocodazole treatment argues strongly that Golgi-associated MTs form at the Golgi independent of the centrosome. However, in steady state the Golgi and the centrosome are in close proximity and thus it is formally possible that MTs formed at the centrosome can be released and anchored the Golgi membranes. In order to unequivocally test whether the Golgi functions as an MTOC, we selectively destroyed GFP-centrin labeled centrosomes by the laser microsurgery technique (Fig. 2A-L). It has been demonstrated that this approach results in complete disappearance of MTs directly associated with the centrosome in a matter of minutes (Khodjakov et al., 2000). However, overall organization of cells lacking centrosomes resembles that of centrosomal cells (Khodjakov and Rieder, 2001). MT-tracking analyses conducted at least 30 min after centrosome ablation revealed that radial arrays of MTs normally growing from the centrosome were absent in cells lacking the centrosome (Fig. 2A-L). In contrast, MTs continued to grow persistently from the Golgi complex revealing that this organelle can function as MTOC even in the absence of centrosomes.
Fig. 2. MT nucleation at the Golgi does not require the centrosome but needs γ-tubulin.
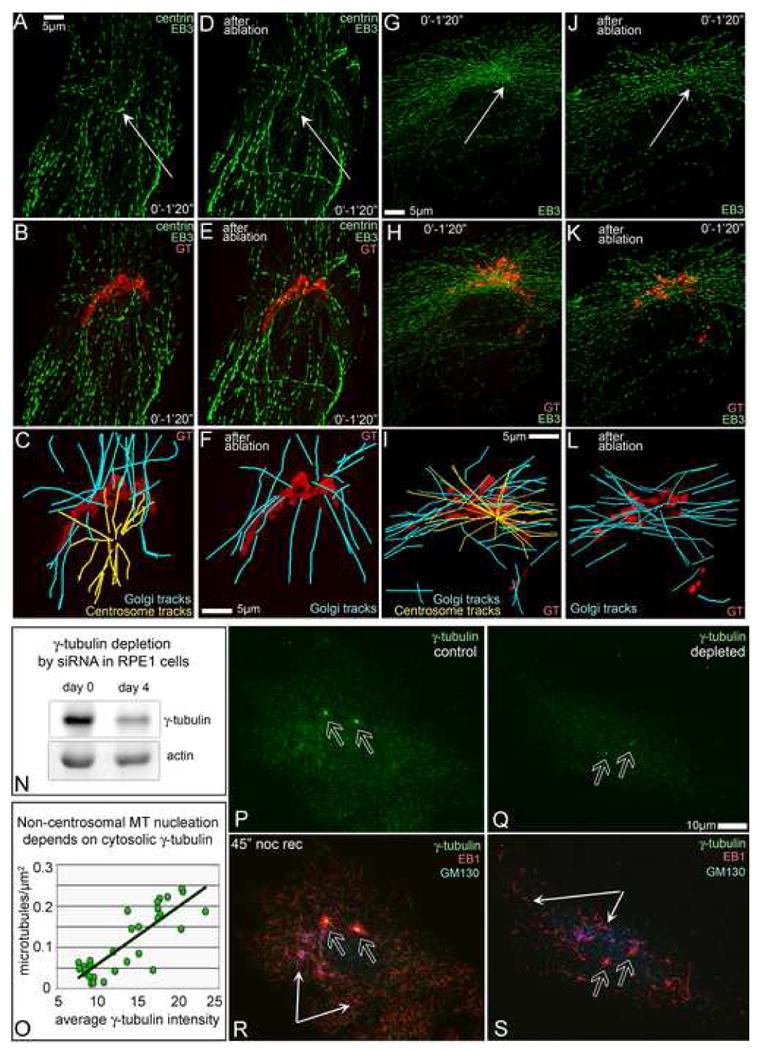
A-F, GFP-EB3 (green), GFP-centrin (green) and mCherry-GT (red) expressing cell. G-L, GFP-EB3 (green) and mCherry-GT (red) expressing cell. A-C, G-I – cells prior to ablation. D-F, J-L – cells 30 min after ablation. Nucleation sites were analyzed as in Fig. 1 A-D. A,B,C,D, EB3 tracks within 1 min 20 sec. Arrows, centrosomes. B,E,H,K, EB3 tracks (green) superimposed on the Golgi image (GT, red). C,F,I,L, centrosomal (yellow) and non-centrosomal (cyan) MT tracks in the cell center and their relation to the Golgi position (GT, red). Centrosomal but not Golgi-originated arrays disappear after centrosome ablation (F, L). N, 50% depletion of γ-tubulin in RPE1 cells by siRNA. Actin, loading control. O, Number of non-centrosomal MTs directly corresponds to intensity of cytosolic γ-tubulin staining. P-S, Fewer centrosomal (hollow arrows) and non-cnetrosomal (thin arrows) MTs is formed 45″ after nocodazole washout in γ̃–tubulin depleted (Q,S) than in control (P,R) cell. γ-tubulin, green (P-S); EB1, red (R,S); GM130, blue (R,S). Immunostaining.
MT nucleation at the centrosome requires molecular templates known as γ-tubulin-ring complexes (γ-TuRC). Previous studies suggested that γ-tubulin may be also associated with the Golgi membranes (Chabin-Brion et al., 2001; Rios et al., 2004). In order to directly test if MT nucleation at the Golgi requires γ-TuRC, we depleted γ-tubulin from RPE1 cells by siRNA (Fig. 2N). The number of newly nucleated in nocodazole washout experiments non-centrosomal MTs depended on the amount of γ-tubulin in the cytoplasm (Fig. 2O-S). Thus, similar to the centrosome, MT nucleation at the Golgi requires γ-TuRC.
MT regulators CLASPs localize at the TGN via association with TGN protein GCC185
To identify molecular players involved in MT formation at the Golgi we investigated plus-end binding proteins CLASPs which are potent regulators of MT dynamics. CLASPs were previously detected at the centrosome and the Golgi (Akhmanova et al., 2001). In mammalian cells, CLASPs exist as two closely related homologues, CLASP1 and CLASP2. In all experiments described below, we visualized both of them using either a mixture of CLASP1 and CLASP2 antibodies or a pan-CLASP antibody (VU-83, Fig.S3).
CLASPs are localized to MT tips, the centrosome and the Golgi. By co-staining for CLASPs and Golgi compartment markers we have found that CLASPs specifically co-localize with trans-Golgi network (TGN) marker TGN46 (Fig. 3AB) but not with cis and mid/trans Golgi markers (Fig.S4). This indicates that CLASPs specifically bind outer subcompartment of trans-Golgi, TGN, and are absent from other Golgi compartments.
Fig. 3. CLASPs specifically localize to TGN membranes via GCC185 binding.
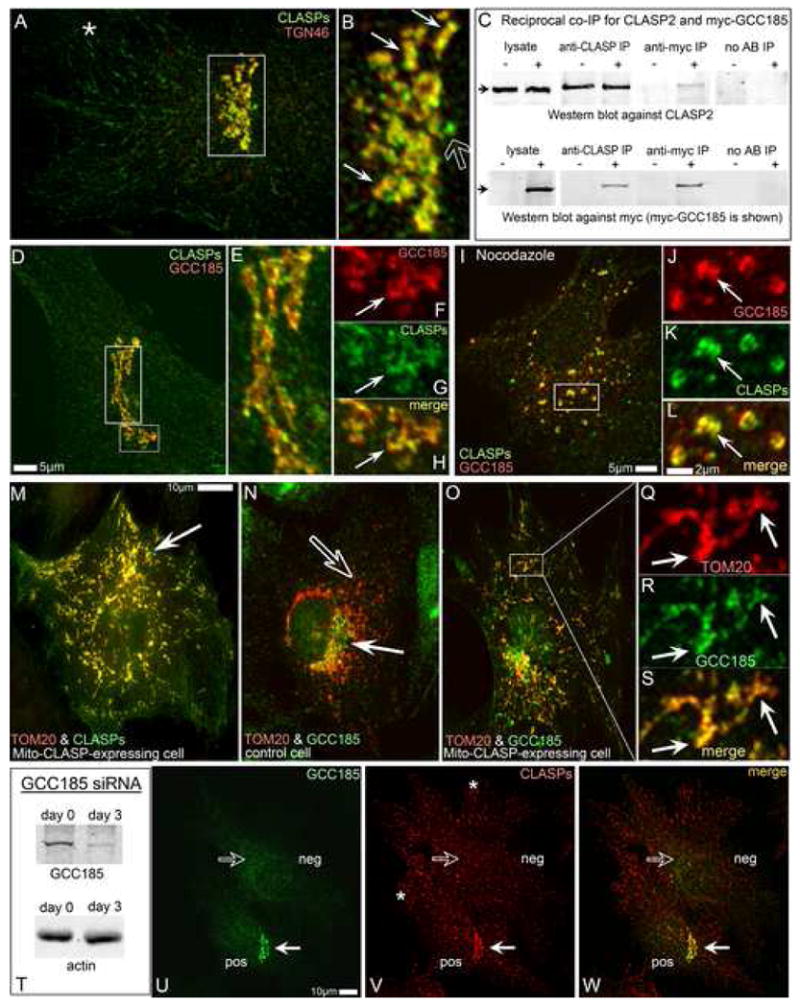
A,B, CLASPs (green) co-localize the TGN46 (red, thin arrows in B) along with the MT tips (star in A) and the centrosome (hollow arrow in B). Box in A is enlarged in B. C, Co-IP of CLASP2 and myc-GCC185 using either anti-CLASP2 or anti-myc antibodies. Upper panel, CLASP2. Lower panel, myc-GCC185. Non-transfected cells are marked as “–”, myc-GCC185-transfected cells as “+”. CLASP2 is co-precipitated from transfected cells by anti-myc antibody (Anti-myc IP). Myc-GCC185 is co-precipitated from transfected cells by anti-CLASP antibody (Anti-CLASP IP). Antibody-free beads used as control (No AB IP). D-H, CLASP co-localizes with ectopically expressed myc-tagged GCC185 at the TGN. Myc (red), CLASP (green). E, Enlarged tall box from D. F-H, Enlarged small box from D. F, myc-GCC185. G, CLASP. H, merge. Arrows, reference point. I-L, association of CLASPs and GCC185 is preserved in nocodazole. J-L, Enlarged small box from I. J, myc-GCC185. K, CLASPs. L, merge. Arrow, reference point. M, CLASP2-dTOM20 chimera (mito-CLASP) and mCherry-dTOM20 (red) co-expressing cell. CLASP staining (green) reveals both mito-CLASP (arrow, co-localized with mCherry-dTOM20 at mitochondria) and endogenous CLASPs. N. GCC185 at the Golgi (green, white arrow) does not co-localize with mCherry-dTOM20 at mitochondria (red, hollow arrow) in control cells. O-S. Mito-CLASP expressing cell. Endogenous GCC185 (green) is recruited to mitochondria (mCherry-dTOM20, red). Box is enarged in Q-S. Q, mCherry-dTOM20. R, GCC185. S, merge. Arrows, reference points. T, GCC185 is 90% depleted from RPE1 cells by siRNA. U-W, Mixed culture of GCC185-depleted (neg) and control (pos) cells. U, GCC185. V, CLASPs. W, merge of U and V. CLASP is localized to the Golgi in control cell (white arrow) but not in GCC185-depleted cell (hollow arrow). MT tip colalization of CLASP is intact in both cells (asterrisks). All images show immunostainings.
In detergent-extraction experiments, CLASPs behave as peripheral membrane proteins (Fig.S5). To search for Golgi components that recruit CLASPs to the Golgi membranes, we used HeLa cells stably expressing LAP-tagged GFP-CLASP1α. Co-immunoprecipitation with anti-GFP antibodies and subsequent mass spectrometry analysis revealed the GRIP-domain containing protein GCC185 (Luke et al, 2003) as a part of a complex together with known CLASPs-interacting partners (Suppl. Table 1). Further, endogenous CLASP2 specifically co-precipitated with ectopically expressed myc-tagged GCC185 from HEK293T cells by either anti-CLASP or anti-myc antibodies (Fig.3 C). This indicates that either CLASP isoform is able to bind GCC185.
GCC185 is a peripheral membrane protein specifically distributed to the TGN. In RPE1 cells, GCC185 shows significant co-localization with CLASPs at the Golgi in untreated (Fig.3 D-H) as well as in nocodazole-treated cells where Golgi ribbon is dispersed into individual mini-stacks (Fig.3 I-L).
To gain further support for the interaction of CLASPs and GCC185 we took advantage of CLASP mis-localization approach. We have produced an expression construct that contains dTOM20 (Drosophila outer mitochondrial protein) fused to CLASP2. Chimeric mito-CLASP localized specifically to the mitochondria when ectopically expressed in RPE1 cells (Fig.3 M). It was recognized by anti-CLASP antibodies in immunofluorescence (Fig.3 M) and Western blotting and sequestered known CLASP partners CLIP170, CLIP115 and EB1 to the mitochondria (not shown). Importantly, GCC185 was readily recruited to mis-targeted mitochondria-bound CLASP2 while normally it does not localize to mitochondria (Fig.3 N-S).
As a membrane-associated GRIP-domain golgin, GCC185 may serve a scaffolding protein for CLASPs at the TGN. To evaluate this possibility, we have depleted GCC185 from RPE1 cells by siRNA, resulting in up to 90% protein loss (Fig 3T). Loss of GCC185 led to exclusion of CLASPs from the TGN while MT plus end-associated CLASPs remained uninfluenced (Fig 3U-W). Expression of non-silenceable GCC185 variant rescued the phenotype (Fig. S6 A-B).
Together, these results demonstrate that GCC185 recruits CLASP to the TGN that is located at the convex side of Golgi stacks. Thus, CLASPs are plausible candidates for facilitating MT assembly at this location.
CLASPs depletion from RPE1 cells impairs MT formation at the Golgi and alters MT pattern
Since the major known function of CLASPs is regulation of MT dynamics, we tested if Golgi-associated CLASPs are involved in MT-organizing activity at the Golgi.
We depleted up to 75% of both CLASP1 and CLASP2 proteins from RPE1 cells as described (Mimori-Kiyosue et al., 2005) (Fig.4 A). Localization of CLASP-binding proteins, EB1 and CLIP170 to MT plus ends was not impaired by CLASP knockdown (Fig. S7).
Nocodazole washout experiments in mixed cultures of CLASP-positive and CLASP-depleted cells revealed a striking difference in the formation of non-centrosomal MTs (Fig.4 B-F). In CLASP-positive cells, numerous non-centrosomal MTs radiated from Golgi mini-stacks (Fig.4 C). In contrast, CLASP-depleted cells contained much lower numbers of non-centrosomal MTs not associated with Golgi mini-stacks (Fig.4 D). Ectopic expression of GFP-CLASP2 in CLASP-depleted cells rescued the phenotype to control levels of non-centrosomal nucleation (Fig.4, Fig. S6 G-J). Live cell imaging of MT recovery in CLASP depleted cells confirmed elimination of Golgi-initiated MT outgrowth (Fig.4 G,H).
In order to determine whether CLASP-dependent MT formation affects interphase MT networks, we used the plus-tip tracking approach illustrated in the Fig. 1 A-D. Upon CLASP depletion, centrosomal MT nucleation was not altered but the number of MTs growing from the Golgi decreased dramatically (Fig.5 A-C,G,H). Similarly, the number of Golgi-emanating MTs was significantly lowed in GCC185-depleted cells that lack CLASP at the Golgi (Fig.5 D-H). Further, we directly compared MT patterns in control, CLASP-depleted and GCC185-depleted cells immunostained for tubulin. Cells depleted of either CLASPs (Fig.5 I,J) or GCC185 (Fig.5 K,L) lacked the dense meshwork of MTs characteristic of the Golgi area, as well as the extensive MT array directed to the leading cell edge.
These data suggest a specific role for CLASPs at the TGN in the formation of Golgi-associated MTs. In contrast, centrosomal MTs were not affected by CLASP or GCC185 depletion.
Non-centrosomal MT seeds are coated with CLASPs and anchored to the Golgi membrane
Our results suggest that CLASPs are required for MT formation at the Golgi but not at the centrosome. Consistent with this idea, immunostaining of steady-state MTs revealed that the amount of CLASP associated with MT tips was significantly higher for MTs growing from the Golgi than for those growing from the centrosome (Fig.6 A,B). Moreover, live-cell imaging showed that GFP-CLASP2 highlighted MTs originated at CLASP-containing TGN membranes (Fig.6 C) but not at the centrosome. At the initial stages of MT outgrowth at the Golgi, GFP-CLASP2 was distributed along the entire MT length (up to 2μm, Fig 6D). At later stages, CLASP2 remained associated with the TGN and with the growing plus end (Fig.6D, 40 sec). Fix-cell (Fig. 6 E-G) or live-cell (Fig.6 I-K) nocodazole washout assays confirmed that non-centrosomal MTs were completely coated with CLASPs at early stages of re-growth. In contrast, only moderate amount of CLASPs could be detected at MTs growing from the centrosome. CLASP-binding MT tip proteins EB1 and CLIP-170 did not display differential association with Golgi-associated versus centrosomal MTs (Fig. 6 B, Fig. S7).
Fig. 6. CLASPs preferentially bind to MTs originated at the Golgi.
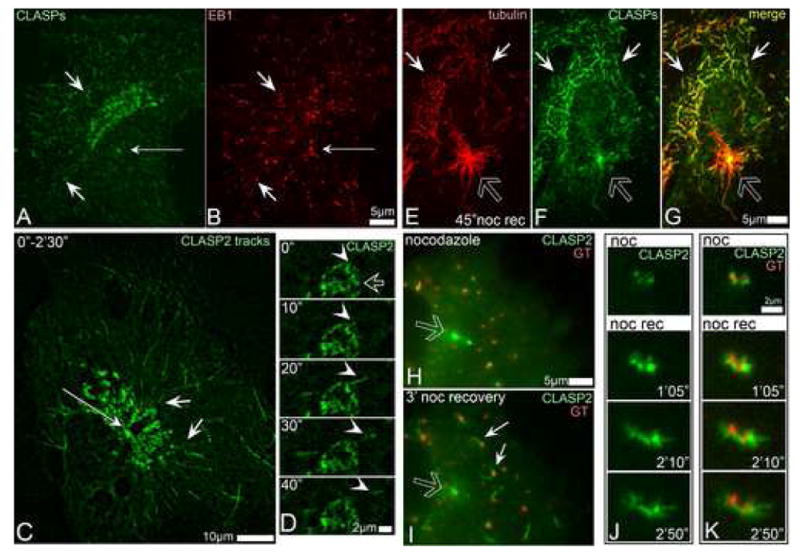
A, CLASPs (green) in RPE1 cells are associated with MT tips close to the Golgi (short arrows), but not around the centrosome (thin arrow). B, EB1 (red) at the MT tips in the cell shown in G. Immunostaining. C, Overlaid live recording of GFP-CLASP2-expressing RPE1 cell within 2.5 min. CLASP2-associated MT tracks (short arrows) radiate from the TGN but not form the centrosome (thin arrow). D, Live sequence illustrating formation of GFP-CLASP2-decorated MT (arrowhead) at CLASP-rich TGN (hollow arrow) in untreated cell. CLASP2 coats whole newly formed MT (10″-20″) but remains only at the tip at a later stage (30″-40″). E-G, A cell fixed 45″ after nocodazole washout. E, tubulin (red). F, CLASPs (green). G, Merge. CLASPs localize to non-centrosomal (white arrows) but not to the centrosomal (hollow arrow) MTs. H-I, GFP-CLASP2 (green) localizations before and after nocodazole washout. In nocodazole (left) CLASP2 associate with Golgi stacks (mCherry-GT, red) and with the centrosome (hollow arrow). 3′ after nocodazole removal (right) CLASP2 is detected at the Golgi-associated MTs (white arrows) but not around the centrosome (hollow arrow). J-K, Video frames illustrating formation of GFP-CLASP2-coated MTs at GFP-CLASP2-enriched Golgi stacks in nocodazole washout. J, GFP-CLASP2. K, GFP-CLASP2 (green) and mCherry-GT (red). Time after nocodazole removal is shown.
Initial coating of Golgi-originated MTs by CLASPs suggests that CLASPs may be directly redistributed to MT seeds from the Golgi membrane and stabilize them. If this hypothesis is true, CLASPs should be able to nucleate microtubules even when they are not associated with the Golgi. To test this prediction, we analyzed MT re-growth after nocodazole washout in cells where CLASPs were displaced from the TGN by GCC185 depletion (Fig. 3 U-W). The number of non-centrosomal MTs formed after nocodazole removal did not significantly differ from controls (Fig.7 A-D, I) indicating that CLASPs can support MT formation also when they are not bound to the Golgi membranes. Similarly, MT numbers after nocodazole washout were not altered by Brefeldin A (BFA) treatment (Fig.7 I) that causes dissociation of CLASPs (Fig.7 E) as well as other peripheral Golgi proteins (Presley et al., 1998) from the membrane. Persistence in non-centrosomal MT formation may be due to the fact that CLASP binding to MT tips as well as CLASP-coating of MT seeds upon nocodazole washout were not affected by either BFA treatment (Fig. 7 F-H) or GCC185 depletion (not shown). Additional depletion of CLASPs resulted in elimination of non-centrosomal nucleation under those conditions (Fig S.8)
Fig. 7. CLASPs dissolved in the cytosol support MT nucleation but not minus end anchoring.
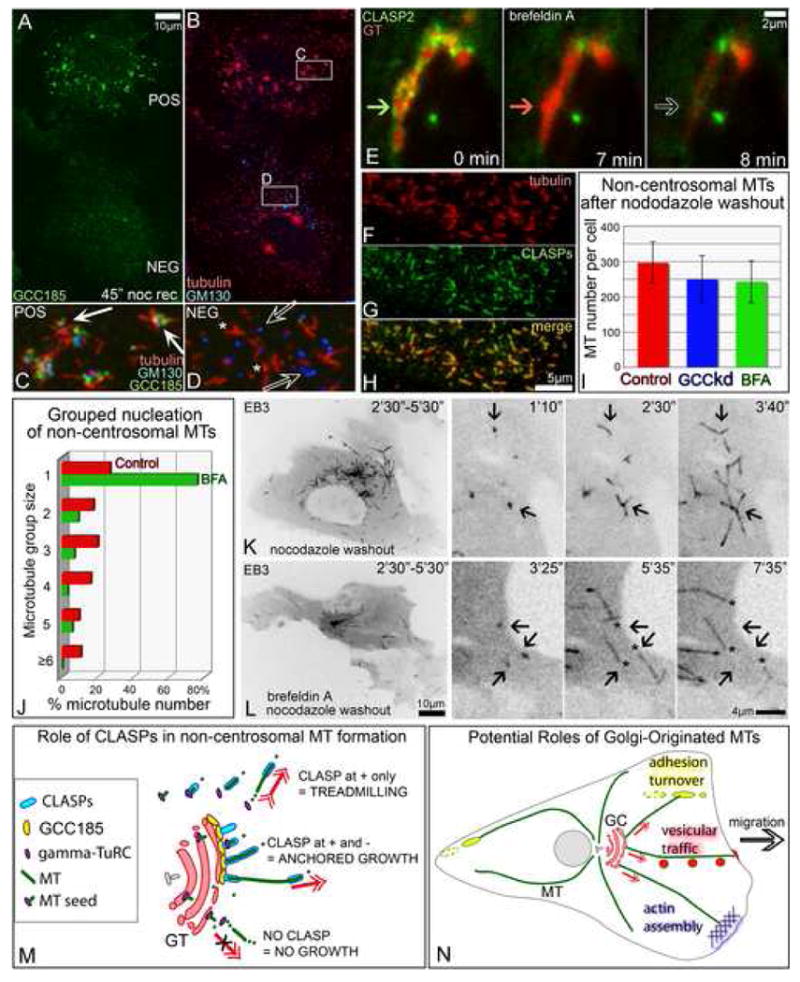
A-D. Mixed culture of GCC185-positive (pos) and GCC185-depleted (neg) cells 45″ after nocodazole washout. Boxes from B enlarged in C and D. GCC185 (green, A,C,D). Tubulin (red, B-D). GM130, (Blue B-D). C, In a control cell, MTs grow from the Golgi stacks. D, In GCC185-depleted cell, Golgi stacks (hollow arrows)are not associated with random single MTs (asterisks). E. In brefeldin A, GFP-CLASP2 (green) dissociates from the Golgi (7′) prior to Golgi-ER fusion (8′). Fusion is visualized by acute loss of mCherry-GT signal (red). Arrow hue illustrates presence of proteins. Time in brefeldin A is shown. F-H, In brefeldin A, CLASPs coat non-centrosomal MTs formed 45″ after nocodazole washout. F, tubulin. G, CLASPs. H, merge. Immunostaining. I, MTs formed in control (red), GCC185-depleted (GCCkd, blue) and Brefeldin A-treated (BFA, green) cells 45″ after nocodazole washout. For each set, 30 cells immunostained for tubulin were analyzed. J, Number of MTs nucleated singly and in groups after nocodazole washout with (BFA, green) and without (red) Brefeldin A. Live recordings of 7 mCherry-EB3-expressing cells for each set were analyzed. K-L. Non-centrosomal MTs in GFP-EB3-expressing RPE cells. Inverted images. Left, EB3 tracks within 3′. Right, enlarged inset video frames. Time after nocodazole removal is shown. K, In a control cell, MTs grow from distinct centers (arrows). L, In brefeldin A treated cell, minus ends of single MTs (asterisks) do not remain at the nucleation sites (arrows). M. Model for mechanisms of CLASP-dependent MT formation. γ-tubulin nucleates MT seeds in cytosol or at the Golgi. Cytoplasmic CLASPs associate with the MT plus end and support polymerization while minus end is unstable and depolymerizes resulting in MT treadmilling (top). CLASPs bound to TGN via GCC185 coat MT portion proximal to the minus end, anchor it to the membrane and prevent it's depolymerization, while CLASP-associated plus end steadily grows (middle). In the absence of CLASPs, MT seeds are unstable (bottom). GC, Golgi complex. N, Potential roles of Golgi-originated MTs in cell migration (model). Front-oriented MTs likely support post-Golgi vesicular trafficking (red), actin polymerization (blue) and focal adhesion turnover behind the leading edge (yellow).
We also investigated whether non-centrosomal MTs were formed at particular nucleation sites when CLASPs were dissociated from the Golgi membranes. Therefore, we registered all loci where non-centrosomal MTs were first detected in nocodazole washout experiments. Unlike controls (Fig.7 J red, K), non-centrosomal MTs formed in the presence of BFA did not arise from distinct centers. Instead, single MTs were randomly distributed in the cytoplasm (Fig.7 J green, L) similar to those in GCC185-depleted cells (Fig 7 A-D, Fig.S6 C-F).
Live-cell video analyses revealed that after non-centrosomal MTs were formed, instead of elongating with their minus ends anchored at the sites of nucleation, short MT fragments relocated in the cytoplasm most likely due to treadmilling (Fig 7 L).
These data indicate that while CLASPs can support MT nucleation both at the Golgi and in cytosol, their association with the Golgi membrane is needed for anchoring and stabilization of the minus ends of non-centrosomal MTs.
Discussion
Our results reveal that CLASPs are critical players in the non-centrosomal MT nucleation at the TGN membranes. As MTs can form in the cytoplasm of GCC185-depleted or BFA treated cells, we conclude that CLASPs are necessary for MT formation while the membrane binding is not. Probably, MTs preferably form at the TGN due to high concentration of CLASP molecules at this location. Since γ-tubulin is likely needed for nucleation per se, we suggest that CLASPs selectively stabilize pre-existing MT seeds. It has been suggested before (Lansbergen et al., 2006) that CLASPs accumulated in the proximity of plasma membrane can stabilize MTs without capping their plus ends. Such stabilization is likely due to the ability of CLASPs to bind the MT lattice (Wittmann and Waterman-Storer, 2005). We propose that a similar mechanism assures selective survival of MT seeds at the TGN membranes (see model, Fig. 7 I). Being MT tip-binding proteins, CLASPs have high affinity for newly polymerized MT regions including freshly nucleated MT seeds. Prominent localization to the Golgi indicates that CLASPs also have a high affinity for Golgi membranes. We suggest that CLASPs at the Golgi membranes are able to anchor very short MT fragments formed in the proximity, and by coating them prevent their disassembly and allow them to serve as seeds for polymerization. This model suggests that re-distribution of CLASP molecules from the Golgi to growing MTs is critical for the CLASP function. Consistent with our model, mito-CLASP chimera that is irreversibly bound to mitochondrial membranes exhibited no potential for MT nucleation (not shown). As MTs elongate, CLASPs remain associated with the structures for which they have high affinity: plus ends and the Golgi membrane.
Interestingly, centrosomal MTs that do not depend on CLASPs also are not coated by CLASPs as they form. Such a deficiency may be due to selective GSK3beta activation at the centrosome (Higginbotham et al., 2006) that can inhibit CLASP binding to the MT lattice (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005). The difference in CLASP binding to the centrosomal and Golgi-associated MTs prompts us to speculate that these two groups of MTs may have different dynamic properties from the start. It is noteworthy in this regard that Golgi-associated MTs are acetylated more rapidly than centrosomal ones (Chabin-Brion et al., 2001) pointing to their elevated stability.
Understanding that CLASPs stabilize pre-existing MT seeds leaves the question where these seeds are formed unanswered. It is possible that high concentration of the seeds at the Golgi is established by recruitment of γ-TuRC to cis-Golgi proteins GMAP210 (Rios et al., 2004) or AKAP350 (Shanks et al., 2002). However, successful growth of these seeds is likely achieved only when they become associated with CLASP-rich TGN membranes. Thus, seeds may be formed at the cis-Golgi membranes, dissociate and bind TGN or, consistent with cisternal maturation model (Mironov et al., 2005), stay associated with maturing cisternae until they become enriched in TGN proteins including GCC185 and CLASPs which then promote MT growth.
Although not needed for the initiation of MT growth, TGN membrane appeared to serve as an anchoring site for non-centrosomal MT minus ends, thereby preventing them from depolymerization. The anchoring may involve yet unidentified molecular players. Alternatively, CLASP bound to the membrane may accomplish the anchoring either by lateral MT binding or due to yet unknown ability of CLASPs to cap MT minus ends. Actually, CLASPs are strongly associated with both minus end anchoring sites: the Golgi and the centrosome. In our experiments, anchoring and release of centrosomal MTs were not altered by CLASP depletion (not shown). We have to admit though that siRNA knockdowns were not efficient enough to fully deplete CLASPs from the centrosome. Thus, the centrosome appears to have very strong affinity for CLASPs suggesting that they may bear an important function at this location, possibly in minus end anchoring.
A striking feature distinguishing the Golgi-associated arrays from centrosomal ones is their asymmetry. We suggest that this asymmetry is due to the selective MT formation only at CLASP-containing TGN membranes and not other Golgi compartments. Being asymmetric, Golgi-originated MT arrays could contribute to cell polarization.
The centrosome localization in front of the nucleus typical for many motile cells is thought to be an important stage in establishing MT asymmetry and, in turn, cell polarization (Raftopoulou and Hall, 2004). The centrosome itself nucleates symmetrically (Bergen et al., 1980; Salaycik et al., 2005). However, dynein-driven transport organizes an asymmetric MTOC, the Golgi, in close proximity to the centrosome. We suggest that centrosome localization influences MT asymmetry indirectly via the positioning of the Golgi complex. This model can explain such recent data as direct correlation between MTOC (centrosome) positioning and formation of asymmetric MT arrays in neurons (de Anda et al., 2005).
Notably, CLASP2 was recently found necessary for persistent directional cell migration (Drabek et al., 2006). As Golgi-associated MT arrays in motile cells are oriented towards the cell front (see model Fig.7 J), they can be a source for pioneer MTs that are implicated in stabilization of protrusion and are important for the directionality of migration (Suter et al., 2004; Waterman-Storer et al., 1999). Additionally, Golgi-originated MTs in motile cell could be facilitate turnover of adhesions behind the leading edge (Rid et al., 2005).
Importantly, Golgi-associated MTs could be involved in vesicular transport to, within or out of Golgi stacks. As Golgi-originated MTs in motile cells directly connect the TGN with the cell front (Fig.7 J) they may support vesicular transport towards the leading edge that is important to maintain high motility rates (Preisinger et al., 2004; Prigozhina and Waterman-Storer, 2004). In polarized epithelial cells, Golgi-dependent nucleation could originate vertical MT arrays that are associated with the Golgi at their minus ends and are responsible for baso-apical vesicular transport (Bacallao et al., 1989; Musch, 2004).
Overall, CLASP-dependent mechanism of non-centrosomal MT formation at the Golgi could be engaged in various cell types where MTs not attached to centrosomes bear diverse functions (Bartolini and Gundersen, 2006).
Experimental Procedures
Cells
Immortalized human pigment epithelial cells hTert-RPE1 (Clontech) were maintained in DMEM/F12, HeLa, MDCK, RIE, BT549 and HEK293T cells in DMEM, all with 10%FBS at 37°C in 5%CO2. IMCE cells were maintained in RPMI with 5% FBS and 1%insulin at 33°C. Cells were plated on fibronectin-coated glass coverslips 24 hours prior to experiments. Mixed culture of control and knockdown cells were obtained by mixing trypsinized cell suspensions in proportion 1:1 before plating.
Treatments
For MT depolymerization, nocodazole (2.5μg/ml) was added to culture media for 2 h. For nocodazole washout fixations, cells were rinsed 5 times with ice-cold medium and then moved to a dish with warm (37°C) medium (time 0 for MT re-growth). For live imaging of MT re-growth, cells were washed with cold medium directly at the microscope stage after initial recording of cells in nocodazole. Temperature was thus slowly raised by the heated stage. Slightly variable time of MT re-growth initiation in video sequences is due to this experimental setup.
BFA (Alexis Biochemicals) was used at 5μg/ml in culture media. For nocodazole washout experiments of BFA treated cells, nocodazole-containing and final wash nocodazole free media were supplemented with BFA.
siRNA and expression constructs
Mixed siRNA oligos against CLASP1 and CLASP2 (Mimori-Kiyosue et al., 2005) were transfected using Oligofectamine (Invitrogen) according to manufacturer's protocol. Experiments were conducted 72 h after transfection as at this time minimal protein levels were detected. siRNA oligos against γ-tubulin were transfected two times within 72 hours as described in (Luders et al., 2006). Anti-GCC185 siRNA oligo (corresponding to nucleotides767-784 in KIAA0336 cDNA) were designed by Ambion. GCC185 from Myc-GCC185 was cloned into XhoI/BamHI restriction sites of Venus-C1 vector (J.Roland, modified from Clontech). Rescue construct was made by site-directed mutagenesis to substitute 6 nucleotides in area recognized by siRNA (siRNA targeted sequence: “G GCT AAT TCT CAG CAT TAC C”; mutant sequence: “G GCC AAC TCC CAA CAC TAT C”). Non-targeting siRNA (Dharmacon) was used for controls. mCherry plasmid used for red fluorescent imaging was kindly provided by Dr. R.Tsien (San Diego). YFP-GT (YFP-Golgi marker, Clontech) and mCherry-GT (modified from Clontech) were used for Golgi visualization. EGFP-EB3 and mCherry-EB3 (kind gift of Dr. J.V.Small, Vienna), mCherry-tubulin (kind gift of Dr. R.Tsien, San Diego), 3GFP-EMTB (kind gift of Dr. J.C.Bulinski, New York) were used for MT plus tip and MT visualization. GFP-centrin (kind gift of Dr. M.Bornens, Paris) was used for centrosome visualization in laser ablation experiments. GFP-CLASP1α, GFP-CLASP2α and non-silenceable rescue construct GFP-CLASP2 were described in (Mimori-Kiyosue et al., 2005). Myc-tagged GCC185 construct was described in (Luke et al., 2003). mCherry-dTOM20 (Drosophila outer mitochondrial protein) was kindly provided by Dr. E.Lee (Nashville). Mito-CLASP (dTOM20 fused with N-terminal end of CLASP2α in a pCS2 vector) was used for CLASP mis-localization to mitochondria. Transient transfections were performed with Fugene6 (Roche) according to manufacturer's protocol.
Antibodies
For rabbit polyclonal antibodies against CLASP2 VU-83 see Supplemental Data and Fig.S3. For guinea pig polyclonal antibodies against GCC185 VU-140 see Supplemental Data and Fig.S9. Rabbit polyclonal antibodies against CLASP2 are described in (Akhmanova et al., 2001). Rabbit polyclonal antibodies against CLIP170 are described in (Coquelle et al., 2002). Rabbit polyclonal antibodies against CLASP1 were kindly provided by Dr. F.Severin (Dresden). For Golgi compartment identification, mouse monoclonal antibody against GM130 (Transduction Laboratories) and rabbit polyclonal antibody against GCC185 (Luke et al., 2003) were used. Myc-tag was identified by monoclonal anti-myc antibody clone 9E10 (Upstate). Mouse monoclonal antibodies against EB1 and p150 Glued were from Transduction Laboratories, rat monoclonal antibody against Tyr-tubulin from AbCam, anti-α-tubulin and anti-γ–tubulin mouse monoclonal and rabbit polyclonal antibody were from Sigma.
Immunofluorescence, confocal and live cell imaging
Wide field fluorescence imaging was performed using Nikon 80I with CFI APO 60X OIL lens, NA 1.4 and CoolSnap ES CCD camera (Photometrics).
Confocal stacks were taken by Yokogawa QLC-100/CSU-10 spinning disk head (Visitec assembled by Vashaw) attached to Nikon TE2000E microscope using CFI PLAN APO VC 100X OIL lens, NA 1.4. with or without 1.5× intermediate magnification and back-illuminated EM-CCD camera Cascade 512B (Photometrics) driven by IPLab software (Scanalytics). 75 mW 488/568 Krypton-Argon laser (Melles Griot) with AOTF was used for 2-color excitation. Custom double dichroic mirror and filters (Chroma) in a filter wheel (Ludl) were used in the emission light path. 0.2 μm Z-steps were driven by Nikon built-in Z-motor.
Live cells plated on MatTech glass bottom dishes were maintained at 37°C by heated stage (Warner Instruments) at Nikon TE2000E inverted microscope equipped with PerfectFocus automated focusing device. Single plane confocal video sequences were taken as described for confocal stacks. Similar setup with pinkel triple filter set (Semrock) was used for nearly-simultaneous 2-color wide field imaging.
For image processing details see Supplemental Data.
Centrosome ablation
Centrosome ablation was carried out as described elsewhere (Khodjakov et al., 2000; La Terra et al., 2005). Briefly, 532-nm 8-ns laser pulses generated by a Q-switched Nd:YAG laser (Diva II; Thales Lasers) were focused on the specimen with a 100X 1.4 NA PlanApo lens.
Centrosomes were identified via centrin-GFP expression and ablated with approximately 10-20 laser pulses (0.5-1.0 s). Fluorescence images were recorded with a back-illuminated EM-CCD camera (Cascade512B, Photometrics) in confocal mode (spinning disk confocal; Perkin-Elmer, Boston, MA). All light sources were shuttered by either fast mechanical shutters (VincentAssociates) or AOTF (Solamere Technology Group), so that cells were exposed to light only during laser operations and/or image acquisition.
Co-immunoprecipitation, LAP tagging, and mass spectrometry
HEK293T cells transiently transfected with myc-GCC185 were used for immuno-precipitation with the affinity purified anti-CLASP antibodies VU-83 or anti-myc monoclonal antibody 9E10. The precipitated protein complexes were analyzed by Western blotting. HeLa cells stably expressing the GFP-CLASP1α (LAP) construct in the pIC113 vector were used for GFP-CLASP1 (LAP) fusion immunoprecipitation by anti-GFP antibodies as described (Cheeseman and Desai, 2005). TEV protease-cleaved CLASP protein was desalted and analyzed using a modified 12-step separation described previously (Washburn et al., 2001). MS/MS spectra were searched with the SEQUEST™ algorithm (Eng et al., 1994). Several step of filtering were applied. Usual contaminants obtained by this technique were excluded from possible CLASP1 interactors. See Supplemental Data for protocol details.
Supplementary Material
Acknowledgments
This study was funded by Development funds of Department of Cell and Developmental Biology, Vanderbilt University Medical Center and by Austrian Science Fund grant #P16066-B to I.K., grant POCI/SAU-MMO/58353/2004 from Fundação para a Ciência e a Tecnologia of Portugal (POCI2010 and FEDER) and grant L-V-675/2005 from Luso-American Foundation for Development Work to H.M, NIH grant P41RR11823-10 to J.Y., NIH grant GM59363 to A.Khod. We thank Mr. Bejan Abtahi and Ms. Jana Tashkova for technical help and Dr. Ethan Lee for helpful discussion.
Abbreviations
- trans-Golgi network
TGN
- microtubule
MT
- γ-tubulin ring complex
γ-TuRC
- Brefeldin A
BFA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EH, Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Bergen LG, Kuriyama R, Borisy GG. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E, Zaal KJ, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005. 2005:l1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, et al. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Pollarolo G, Da Silva JS, Camoletto PG, Feiguin F, Dotti CG. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack A, Yates J. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Tanaka T, Brinkman BC, Gleeson JG. GSK3beta and PKCzeta function in centrosome localization and process stabilization during Slit-mediated neuronal repolarization. Mol Cell Neurosci. 2006;32:118–132. doi: 10.1016/j.mcn.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova YA, Vorobjev IA, Borisy GG. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci. 2002;115:3527–3539. doi: 10.1242/jcs.115.17.3527. [DOI] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Wu CC, McIntosh S, McIntosh JR, Howell KE. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell. 2002;13:2810–2825. doi: 10.1091/mbc.01-12-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, Akhmanova A. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Luders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Luke MR, Kjer-Nielsen L, Brown DL, Stow JL, Gleeson PA. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J Biol Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- Malikov V, Kashina A, Rodionov V. Cytoplasmic dynein nucleates microtubules to organize them into radial arrays in vivo. Mol Biol Cell. 2004;15:2742–2749. doi: 10.1091/mbc.E03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV, Polishchuk RS, Trucco A. Intra-Golgi transport: a way to a new paradigm? Biochim Biophys Acta. 2005;1744:340–350. doi: 10.1016/j.bbamcr.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Musch A. Microtubule organization and function in epithelial cells. Traffic. 2004;5:1–9. doi: 10.1111/j.1600-0854.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Smith C, Hirschberg K, Miller C, Cole NB, Zaal KJ, Lippincott-Schwartz J. Golgi membrane dynamics. Mol Biol Cell. 1998;9:1617–1626. doi: 10.1091/mbc.9.7.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina NL, Waterman-Storer CM. Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr Biol. 2004;14:88–98. doi: 10.1016/j.cub.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Reilein A, Nelson WJ. APC is a component of an organizing template for cortical microtubule networks. Nat Cell Biol. 2005;7:463–473. doi: 10.1038/ncb1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rid R, Schiefermeier N, Grigoriev I, Small JV, Kaverina I. The last but not the least: the origin and significance of trailing adhesions in fibroblastic cells. Cell Motil Cytoskeleton. 2005;61:161–171. doi: 10.1002/cm.20076. [DOI] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation, growth and dynamics in wound-edge cells. J Cell Sci. 2005;118:4113–4122. doi: 10.1242/jcs.02531. [DOI] [PubMed] [Google Scholar]
- Shanks RA, Steadman BT, Schmidt PH, Goldenring JR. AKAP350 at the Golgi apparatus. I. Identification of a distinct Golgi apparatus targeting motif in AKAP350. J Biol Chem. 2002;277:40967–40972. doi: 10.1074/jbc.M203307200. [DOI] [PubMed] [Google Scholar]
- Suter DM, Schaefer AW, Forscher P. Microtubule dynamics are necessary for SRC family kinase-dependent growth cone steering. Curr Biol. 2004;14:1194–1199. doi: 10.1016/j.cub.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon AM, Wadsworth P. Non-centrosomal microtubule formation and measurement of minus end microtubule dynamics in A498 cells. J Cell Sci. 1997;110(Pt 19):2391–2401. doi: 10.1242/jcs.110.19.2391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


