Abstract
Cells from patients with Cockayne syndrome (CS) are hypersensitive to DNA-damaging agents and are unable to restore damage-inhibited RNA synthesis. On the basis of repair kinetics of different types of lesions in transcriptionally active genes, we hypothesized previously that impaired transcription in CS cells is a consequence of defective transcription initiation after DNA damage induction. Here, we investigated the effect of UV irradiation on transcription by using an in vitro transcription system that allowed uncoupling of initiation from elongation events. Nuclear extracts prepared from UV-irradiated or mock-treated normal human and CS cells were assayed for transcription activity on an undamaged β-globin template. Transcription activity in nuclear extracts closely mimicked kinetics of transcription in intact cells: extracts from normal cells prepared 1 h after UV exposure showed a strongly reduced activity, whereas transcription activity was fully restored in extracts prepared 6 h after treatment. Extracts from CS cells exhibited reduced transcription activity at any time after UV exposure. Reduced transcription activity in extracts coincided with a strong reduction of RNA polymerase II (RNAPII) containing hypophosphorylated C-terminal domain, the form of RNAPII known to be recruited to the initiation complex. These results suggest that inhibition of transcription after UV irradiation is at least partially caused by repression of transcription initiation and not solely by blocked elongation at sites of lesions. Generation of hypophosphorylated RNAPII after DNA damage appears to play a crucial role in restoration of transcription. CS proteins may be required for this process in a yet unknown way.
Nucleotide excision repair (NER) is the versatile process by which a wide variety of helix-distorting DNA lesions are removed from the genome, including UV-induced photolesions, i.e., cyclobutane pyrimidine dimers and 6–4 photoproducts. Two NER subpathways have been identified, namely global genome repair, responsible for the removal of lesions throughout the genome, and transcription-coupled repair. The latter pathway is responsible for the accelerated removal of lesions from the template strand of genes undergoing transcription (1, 2).
Three hereditary human disorders are associated with photosensitivity: xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy. Cells derived from patients with these disorders exhibit enhanced UV sensitivity because of deficiencies in the repair of UV-induced damage. Cell fusion studies have revealed seven XP complementation groups (XPA to XPG) and two CS complementation groups (CSA and CSB); in addition, some patients belonging to XP groups B, D, and G exhibit combined XP/CS (3, 4).
Clinically, CS is a complex disorder characterized by growth retardation, skeletal and neurological abnormalities, photosensitivity, and premature aging (5). At the cellular level, the hallmark of CS is the inability of cells to restore inhibited RNA synthesis after exposure to DNA-damaging agents (6). Also, cells from patients expressing both XP and CS symptoms exhibit a more severe impairment of transcription recovery after UV exposure than cells from patients with XP symptoms only (7). The inability of CS cells to resume inhibited transcription has been attributed to impaired transcription-coupled repair (TCR) of DNA lesions in transcriptionally active genes, including UV-induced photolesions and oxidative damage (8–10). In the case of UV damage, impaired TCR abolishes the accelerated repair of cyclobutane pyrimidine dimers in the transcribed strand of transcriptionally active genes, resulting in a repair rate indistinguishable from that of the nontranscribed strand (9). These observations have led to the concept that efficient TCR of transcription-blocking DNA lesions in cells exposed to DNA damage accounts for the restoration of RNA synthesis. In this process, CS gene products may fulfill a role as transcription-repair coupling factors by stimulating repair of DNA lesions at the site of stalled transcription (11). Indeed, the CSB protein has been shown to be associated with RNA polymerase II (RNAPII) in cellular extracts of human cells (12). However, this model has been challenged by results of studies with the carcinogen Nacetoxy-2-acetylaminofluorene (NA-AAF) (13). Although CS cells were clearly sensitive to the effects of NA-AAF, there was no obvious difference between CS and normal cells in the repair kinetics for NA-AAF-induced lesions in transcriptionally active genes. Yet, after NA-AAF treatment, the CS cells exhibited prolonged repression of RNA synthesis in comparison to normal cells. Also the ability and inability of UV-irradiated XPD and XPD/CS cells, respectively, to restore transcription while exhibiting equal levels of repair of transcriptionally active DNA point to other mechanisms underlying transcription inhibition (7). Together, these observations have led to the hypothesis that the sensitivity of CS cells to DNA-damaging agents is caused not by any repair defect but by failure to recover RNA synthesis after DNA damage, pointing to a defect in transcription initiation.
Experimental evidence for inhibition of transcription by transacting factors is scarce. In support of such a mechanism are results from microbeam UV-irradiation studies: induction of local damage in the nucleus led to transcription inhibition throughout the nucleus (14). The mechanism of inactivation of transcription might be related to the recruitment by NER of factors essential for the transcription process itself (3). A key factor in the transcription response to DNA damage is the basal transcription factor IIH (TFIIH), required for initiation and early elongation of transcription (15). The TFIIH complex contains the XPB and XPD proteins and plays an essential role in repair throughout the genome. The requirement of TFIIH for repair of DNA lesions might provide a mechanistic link to transcription inhibition at the initiation level. Evidence has been obtained in yeast that different forms of TFIIH for transcription (holo-TFIIH) and DNA repair (repairosome) may exist (16). Experiments aimed at assessing directly the inhibitory effect of NER on transcription have generated conflicting results. Human cell extracts supporting both RNAPII transcription, and NER showed no competition between both processes (17), whereas in yeast extracts, NER can inhibit transcription. This inhibition could be complemented with holo TFIIH, suggesting that inhibition of transcription was mediated by sequestration of TFIIH for NER in the presence of DNA damage (18).
A commonly used technique to monitor RNA synthesis in intact mammalian cells is to measure incorporation of radiolabeled uridine; however, this system cannot differentiate between inhibition of transcription initiation and inhibition of elongation at sites of DNA lesions. To distinguish between initiation and elongation inhibition, we have assessed the effects of UV irradiation on transcription by using an in vitro assay. Transcription measured in an in vitro transcription system must include initiation events, and therefore this approach was taken to examine transcription initiation in nuclear extracts prepared from UV-irradiated human cells. The transcription activity of extracts was analyzed by using an undamaged template, which rules out that transcription elongation is inhibited by DNA lesions. A cosmid containing the entire chick β-globin locus was used as a template for transcription (19), enabling the investigation of transcription initiation in a system containing complete promoter/enhancer sequences for a single gene. Nuclear extracts from normal human, CS (complementation groups A and B), and XPA cell lines were analyzed for their transcription activities. The results show that in vitro transcription activity closely mimics the kinetics of inhibition and recovery of RNA synthesis in UV-irradiated intact cells, despite the fact that the transcription template is deprived of DNA photolesions. This provides evidence that transcription inhibition after UV occurs not only by elongation blockage but also by impaired initiation. Western blot analysis of the extracts revealed a tight correlation between transcription competence of nuclear extracts and the presence of the hypophosphorylated RNAPII large subunit known to be involved in transcription initiation. Our results demonstrate that successful restoration of transcription after DNA damage depends on the regeneration of the hypophosphorylated RNAPII large subunit.
Materials and Methods
Cell Culture and Irradiation Conditions.
Simian virus 40-immortalized normal human (MRC5), Cockayne syndrome group A (CS3BE) and B (CS1AN), as well as XP group A (XP2OS) fibroblasts were grown in Ham's F-10 medium (Life Technologies, Paisley, Scotland) supplemented with 15% FCS and antibiotics in 5% CO2 at 37°C.
Cells were seeded in dishes and grown to confluence. Before irradiation, cells were washed with PBS and irradiated with UV light [Philips (Eindhoven, The Netherlands) TUV lamp, 254 nm) at a dose rate of 0.2 J/m2/s. After irradiation, cells were grown in fresh medium for various time periods.
RNA Synthesis in Intact Cells.
14C-thymidine-prelabeled confluent cells were washed with PBS, irradiated with UV, and incubated in fresh medium for various periods of time. Subsequently, the medium was replaced by medium containing 3H-uridine (20 Ci/ml, 43 Ci/mmol), and cells were incubated for 30 min before being processed for liquid scintillation counting (7). The 3H/14C ratio was taken as a measure of RNA synthesis.
Nuclear Extract Preparation.
Nuclear extracts were prepared as described by Barton et al. (19). All procedures were performed on ice or at 4°C. Briefly, 108 fibroblasts were harvested and washed in PBS and subsequently in hypotonic buffer (10 mM Hepes⋅KOH, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.5 mM DTT/0.2 mM PMSF). Cells were incubated in 3 (cell) volumes of hypotonic buffer for 10 min, dounced until 90% were broken, and the nuclei were pelleted.
The nuclei were resuspended in 0.5 (nuclear) volumes of nuclear buffer (25% glycerol/20 mM Hepes⋅KOH, pH 7.9/1.5 mM MgCl2/0.5 mM DTT/1 mM PMSF) containing 0.02 M KCl. Subsequently, 0.5 (nuclear) volume of nuclear buffer containing 1.2 M KCl was added dropwise, and nuclei were extracted for 30 min. Nuclei were then removed by centrifugation (10,000 × g, 30 min), and the supernatant was dialyzed for 2 h against 300 ml of dialysis buffer (20% glycerol/20 mM Hepes⋅KOH, pH 7.9/100 mM KCl/0.2 M EDTA/1 mM DTT/1 mM PMSF). Subsequently, the extract was centrifuged [8,000 rpm, 5 min (Biofuge A, Hereaus Sapatech)] and stored at −120°C.
In Vitro RNA Synthesis.
In vitro transcription assays were performed as described by Barton et al. (19) by using 0.5 μg of chick β-globin cosmid (Cos5βA1) encompassing the entire β-type globin locus and nuclear extract (125–150 μg protein). Briefly, extracts were combined with the cosmid in reaction buffer (20 mM Hepes⋅KOH, pH 7.9/10 mM MgCl2/40 mM KCl/25 mg/ml BSA/4 mM DTT/25 mM NTP/10 mM phosphocreatine/0.5 units creatine phosphokinase) and incubated for 1 h at 30°C. The RNA was purified by phenol extraction and annealed to a 32P-end-labeled primer with sequence homology to the β-globin gene 120 bp downstream of the transcription start site (5′-CCT CAG CAG TCC AGT GCA CCA-3′). The annealing reaction was performed for 60 min at 60°C in annealing buffer (10 mM Tris⋅HCl. pH 8.0/1 mM EDTA, pH 8.0/1.25 M KCl). Subsequently, 22.5 μl reverse transcriptase mix (20 mM Tris⋅HCl, pH 8.7/10 mM MgCl2/100 μg/ml actinomycin D/0.33 mM dNTP/5 mM DTT) and 100 units of Superscript II Reverse Transcriptase (Promega) was added to each reaction, and samples were incubated for 60 min at 42°C. Reverse transcriptase products were analyzed by electrophoresis on a polyacrylamide gel (8%, 7 M urea) and quantified by using an Instant Imager Electronic Autoradiography System (Packard). In vitro complementation assays were performed by addition of purified CSB protein to the nuclear extract or by mixing nuclear extracts before the transcription reaction.
All extracts were analyzed for their transcription activities, at least in duplo, exhibiting less than 15% variation. At least two series of extracts were prepared from all cell lines used in this study to assess the effect of UV on transcription activities.
Western Blot Analysis.
Nuclear extracts (1 mg/ml) were heated in Laemmli buffer (10% glycerol/5% β-mercaptoethanol/3% SDS/100 mM Tris⋅HCl, pH 6.8/bromophenol blue), subjected to SDS/PAGE, and transferred to a nitrocellulose membrane. Aspecific sites were blocked in sterilized skimmed milk in the presence of 0.05% Tween-20, and the membrane was incubated with an appropriate antibody. We used mono- and polyclonal antibodies raised against RNAPII LS (H14) (20), p62 (a gift of J.-M. Egly, Institut de Génétique et de Biologie Moléculaire et Céllulaire, Université Louis Pasteur, Illkirch Cedex, France), CSB (12), and TATA box-binding protein (TBP) (Promega), followed by incubation with horseradish peroxidase-conjugated antibody. Finally, the membrane was incubated for 10 min in Supersignal West Dura solution (Pierce) and exposed to x-ray film.
Results
Transcription Inhibition in Normal Human and CS Fibroblasts.
Uridine incorporated during pulse labeling represents predominantly RNAPII-driven transcription. Fig. 1 shows that 5 J/m2 UV irradiation caused a profound inhibition of RNA synthesis in confluent human fibroblasts, and that CS cells were unable to restore UV-inhibited RNA synthesis. Expression of ERCC6 c-DNA in CS1AN cells, however, complemented the transcription defect.
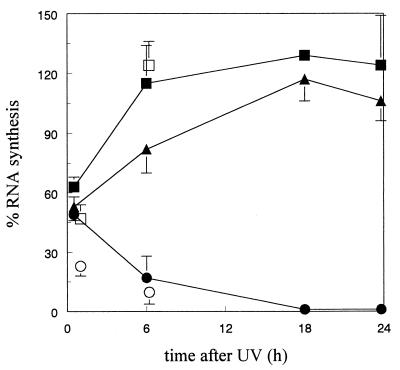
Figure 1.
Recovery of UV-inhibited RNA synthesis. RNA synthesis was measured as the relative incorporation of 3H-uridine in 5 J/m2-irradiated cells compared with unirradiated cells (100%) (average values of three experiments). MRC5 (■), CS1AN (●), E61AN transfectant (CS1AN cells carrying the ERCC6 gene) (▴). The relative transcription capacity of nuclear extracts from UV-irradiated cells is indicated as well (average values of two experiments): MRC5 (□), CS1AN (○). Bars represent standard errors.
Transcription Activities in Nuclear Extracts.
To investigate whether the reduction in RNA synthesis observed in intact cells after UV exposure is caused by inhibition of transcription initiation or elongation, we assessed the transcription capacity of nuclear extracts prepared from human cell lines at various times after UV exposure. The rationale is that an in vitro transcription system must include initiation and elongation events to generate transcription products, but that the use of an undamaged template eliminates inhibition of transcription elongation by DNA damage. Monolayer cultures were grown under conditions identical to those used for RNA synthesis measurements in intact cells. Transcription activity of extracts was assayed on a cosmid containing the entire β-globin gene cluster. For quantification, RNA was converted into a 120-bp DNA fragment by reverse transcriptase reaction.
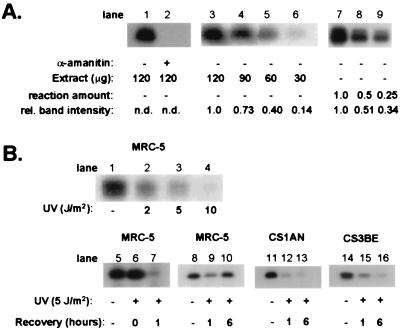
Previous experiments (19) investigating transcription of the β-globin gene involved the use of extracts from exponentially growing HeLa cells expressing high levels of transcription activity. Fig. 2A, lane 1, shows that extracts of HeLa cells were proficient in transcription of the β-globin gene. RNAPII inhibitor α-amanitin completely blocked RNA synthesis, indicating that the assay fully depends on RNAPII-driven transcription (Fig. 2A, lane 2). Next, we analyzed the transcription activity of extracts of human (MRC5) and CS1AN cells grown to confluence. On the basis of equal protein concentration, nuclear extracts from the monolayer fibroblasts exhibited a 2-fold lower transcription activity compared with the extract from HeLa cells. No differences in transcription activity were found between extracts of MRC5 and CS1AN cells.
Figure 2.
(A) Characterization and quantification of in vitro transcription of the β-globin gene by nuclear extracts from unirradiated cells. (i) Extracts from HeLa cells were assayed for transcription activity in the presence or absence of α-amanitin (lanes 1, 2). (ii) Lanes 3–6 show that the amount of 120-bp DNA fragment varies proportionally to the amount nuclear extract: different quantities (120, 90, 60, and 30 μg) of nuclear extracts from confluent MRC5 cells revealed relative transcription activities of 1.0, 0.73, 0.40, and 0.14 respectively. (iii) Lanes 7–9 show that the amount of 120-bp DNA fragment varies proportionally to the amount of RNA synthesized by MRC5 extracts: different quantities of transcription reactions (1.0, 0.5, and 0.25 vol) revealed relative amounts of transcription product of 1.0, 0.51, and 0.34. (B) In vitro transcription of the β-globin gene by nuclear extracts from UV-irradiated cells. (i) Transcription activity by nuclear extracts from MRC5 cells mock treated or UV irradiated with 2, 5, and 10 J/m2 (lanes 1–4). (ii) Transcription response at different time periods after UV [5 J/m2; nuclear extracts were prepared from MRC5 immediately and 1 h after UV treatment (lanes 5–7) and from MRC5, CS1AN, or CS3BE cells 1 or 6 h after UV treatment (MRC5, lanes 8–10; CS1AN, lanes 11–13; CS3BE, lanes 14–16)].
Quantitative comparison of the transcription capacity of nuclear extracts requires that the amount of 120-bp DNA fragment produced by reverse transcriptase is proportional to the amount of β-globin transcript made in the transcription reaction. We checked the proportionality of the assay by varying the amount of extract added to the reaction as well as by taking different-sized aliquots of a single transcription reaction mix (Fig. 2A, lanes 3–9). The results obtained in two independent experiments show clearly that the amount of 120-bp DNA fragment varied proportionally to the amount of reaction mix or nuclear extract added to the reaction.
Transcription Activity in Extracts from UV-Irradiated Cells.
Transcription in human fibroblasts is inhibited by UV in a dose-dependent manner, reaching complete inhibition at approximately 10 J/m2. To assess the effect of UV on transcription activity of nuclear extracts, MRC5 cells were grown to confluence and irradiated with different doses of UV. One hour after UV exposure, nuclear extracts were isolated and assayed for transcription activity. A dose-dependent decrease in transcription activity was observed (Fig. 2B, lanes 1–4) despite the fact that the template for RNA synthesis is undamaged. Transcription activity in extracts prepared from MRC5 cells immediately after UV-exposure (5 J/m2) was similar to that of unirradiated cells, indicating that repression of transcription does not occur instantly (Fig. 2B, lanes 5–6).
In intact normal human cells exposed to 5 J/m2, transcription was restored 6 h after irradiation (Fig. 1). To determine whether in vitro transcription activity increased with time, nuclear extracts were prepared from unirradiated MRC5 and CS1AN cells as well as from cells exposed to 5 J/m2 and kept for 1 or 6 h under normal growth conditions before extract preparation. Extracts were tested for their transcription activity with equal quantities of protein in each reaction (Fig. 2B, lanes 8–16). The transcription activity of extracts prepared 1 h after irradiation was significantly reduced for MRC5 cells as well as for CS1AN and CS3BE cells. We found consistently that transcription activity in extracts of CS cells was more reduced (80%) than in extracts from MRC5 cells (50%). Extracts prepared from MRC5 cells at 6 h after UV exposure showed a significantly higher level of transcription activity compared with the extract prepared 1 h after treatment, amounting to approximately 90% of the level found in extracts from mock-treated cells. In contrast, reduced levels of transcription activity were seen in extracts from UV-irradiated CS1AN and CS3BE cells prepared 6 h after UV treatment. Fig. 1 shows that relative transcription activity in extracts of UV-irradiated normal human and CS cells closely resembled the transcription response measured in intact cells by radiolabeled uridine incorporation.
Additional experiments indicated that extracts from MRC5, CS1AN, and XP2OS (XPA) cells prepared 24 h after 2 J/m2 UV light showed full recovery of in vitro transcription of the β-globin gene (data not shown). Nuclear extracts of XP2OS (Fig. 4) and CS1AN (data not shown) cells exposed to 10 J/m2 of UV light exhibited a more than 90% inhibition of transcription up to 24 h after UV exposure.
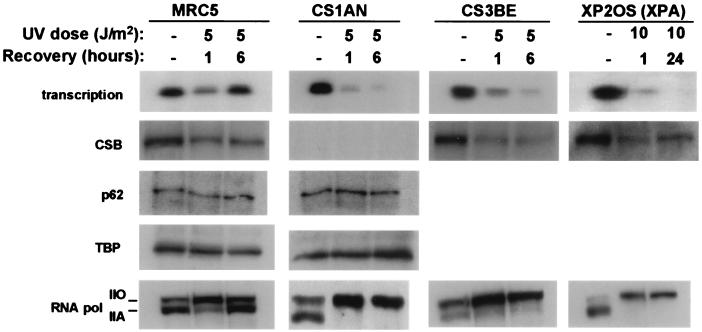
Figure 4.
Western blot analysis of nuclear extracts from mock-treated and UV-irradiated MRC5, CS1AN, CS3BE, and XP2OS cells. All cells were irradiated with 5 J/m2 except XP2OS (10 J/m2). Transcription activity is indicated as well.
Complementation Studies.
UV-irradiated CS1AN cells transfected with the ERCC6 gene exhibited a transcription response similar to that seen in normal human cells, indicating that the CSB gene product corrects the deficiency in RNA synthesis recovery in CS1AN cells (Fig. 1). We attempted to complement the reduced transcription activity in CS1AN nuclear extracts prepared 6 h after UV exposure by addition of purified hemagglutinin-tagged CSB protein. Fig. 3A shows that different amounts of purified CSB protein (3 and 30 ng) were unable to complement the transcription activity of the extracts. This lack of complementation was seen under all conditions tested, including stimulation of ATPase activity of the CSB protein by preincubation of the protein and extract at 30°C in the presence of ATP before the transcription reaction. The purified CSB protein exhibited ATPase activity in vitro and was able to restore RNA synthesis after microinjection into UV-irradiated CS cells (21), indicating that the protein is functionally active.
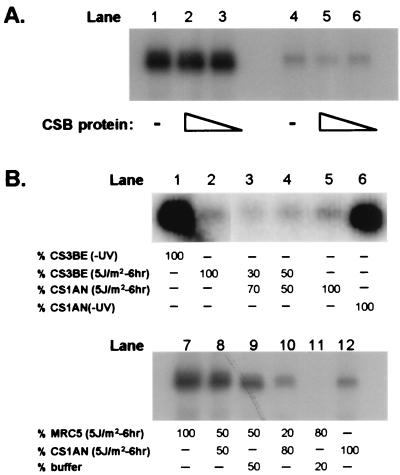
Figure 3.
(A) Complementation studies with purified CSB protein. Before the transcription reaction, no (lanes 1, 4) or increasing amounts of purified CSB protein [3 ng (lanes 3, 6) and 30 ng (lanes 2, 5)] were added to a nuclear extract prepared from mock-treated or UV-irradiated (5 J/m2) CSIAN cells 6 h after UV exposure. (B) Transcription activity in mixed extracts. (i) Extracts from UV-irradiated CS1AN and CS3BE cells prepared 6 h after UV exposure were mixed and assayed for transcription activity (lanes 1 and 6, nonirradiated CS1AN and CS3BE, respectively; lanes 2 and 5, UV irradiated (5 J/m2 CS1AN and CS3BE, respectively); and lanes 3 and 4, mix of equal amounts of UV-irradiated CS1AN and CS3BE). (ii) Different quantities of MRC5 and CS1AN extracts [prepared 6 h after UV treatment (5 J/m2)] were mixed and assayed for transcription activity (lanes 7 and 12, equal amounts of UV-irradiated MRC5 and CS1AN, respectively; lanes 8–11, mix of various amounts of MRC5, CS1AN, and buffer).
The lack of restoration of in vitro transcription by the purified CSB protein prompted us to assess whether extracts of CS1AN and CS3BE could complement each other. Mixtures of nuclear extracts (based on equal protein concentration) from UV-irradiated CS1AN and CS3BE did not exhibit significant restoration of reduced transcription activity (Fig. 3B, lanes 1–6). A possible explanation for this failure would be the presence of factors in CS extracts that are inhibitory to transcription initiation. To rule out this possibility, we mixed extracts from CS1AN and MRC5 cells prepared 6 h after UV irradiation in different ratios and assayed for their transcription activity. We did not observe reduction of transcription activity in MRC5 (Fig. 3B, lanes 7–12), indicating that CS1AN extracts are devoid of freely exchangeable inhibitors of transcription activity. We also noted that the transcription activity in these mixtures of MRC5 and CS1AN extracts was additive rather than synergistic: addition of MRC5 extracts to CS1AN extracts in different ratios did not complement the impaired transcription activity seen in CS1AN nuclear extracts.
Immunoblot Analysis of Nuclear Extracts.
We examined the extracts by immunoblot analysis for the presence of proteins that have key functions in the initiation of transcription and/or might be involved in the recovery of inhibited transcription on infliction of DNA damage.
Immunoblot analysis revealed that the amount of CSB protein was significantly reduced (approximately 50%) in nuclear extracts prepared from UV-irradiated MRC5 cells. An even more pronounced reduction was obtained for CS3BE (approximately 70%) and XP2OS cells (approximately 60%) (Fig. 4). Preliminary results indicate that the recovery of transcription activity in nuclear extracts of MRC5 cells (6 h after UV) may not coincide with the restoration of the CSB protein level. Thus, alterations in transcription activity in MRC5 nuclear extracts do not correlate with levels of CSB protein. Next, we investigated transcription factors that have been implicated in inhibition of transcription, e.g., TBP (TBP/TFIID) and basal transcription factor TFIIH. TBP has been shown to bind to UV-induced photolesions, and it has been proposed that UV-induced photolesions compete with promotor sequences for TBP, thereby inhibiting transcription (22). Using a monoclonal antibody against TBP, we could find no depletion of TBP in the extracts from UV-irradiated cells. Also, no reduction of TFIIH in nuclear extracts was observed by using antibodies against the core proteins p62 (Fig. 4) and p89 (XPB) (data not shown).
Studies by Laybourn and Dahmus (23) have indicated that the large subunit of RNAPII containing a hypophosphorylated C-terminal domain (CTD) is recruited to the transcription initiation complex, whereas the elongating RNAPII is associated with a phosphorylated CTD. When extracts were examined for the presence of RNAPII with an antibody recognizing its CTD, we observed that RNAPII in the extracts of mock-treated MRC5 and CS1AN cells resided mainly in the hypophosphorylated state (RNAPIIa) (Fig. 4). In contrast, extracts from cells exposed to UV and prepared 1 h after exposure revealed a profound increase of the hyperphosphorylated form of RNAPII (RNAPIIo) and a decrease of RNAPIIa. This effect was most pronounced in the CS1AN extracts, exhibiting the lowest in vitro transcription activity. A distinct difference in the state of phosphorylation of RNAPII between MRC5 and CS1AN was seen in extracts prepared 6 h after exposure to UV: MRC5 extracts exhibited a reappearance of RNAPIIa, whereas in extracts prepared from UV-irradiated CS1AN cells, RNAPIIa was virtually absent. A similar observation was made for extracts from CS3BE fibroblasts belonging to complementation group A. The lack of recovery of RNAPIIa was not specific for CS cells, because extracts of UV-irradiated XPA cells also revealed a depletion of RNAPIIa.
In more than 40 independently prepared extracts investigated so far, low levels or absence of transcription activity correlated with reduced levels of RNAPIIa. To assess whether the depletion of RNAPIIa was because of its trapping in the chromatin fraction after UV, we examined the chromatin fraction for the presence of RNAPII. Only RNAPIIo was recovered from the chromatin fraction, suggesting that RNAPIIa was absent in the isolated nuclei.
Discussion
Previously, we have shown that CS cells are defective in the restoration of inhibited RNA synthesis after NA-AAF treatment, whereas normal human cells do recover inhibited RNA synthesis (13). Despite this inhibition, CS cells exhibited no detectable defect in the repair of NA-AAF-induced DNA lesions in transcriptionally active genes. We argued that the enhanced sensitivity of CS cells to DNA damage is not caused by impaired transcription coupled repair of transcription elongation blocking DNA lesions, but rather is a consequence of a defect in transcription initiation in the presence of DNA damage. We designed an experimental system that enabled us to dissect effects of DNA damage on transcription initiation events by using nuclear extracts from UV-irradiated and nonirradiated cells and a nondamaged DNA template.
The results obtained in the current study unambiguously demonstrate that reduction of transcription activity can be achieved in the absence of DNA photolesions in the transcription template. Efficient in vitro transcription requires initiation, and thus the poor transcription activity in nuclear extracts from UV-irradiated cells is most likely the result of impaired initiation. The analysis of the phosphorylation state of the largest subunit of RNAPII strongly supports the notion that the initiation function is impaired. RNAPII harbors a unique structure at its C terminus known as the CTD. At least two forms of RNAPII have been detected in cells: one form contains a hypophosphorylated CTD (IIa), the second contains a hyperphosphorylated CTD (IIo) (23, 24). The phosphorylation state correlates to distinct functions of the two forms as the hypophosphorylated form of RNAPII is recruited to the initiation complex, whereas the elongating RNA polymerase complex contains the hyperphosphorylated CTD. Western blot analysis of the nuclear extracts revealed that in vitro transcription activity correlated quantitatively with the presence of the hypophosphorylated form of RNAPII. It is tempting to speculate that the depletion of RNAPIIa might be caused by conversion of IIa into IIo after UV-irradiation. First, RNAPIIa was absent in the pelleted chromatin fraction from UV-irradiated cells, excluding the possibility that the RNAPIIa is trapped by photolesions in the DNA. Also, Western blot analysis of whole-cell lysates from primary human fibroblasts by Ratner et al. (25) revealed the absence of RNAPIIa after UV irradiation. Second, the depletion of RNAPIIa in nuclear extracts goes along with an increase in RNAPIIo. Thus, these data are in agreement with the notion that repression of transcription initiation in nuclear extracts from UV-irradiated cells is mediated by depletion of RNAPIIa. In nonirradiated cells, the vast majority of RNAPIIo is associated with the chromatin pellet, and this fraction is generally assumed to include elongating RNAPII firmly attached to DNA (26). On UV irradiation, a distinct increase of RNAPIIo in the soluble fraction is observed, both in normal cells and in CS and XPA cells, whereas the amount of IIo in the pellet remained unchanged. This suggests the following scenario: on UV irradiation, the RNAPIIa is consumed rapidly by transcription initiation, phosphorylated, and subsequently during elongation, blocked at sites of DNA lesions. We presume that these stalled RNAPIIo complexes are no targets for recycling into RNAPIIa. In UV-irradiated CS cells, the consumption of RNAPIIa continues with time, ultimately resulting in its complete depletion. The elongating RNAPIIo complexes stalled at the sites of UV photolesions either might fall off the template or are unstable, resulting in their detachment during extract preparation. We note here that other mechanisms might lead to depletion of RNAPIIa as well. For example, induction of kinase activity by UV might lead to alteration of the amount of RNAPIIa without its recruitment to the transcription initiation complex.
Currently, we can only speculate on the cellular mechanisms that cause RNAPIIa to regenerate in UV-exposed cells. One possibility is that the conversion of RNAPIIo to RNAPIIa might be part of a RNAPII recycling process involving CTD phosphatase activity such as first identified in HeLa cells by Chambers and Dahmus (26). In the yeast Saccharomyces cerevisiae, mutations in the FCP1 gene encoding a CTD phosphatase essential for dephosphorylation of RNAPII revealed that transcription in vivo requires CTD phosphatase (27). It is conceivable that this process is somehow disturbed in CS cells exposed to DNA damage. On the other hand, the reappearance of RNAPIIa after UV exposure may depend on new protein synthesis. Ratner et al. (25) showed that recovery of RNAPIIa to the baseline level in UV-irradiated human fibroblasts was inhibited in the presence of the protein synthesis inhibitor cycloheximide. However, inhibition of protein synthesis might affect the completion of transcripts possibly necessary for dephosphorylation of RNAPII.
Our complementation studies rule out that inhibitors account for reduced transcription activity in extracts from UV-irradiated CS cells. That we were unable to complement the transcription defect in extracts from UV-irradiated CS cells by addition of purified and functionally active CS protein or by mixing of extracts from CSA and B cells indicates that the CS proteins are not sufficient to activate transcription incompetent extracts and that additional factors are missing or that formation of functional complexes does not occur in nuclear extracts. The basal transcription factor TFIIH contains CTD kinase activity, and this activity is efficient after RNAPII has associated with promotor sequences (28). Our study demonstrates that transcription was not impaired because of gross depletion of TFIIH from the extracts. However, we cannot distinguish between functionally different forms of TFIIH and/or possible UV-induced posttranslational modifications. It is possible that after UV irradiation, nuclear extracts lack the transcriptionally active form of TFIIH, as different forms of TFIIH for transcription and DNA repair may exist (16).
Another transcription factor that has been implicated in DNA damage response is TBP (TBP/TFIID). In a cell-free system, TBP has been shown to bind to UV-induced photolesions, and it has been proposed that UV-induced photolesions compete with promotor sequences for the binding of TBP, thereby inhibiting transcription (22). However, the amount of TBP in extracts from UV-irradiated cells is indistinguishable from that in transcription competent extracts, and thus the inhibition of transcription activity in the extracts cannot be because of depletion of TBP.
The reduction of transcription activity in extracts of UV-irradiated cells and the concomitant depletion of RNAPIIa were found for both CS and XPA cells, indicating that the phenomenon is not specific for CS. The XPA cells are severely UV sensitive and exhibit poor recovery of transcription even at relatively low UV doses (6). Our results suggest that also in XPA cells, the lack of transcription recovery is related to impaired initiation events, but that the mechanisms leading to repressed transcription initiation might be different for CS and XPA. Notably, in XPA cells deficient in repair, persistent lesions in transcribed genes might prevent completion of transcripts, thereby preventing recycling of RNAPIIo.
Acknowledgments
This study was supported by the European Union (Contracts FI3P-CT92–0007 and QLG1-CT1999–00181) and by the Research Council for Earth and Life Sciences (ALW-NWO).
Abbreviations
- RNAPII
RNA polymerase II
- RNAPIIa
hypophosphorylated RNAPII
- RNAPIIo
hyperphosphorylated RNAPII
- CTD
C-terminal domain of RNAPII
- XP
xeroderma pigmentosum
- CS
Cockayne syndrome
- TBP
TATA box-binding protein
- NER
nucleotide excision repair
- NA-AAF
N-acetoxy-2-acetylaminofluorene
- TFIIH
transcription factor IIH
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180169797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180169797
References
- 1.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 2.Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 3.Bootsma D, Hoeijmakers J H J. Nature (London) 1993;363:114–115. doi: 10.1038/363114a0. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann A R. BioEssays. 1998;20:146–155. doi: 10.1002/(SICI)1521-1878(199802)20:2<146::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 5.Nance M A, Berry S A. Am J Med Gen. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 6.Mayne L V, Lehmann A R. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 7.van Hoffen A, Kalle W H J, de Jong-Versteeg A, Lehmann A R, van Zeeland A A, Mullenders L H F. Nucleic Acids Res. 1999;27:2898–2904. doi: 10.1093/nar/27.14.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venema J, Mullenders L H F, Natarajan A T, van Zeeland A A, Mayne L V. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Hoffen A, Natarajan A T, Mayne L V, van Zeeland A A, Venema J, Mullenders L H F. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leadon S A, Cooper P K. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanawalt P C. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 12.Van Gool A J, Citterio E, Rademakers S, Van Os R, Vermeulen W, Constantinou A, Egly J-M, Bootsma D, Hoeijmakers J H J. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Oosterwijk M F, Versteeg A, Filon R, van Zeeland A A, Mullenders L H F. Mol Cell Biol. 1996;16:4436–4444. doi: 10.1128/mcb.16.8.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda S, Naruse S, Yatani R. Nature (London) 1967;213:696–697. doi: 10.1038/213696a0. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H J, Chambon P, Egly J-M. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 16.Svejstrup J Q, Wang Z, Feaver W J, Wu X, Bushnell D A, Donahue T, Friedberg E C, Kornberg R D. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 17.Sato M S, Hanawalt P C. Nucleic Acids Res. 1996;24:3576–3582. doi: 10.1093/nar/24.18.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You Z, Feaver W J, Friedberg E C. Mol Cell Biol. 1998;18:2668–2676. doi: 10.1128/mcb.18.5.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barton M C, Madani N, Emerson B M. Genes Dev. 1993;7:1796–1809. doi: 10.1101/gad.7.9.1796. [DOI] [PubMed] [Google Scholar]
- 20.Bregman D B, Du L, van der Zee S, Warren S L. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citterio E, Rademakers S, van der Horst G T J, van Gool A J, Hoeijmakers J H J, Vermeulen W. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 22.Vichi P, Coin F, Renaud J-P, Vermeulen W, Hoeijmakers J H J, Moras D, Egly J-M. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laybourn P J, Dahmus M E. J Biol Chem. 1989;264:6693–6698. [PubMed] [Google Scholar]
- 24.Dahmus M E. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 25.Ratner J N, Balasubramania B, Coreden J, Warren S L, Bregman D B. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 26.Chambers R S, Dahmus M E. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- 27.Kobor M S, Archambault J, Lester W, Holstege F C, Gileadi O, Jansma D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Nature (London) 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]