Abstract
The RNase III endonuclease Dicer plays a key role in generation of microRNAs (miRs). We hypothesized that Dicer regulates cancer cell susceptibility to immune surveillance through miR processing. Indeed, Dicer disruption up-regulated intercellular cell adhesion molecule (ICAM)-1 and enhanced the susceptibility of tumor cells to antigen-specific lysis by cytotoxic T-lymphocytes (CTLs), while expression of other immunoregulatory proteins examined was not affected. Blockade of ICAM-1 inhibited the specific lysis of CTLs against Dicer-disrupted cells, indicating a pivotal role of ICAM-1 in the interaction between tumor cells and CTL. Both miR-222 and -339 are down-regulated in Dicer-disrupted cells and directly interacted with the 3′ untranslated region (UTR) of ICAM-1 mRNA. Modulation of Dicer or these miRs inversely correlated with ICAM-1 protein expression and susceptibility of U87 glioma cells to CTL-mediated cytolysis while ICAM-1 mRNA levels remained stable. Immunohistochemical and in situ hybridization analyses of 30 primary glioblastoma tissues demonstrated that expression of Dicer, miR-222, or miR-339 was inversely associated with ICAM-1 expression. Taken together, Dicer is responsible for the generation of the mature miR-222 and -339, which suppress ICAM-1 expression on tumor cells, thereby down-regulating the susceptibility of tumor cells to CTL-mediated cytolysis. This study suggests development of novel miR-targeted therapy to promote cytolysis of tumor cells.
Keywords: Dicer, glioma
MicroRNAs (miRs) are 19- to 25-nucleotide noncoding RNA molecules that regulate gene expression at the level of transcription and translation. The RNase III endonuclease Dicer plays a key role in generation of miRs in cells (1). In cancer, Dicer expression levels have been reported to either positively or inversely correlate with malignant behavior of tumors, depending upon the cancer type (2–4), and miRs can act either as oncogenes, as tumor suppressor genes, or sometimes as both (4–6). It is therefore important to determine how Dicer and miRs regulate biological properties of cancers. As a way to elucidate the specific impact of Dicer disruption in cancer cells, Vogelstein's group has established human colorectal cancer cell lines in which exon 5 of Dicer gene was disrupted (ex5−/−) (7). Of 97 known miRs detected in wild-type and (ex5−/−) HCT116 cells, 55 were differentially expressed, and for 53 of these 55 there was an average 7-fold reduction of miR levels in Dicer (ex5−/−) cells compared with wild-type cells. These observations indicate that Dicer is required for the biogenesis of a subset of miRs.
With regard to the roles of Dicer in immune cells, Dicer-generated RNAs appear to be necessary for development of regulatory T cells (8). Dicer is also required for Type-2 T cell response (9), which counteracts antitumor immunity (10). These observations regarding the roles of Dicer in tumor cells and the immune system led us to hypothesize that expression of Dicer in cancer cells might regulate immune surveillance through processing of miRs.
Malignant gliomas, such as glioblastoma multiforme (GBM), represent the most common and aggressive primary brain tumors. Over 12,000 new cases are diagnosed in the United States annually (11), with a median survival rate of approximately 15 months (12). Development of novel, molecularly-targeted, multimodal therapeutic approaches is critical to further improve the outcome of these deadly tumors. Recent studies have developed an attractive vehicle for in vivo miR-targeting with the use of antisense “antagomir” oligonucleotides (13). The antagomir-mediated silencing of disease-associated miRs may enable development of novel cancer therapies.
Here we demonstrate that Dicer-regulated miRs, miR-222 and miR-339, are expressed in cancer cells, including glioma, and negatively regulate intercellular cell adhesion molecule (ICAM)-1 by direct interaction with its 3′ UTR. Up-regulation of ICAM-1 expression by inhibition of Dicer or miR-222 or -339 in cancer cells leads to increased susceptibility of cancer cells to antigen-specific cytotoxic T-lymphocytes (CTLs). In primary GBM tissues, expression of Dicer or miR-222 or -339 was inversely associated with ICAM-1 expression. Identification of miRs that affect immune recognition with regulation of ICAM-1 expression has not been previously reported and extends the importance of miRs in inflammation and cancer.
Results
Dicer-Disrupted Cells Exhibit Up-Regulated ICAM-1 Expression and Enhanced Susceptibility to CTL-Mediated Cytolysis.
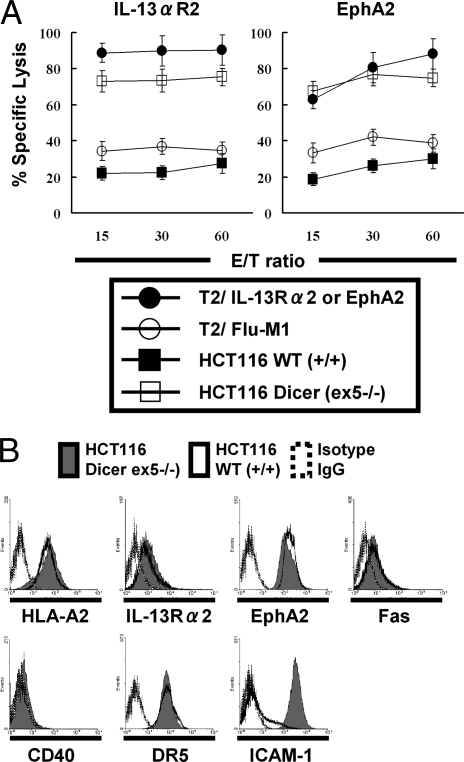
To evaluate effects of altered Dicer expression status, we obtained 3 human colorectal cancer cell lines HCT116, DLD-1, and RKO, in each of which exon 5 of the human Dicer gene was disrupted (7). Among them, wild-type HCT116 and HCT116 [ex5−/−] cells expressed HLA-A2 as well as the nominal tumor antigens interleukin-13 receptor α2 (IL-13Rα2) and EphA2 at similar levels (Fig. 1B) (14, 15), allowing us to evaluate HLA-A2-restricted, antigen-specific lytic activity of CTLs against these tumor cells. CTLs were raised against IL-13Rα2 (345–353) or EphA2 (883–891) by in vitro stimulation of HLA-A2+ healthy donor-derived peripheral blood mononuclear cells (PBMCs) with the corresponding peptides. As demonstrated in Fig. 1A, in 51Cr-release assays, CTLs efficiently lysed T2 cells pulsed with relevant [IL-13Rα2 (345–353) or EphA2 (883–891)] peptides but not T2 cells pulsed with irrelevant Flu-M1 [58–66] peptide (16), demonstrating specificity and activity of the CTL lines. HCT116 (ex5−/−) cells were remarkably more susceptible to cytolysis by 2 established CTL lines when compared with wild-type HCT116 cells (Fig. 1A). These results suggested that Dicer might regulate expression of molecules that mediate tumor-CTL interaction.
Fig. 1.
Dicer (ex5−/−) cells are more sensitive to CTL-mediated cytolysis association with up-regulation of ICAM-1. (A) CTLs raised against IL-13Rα2345–353:1A9V (Left) or EphA2883–891 (Right) were tested for their lytic ability against human colorectal cancer HCT116 cells (solid square, HLA-A2+, EphA2+, IL-13Rα2+), HCT116 Dicer (ex5−/−) cells (hollow square, HLA-A2+, EphA2+, IL-13Rα2+), or T2 cells loaded with IL-13Rα2345–353:1A9V (solid circle on Left), EphA2883–891 (solid circle on Right) or Influenza M158–66 (hollow circle) using 4-hour 51Cr-release assays. Values indicate averages of triplicate samples. Bars indicate SD. (B) ICAM-1 was up-regulated in HCT116 Dicer (ex5−/−) cells. Flow cytometric analyses were performed on wild-type or Dicer (ex5−/−) HCT116 cells for expression of a panel of proteins that are known to mediate CTL-tumor cell interactions. Open and shaded histograms represent wild-type and Dicer (ex5−/−) HCT116 cells, respectively. Dashed lines represent control cells stained with isotype control IgG.
To identify causative molecules that promoted the susceptibility of HCT116 (ex5−/−) cells, we examined expression of a panel of proteins that are known to mediate tumor-CTL cell interactions, including HLA class I, ICAM-1, Fas, CD40, DR5, the antigen processing machinery components (β2 microglobulin, TAP, tapasin, calreticulin, LMP, ERp57, and calnexin) (17) as well as tumor antigens IL-13Rα2 and EphA2 in the ex5−/− and wild-type HCT116 cells (Fig. 1B and data not shown). Among these, ICAM-1 was up-regulated in the ex5−/− cells, while none of the other molecules evaluated were differentially expressed in ex5−/− vs. wild-type HCT116 cells. Up-regulation of ICAM-1 in ex5−/− cells (vs. wild-type cells) was also confirmed in 2 other colorectal cancer cell lines, DLD-1 and RKO (supporting information (SI) Fig. S1).
ICAM-1 Promotes Formation of Tumor-CTL Conjugates and Cytolysis of Dicer (ex5−/−) Cells.
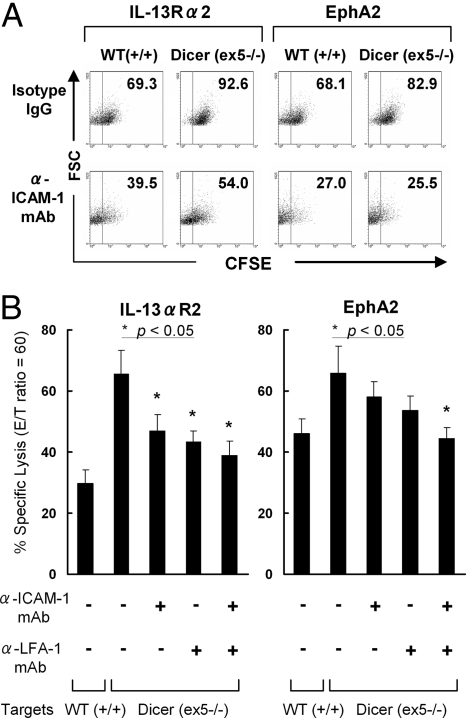
The binding of lymphocyte function-associated antigen-1 (LFA-1) to its ligand, ICAM-1, is a crucial step in the initial interaction of T-cells (18) during generation of tumor-specific CTL responses (19). Enhanced ICAM-1 expression on ex5−/− vs. wild-type cells suggested increased ability of the CTLs to engage tumor cells. Therefore, we performed conjugate assays to test the hypothesis that ex5−/− cells formed more conjugates with the established CTL lines (Fig. 2A). When ex5−/− or wild-type HCT116 cells were labeled with CFSE and incubated with CTLs primed against the IL-13Rα2- or EphA2-derived CTL epitope peptides, HCT116 (ex5−/−) cells showed a remarkably higher frequency of conjugate formation with the CTLs compared with that mediated by wild-type HCT116 cells (Fig. 2A Upper). Blockade of LFA-1-ICAM-1 interaction using an anti-ICAM-1 monoclonal antibody (mAb) resulted in an apparent inhibition of conjugate formation for both ex5−/− and wild-type HCT116 cells (Fig. 2A Lower). Furthermore, blockade of the ICAM-1-LFA-1 interaction by either anti-ICAM-1 or anti-LFA-2 mAb, or by both mAbs, inhibited the specific lysis of the CTLs against HCT116 (ex5−/−) cells (Fig. 2B), indicating a pivotal role of this interaction. Thus, disruption of Dicer in the ex5−/− cells confers an enhanced capacity to form stable conjugates with the CTLs through up-regulation of ICAM-1, thereby promoting the susceptibility of ex5−/− cells to CTL-mediated lysis.
Fig. 2.
Up-regulated ICAM-1 on Dicer (ex5 −/−) cells mediates enhanced formation of tumor-T cell conjugates and susceptibility to antigen-specific CTLs. (A) CFSE-labeled HCT116 wild-type cells (Left) or Dicer (ex5−/−) cells (Right) with (Lower) or without (Upper) mAb-mediated ICAM-1 blockade were stained with TriColor (TC)-anti-CD8 mAb. Tumor-T cell conjugates were identified by flow cytometry in the electronically gated CD8+ T cell population based on increased FSC and CFSE fluorescence of conjugated cells. The percentage of CD8+ T cells that have formed conjugates with the CFSE+ tumor cells in the gated CD8+ cells is indicated in each dot plot. (B) Bar graphs demonstrating the percentage of specific lysis of HCT116 Dicer (ex5−/−) cells by CTLs primed against the IL-13Rα2- (Left) or EphA2- (Right) derived epitope peptide in the presence of anti-ICAM-1, anti-LFA-1 mAb, or isotype IgG [at the Effector:Target (E:T) ratio of 60:1]. The specific lysis of wild-type HCT116 by the CTLs was shown as a control. The data represent the means of the values from triplicate samples ± SD. *, P < 0.05 for the specific lysis of HCT116 (ex5−/−) cells in the presence of anti-ICAM-1, anti-LFA-1, or both antibodies, compared with the specific lysis of HCT116 (ex5−/−) cells pretreated with isotype IgG antibodies.
MiR-222 and -339 Have Predicted Targets in the 3′- UTR of ICAM-1 mRNA.
We next sought to identify specific miRs that could modulate ICAM-1 expression. Among miRs that were down-regulated in HCT116 (ex5−/−) cells compared with wild-type HCT116 cells (7), miRs 222 and 339 were predicted to bind to ICAM-1 based on the miRBase algorithm (http://microrna.sanger.ac.uk). Indeed, quantitative RT-PCR analysis showed the decrease of miRs-222 and -339 in all 3 ex5−/− cell lines compared with corresponding wild-type cells (Fig. S2A). The “seed” sequences in miR-222 (5′-GCUACA-3′, nucleotides 2–7) and -339 (5′-CCCUGUCCUCC -3′, nucleotides 2–12) matched nucleotides 2067–2072 and nucleotides 1773–1783, respectively, in the human ICAM-1 3′-UTR (NM_000201) (Fig. S2B).
SiRNA-Mediated Inhibition of Dicer Suppress miRs-222 and -339 in Human Malignant Glioma Cells.
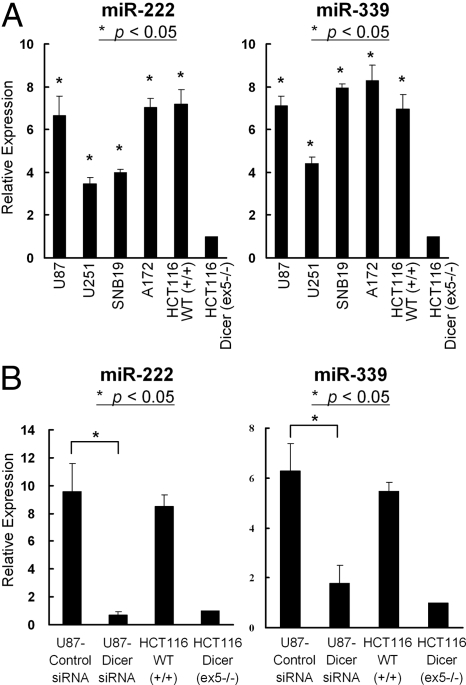
Development of novel and molecularly targeted therapeutic approaches is critical to improve the outcome of patients with GBM. We next evaluated expression of Dicer and miRs-222 and -339 in human GBM cell lines. A recent study by others (20) and our analysis (shown in Fig. S3) suggested expression of Dicer protein in human GBM cell lines, U87, A172, SNB19 and U251. Moreover, 4 GBM cell lines (U87, U251, SNB19, and A172) had elevated expression levels of miRs-222 and -339 when compared to Dicer-disrupted (ex5−/−) HCT116 cells (Fig. 3A). SiRNA-based interference of Dicer significantly inhibited the endogenous levels of miRs-222 and -339 in a glioma cell line U87 (Fig. 3B). These results suggest that Dicer-mediated regulation of miRs-222 and -339 is not limited to colorectal cancer cells but may represent a general mechanism in other cancers, including GBM.
Fig. 3.
Human GBM cell lines express Dicer-mediated miR-222 and -339. (A) Relative expression of miR-222 (Left) and miR-339 (Right) in 4 human glioma cell lines, HCT116 WT (+/+) cells and HCT116 Dicer (ex5 −/−) cells by TaqMan analyses. *, P < 0.05 for the relative miR expression in glioma cell lines compared with that in HCT116 Dicer (ex5 −/−) cells. (B) U87 cells were transfected with either a control RNA duplex or a RNA duplex targeting Dicer mRNA. Error bars indicate SD. *, P < 0.05 for the relative miR expression in Dicer siRNA-transfected U87 cells compared with that in control siRNA-transfected U87 cells. (A and B) The relative miR expression level for each cell line was normalized to the small nuclear RNA RNU43 level and calculated as the relative threshold cycle (CT) value to that of the HCT116 Dicer (ex5 −/−) cells. The results represent mean ± SD of triplicate samples.
MiRs-222 and -339 Inhibit ICAM-1 Posttranscriptionally via Direct Interaction with the ICAM-1 3′-UTR.
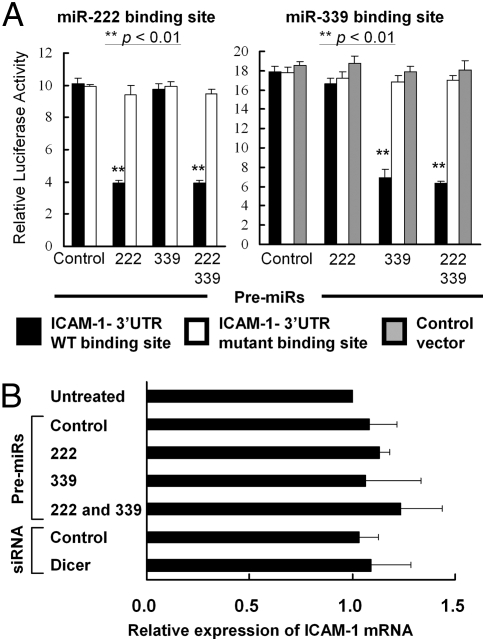
To directly demonstrate that miR-222 and -339 functionally target the 3′UTR of ICAM-1 mRNA, we performed luciferase reporter assays. The psiCHECK-2 reporter plasmids contained the ICAM-1 3′UTR corresponding to each putative binding site for miR-222 (psiCHECK-2 miR-222 WT) or miR-339 (psiCHECK-2 miR-339 WT) (Fig. S2B) downstream of renilla luciferase cDNA. We created additional psiCHECK-2 reporter plasmids, in which mutations were introduced in the putative miR-222 binding site (psiCHECK-2 miR-222 MT) or the putative miR-339 binding site (psiCHECK-2 miR-339 MT) (see Materials and Methods). Each of these psiCHECK-2 vectors or a control luciferase vector without the 3′UTR (psiCHECK-2) was cotransfected into U87 cells with miR-222 or -339 precursor (premiR). Ectopic expression of premiR-222 inhibited the relative luciferase activity in U87 cells transfected with psiCHECK-2 miR-222 WT as compared with the control premiR-transfected cells (Fig. 4A Left and Fig. S4A), but had no effects on U87 cells transfected with psiCHECK-2 miR-222 MT. Transfection of premiR-339 decreased the relative luciferase activity in U87 cells transfected with psiCHECK-2 miR-339 WT as compared with the control premiR-transfected cells but had no effects on U87 cells transfected with the control luciferase vector or psiCHECK-2 miR-339 MT (Fig. 4A Right and Fig. S4A).
Fig. 4.
MiR-222 and miR-339 suppress gene expression by targeting the ICAM-1 3′-UTR. (A) U87 cells transfected with premiR for miR-222, miR-339, or both were cotransfected with psiCHECK-2 miR-222 WT (black columns on the Left), psiCHECK-2 miR-222 MT (white columns on the Left), psiCHECK-2 miR-339 WT (black columns on the Right), psiCHECK-2 miR-339 MT (white columns on the Right) or the control backbone psiCHECK-2 (gray columns on the Right). Renilla luciferase activity was normalized on the constitutive activity of firefly luciferase. The data represent mean ± SD of triplicate samples. **, P < 0.01 for U87 cells transfected with premiR-222/339 alone or ones cotransfected with both of premiR-222 and -339 compared with control premiR-transfected U87 cells. (B) ICAM-1 mRNA levels were assessed by quantitative RT-PCR in wild-type and U87 cells transfected with premiR-222, premiR-339, both premiRs, Dicer-siRNA, or appropriate negative control constructs. The relative ICAM-1 mRNA level for each sample was normalized to the β-actin mRNA level in each sample and calculated as the relative CT value to that of untreated U87 cells. The results represent mean ± SD of triplicate samples.
We predicted that miRs-222 and -339 would down-regulate ICAM-1 expression at the level of translation, because ICAM-1 does not contain an exact match to either miR-222 or miR-339 in its 3′-UTR (Fig. S2B). To corroborate this prediction, we measured ICAM-1 mRNA expression levels in U87 cells transfected with Dicer siRNA or premiRs. Neither premiR-222, premiR-339, nor Dicer siRNA altered the ICAM-1 mRNA expression in these cells (Fig. 4B). These data suggest that miR-222 and -339 down-regulate ICAM-1 expression at posttranscriptional levels by binding to the ICAM-1 3′UTR.
Inhibition of miRs-222 or -339 Leads to Recovery of ICAM-1 Expression in Human Malignant Glioma Cells and Promotes Their Susceptibility to CTL-Mediated Cytolysis.
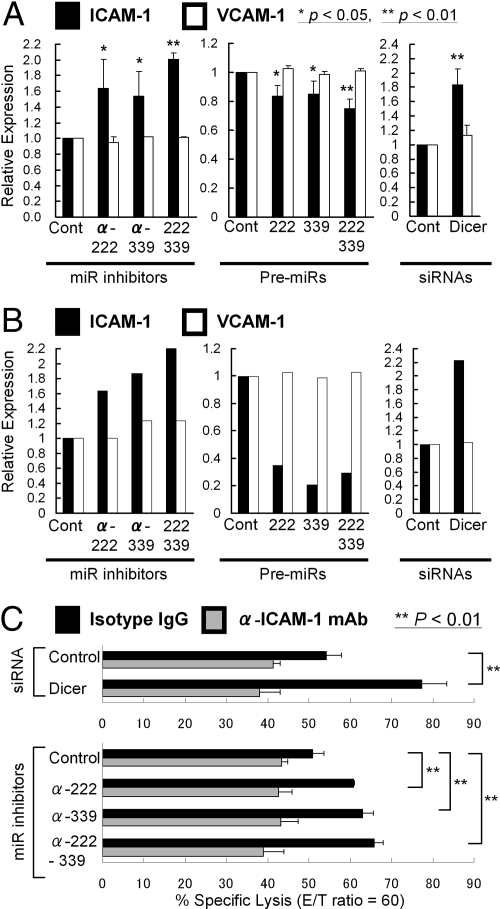
We next assessed the effect of Dicer and miRs-222 and -339 on ICAM-1 expression. U87 GBM cells were transfected with Dicer siRNA, precursors, or inhibitors for the miRs, or appropriate control constructs. Cell surface ICAM-1 expression on the transfectants was quantified by flow-cytometric analyses (Fig. 5A and Fig. S5A), and the total expression in whole cell lysates was measured by immunoblotting (Fig. 5B and Fig. S5B). U87 cells transfected with Dicer siRNA showed increased ICAM-1 expression when compared to control RNA duplex-transfected cells (Fig. 5 A and B and Fig. S5 A Upper and B, top lane). Suppression of miR-222, miR-339, or both by inhibitors led to an increase in ICAM-1 expression (Fig. 5 A and B and Fig. S5 A Upper and B, top lane) without affecting vascular cell adhesion molecule 1 (VCAM-1) that does not have putative miR-222 or -339 binding site (Fig. 5 A and B and Fig. S5 A Lower and B, middle lane). Conversely, overexpression of miR-222, miR-339, or both suppressed ICAM-1 expression (Fig. 5 A and B and Fig. S5 A Upper and B, top lane).
Fig. 5.
Inhibition of Dicer, miRs-222 or -339 recovered ICAM-1 expression in human malignant glioma cells and promoted their susceptibility to CTL. (A and B) Relative ICAM-1 expression (black columns) was evaluated by flow cytometric analyses (A) and Western blotting (B) in U87 cells, in which miR-222, miR-339, or both were inhibited (Left), overexpressed (Middle), or Dicer was inhibited by specific siRNA (Right). VCAM-1 (white columns) was evaluated to determine whether changes in expression levels were specific to ICAM-1. (A) The relative expression level for each cell group was calculated as the relative mean fluorescence intensity (MFI) value to that of U87 cells transfected with the appropriate negative control. Results represent the means of the values from 3 independent experiments. Bars indicate SD. * and ** refer to statistical significance between groups (P < 0.05 and P < 0.01), respectively. (B) The intensity of each specific band was quantified using ImageJ software (see SI Text). The relative expression level of ICAM-1 and VCAM-1 in each cell group was normalized to the β-actin level and calculated as the relative intensity value to that of U87 cells transfected with the appropriate negative control. (C) U87 cells were transfected with control siRNA duplex, Dicer-siRNA duplex, control miR inhibitor, miR-222 inhibitor, miR-339 inhibitor, or both inhibitors before 4 h 51Cr-release assays with PBMCs primed against EphA2883–891 peptide (effector: target ratio was 60:1). Bars represent the percentage of specific lysis in the presence of anti-ICAM-1 mAb (gray bars) or isotype IgG (black bars). The data represent the means of the values from triplicate samples ± SD. **, P < 0.01 for Dicer-siRNA duplex-transfected U87 cells compared with control siRNA-transfected U87 cells; and miR-222 inhibitor-transfected or miR-339 inhibitor-transfected U87 cells compared with control miR inhibitor-transfected U87 cells.
To determine the functional relevance of Dicer and miRs-222 and -339 to CTL-mediated antitumor effects, we performed CTL assays using these genetically engineered U87 cells as target cells for CTLs raised against EphA2 (883–891) epitope. Inhibition of Dicer and antagonism of either miRNA-222 or -339 in HLA-A2+ U87 cells significantly enhanced their susceptibility to the CTLs compared with relevant control groups (Fig. 5C). Moreover, blockade of ICAM-1 with a specific antibody restrained the susceptibility of these genetically engineered U87 cells. These results indicate that the Dicer-regulated miRs-222 and -339 target ICAM-1, leading to enhanced CTL-mediated antitumor effects.
Expression of Dicer and miR-222 and miR-339 Was Inversely Associated with ICAM-1 Expression in Primary GBM Tissues.
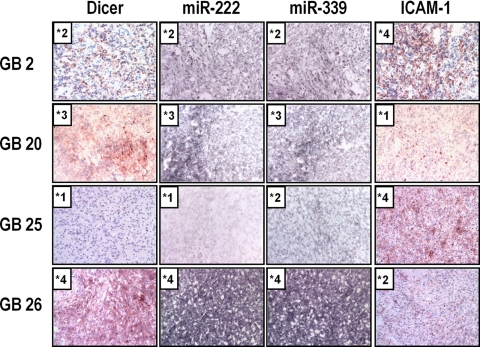
These results with in vitro cultured cells led us to hypothesize that the status of Dicer and the miR expression might inversely correlate with ICAM-1 expression in primary GBM tissues. We performed in situ hybridization of miR-222 and -339, and immunohistochemical analyses on Dicer and ICAM-1 in 30 primary GBM tissues. Table 1 summarizes degrees of miR-222/339 expression and Dicer/ICAM-1 immunopositivity in these cases, and Fig. 6 demonstrates miR expression and Dicer/ICAM-1 staining in 4 cases (GB 2, 20, 25, and 26) as examples. Dicer staining and expression of miR-222 or miR-339 were inversely associated with ICAM-1 staining, with Spearman's correlation coefficients of −0.452 (P = 0.013), −0.532 (P = 0.004) and −0.427 (P = 0.021), respectively. A positive correlation coefficient between miR-222 expression (r = 0.388, P = 0.037) or miR-339 expression (r = 0.385, P = 0.038) and Dicer immunopositivity was observed. These data support our hypothesis that Dicer and Dicer-mediated miRs may have a role in regulation of ICAM-1 in vivo.
Table 1.
Expression of Dicer, miRNAs, and ICAM-1 in clinical samples of GBM patients
| Case | Sex | Age | *miR-222 | *miR-339 | *Dicer | *ICAM-1 |
|---|---|---|---|---|---|---|
| 1 | F | 52 | +2 | +3 | +1 | +2 |
| 2 | M | 53 | +2 | +2 | +2 | +4 |
| 3 | M | 45 | +1 | +1 | +1 | +3 |
| 4 | M | 33 | +2 | +1 | +2 | +1 |
| 5 | F | 44 | +2 | +2 | +1 | +2 |
| 6 | M | 43 | +2 | +3 | +2 | +1 |
| 7 | M | 58 | +2 | +1 | +1 | +2 |
| 8 | F | 62 | +1 | +2 | +2 | +1 |
| 9 | M | 65 | +1 | +1 | +1 | +2 |
| 10 | F | 75 | +2 | +2 | +2 | +1 |
| 11 | M | 69 | +4 | +3 | +3 | +1 |
| 12 | M | 43 | +2 | +2 | +1 | +2 |
| 13 | M | 64 | +2 | +2 | +1 | +2 |
| 14 | M | 37 | +2 | +2 | +3 | +1 |
| 15 | M | 44 | +1 | +1 | +2 | +3 |
| 16 | F | 55 | +4 | +4 | +1 | +1 |
| 17 | F | 69 | +2 | +1 | +1 | +2 |
| 18 | F | 81 | +1 | +2 | +1 | +3 |
| 19 | F | 53 | +3 | +2 | +3 | +1 |
| 20 | F | 69 | +3 | +3 | +3 | +1 |
| 21 | M | 65 | +1 | +1 | +1 | +2 |
| 22 | F | 43 | +2 | +1 | +1 | +3 |
| 23 | F | 68 | +3 | +4 | +2 | +1 |
| 24 | M | 65 | +2 | +1 | +1 | +4 |
| 25 | F | 48 | +1 | +2 | +1 | +4 |
| 26 | M | 45 | +4 | +4 | +4 | +2 |
| 27 | F | 56 | +2 | +2 | +1 | +2 |
| 28 | M | 66 | +1 | +1 | +2 | +3 |
| 29 | F | 44 | +2 | +2 | +1 | +1 |
| 30 | M | 23 | +2 | +1 | +1 | +2 |
*0, negative staining; +1, weakly positive staining; +2, moderately positive staining; +3, strongly positive at focal areas; +4, strongly and diffusely positive throughout the lesion.
Fig. 6.
Dicer and Dicer-regulated miRs expressed in primary human GBM tissues are inversely associated with ICAM-1 expression. Representative examples of Dicer/ICAM-1 expression by immunohistochemistry and miR expression by in situ hybridization. Expression of Dicer, miRs and ICAM-1 was graded as follows: grade 0, negative staining; +1, weakly positive staining; +2, moderately positive staining; +3, strongly positive at focal areas; +4, strongly and diffusely positive throughout the lesion. Original magnification was ×20.
Discussion
This is the first report on roles of miRs-222 and -339 in regulation of ICAM-1 at posttranscriptional levels. MiR-regulated ICAM-1 mediates cancer cells' susceptibility to CTLs, and levels of Dicer and miR-222 and -339 are inversely associated with ICAM-1 expression not only in vitro cultured cells but also in primary GBM tissues. The significance of Dicer expression levels on prognosis of cancer patients is still controversial (2, 3, 6). Because miRs can act either as oncogenes, as tumor suppressor genes, or sometimes as both (5), alteration of Dicer levels can probably influence the malignant behavior of cancers either positively or negatively depending upon the profiles of miR expression status in each cancer type. In the current study with primary GBM tissues, Dicer was expressed at varying levels, and it would be intriguing to know whether Dicer expression in GBM correlates with prognosis of patients. Interestingly, miR let-7, whose reduced expression was associated with shorter patient survival (4), inhibits Dicer expression by directly targeting the 3′ UTR of Dicer (21). Therefore, let-7 serves as a prototypic “tumor-suppressor-miR,” functioning as a key microRNA in a novel regulatory loop limiting Dicer expression. Although recent studies have suggested a general down-regulation of miRs in tumors compared with normal tissues (22, 23), these studies also stress the significance of miR-profiling for dictating tumor-tissue origins and differentiation status.
There are groups of miRs that are known to be overexpressed in solid cancers. Some of these miRs, such as miR-17–5p, miR-20a, miR-21, miR-92, miR-106a, and miR-155, are well characterized for their biological properties contributing to malignant behavior (24). With regard to miR-222 and -339 in cancers, miR-221 and -222 are up-regulated in human thyroid papillary carcinomas in comparison with normal thyroid tissue (25). miR-221 and -222 directly target the p27(Kip1) protein, a key regulator of cell cycle, thereby inducing progression to the S phase of the cell cycle (25). Although cell-cycle analysis was not within the scope of our current study, it would be intriguing to address whether modulation of Dicer and miR-222 can influence the proliferation of colon cancer and glioma cells. A more recent study with melanoma has identified c-KIT receptor as another target of overexpressed miR-222, leading to blockade of differentiation in melanoma cells (26). In GBMs, up-regulation of miR-222/221 has been reported (20, 27, 28), as well as miR-21 (27, 29). Since miR-221 was not predicted to bind to ICAM-1, we did not evaluate miR-221 in the current study. Taken together, information from recent studies suggests miR-222 may be a suitable therapeutic target to suppress the proliferation of poorly-differentiated tumor cells, and to enhance immunoreactivity against CTL-based immunotherapy. There is paucity of information in the literature regarding the roles of miR-339 in neoplastic cells. Further studies are warranted to determine additional roles of miR-339 in neoplasms.
Our data indicated that ICAM-1 has a pivotal role in physical and functional interaction between tumor cells and CTLs. The binding of LFA-1 to its ligand, ICAM-1, is a crucial step in the migration of leukocytes and T cell activation (18). ICAM-1/LFA-1 interaction is required to generate tumor-specific CTL responses (19). These observations suggest that the reduced expression of ICAM-1 on tumor cells could prevent efficient association of CTLs to tumor cells, contributing, at least partially, to tumor escape from the host immune surveillance. Indeed, reduced expression of ICAM-1 is associated with poor prognosis in various cancers, such as melanoma (30) and head and neck (31), breast (32), colorectal (33), and ovarian cancers (34). These previous studies also point to the significance of identifying miRs that regulate ICAM-1. Further studies are warranted to determine the possible predictive value of ICAM-1 on prognosis of patients with GBM.
It has been implicated that the ICAM-1/LFA-1 interaction also stimulates NK cells' cytotoxic activity (35). Accordingly, in our preliminary data (Fig. S6), Dicer (ex 5−/−) cells exhibited greater sensitivity to NK cell-mediated lysis. However, a recent study indicated that inhibition of Dicer leads to up-regulation of ligands for the NKG2D receptor, MICA, and MICB via DNA damage (36). Hence, a variety of immunological mechanisms may operate in increased susceptibility of Dicer (ex5 −/−) cells against immune surveillance.
Recent studies have developed an attractive vehicle for in vivo miR-targeting by the use of antisense “antagomir” oligonucleotides (13). Based on the present study, targeted therapies suppressing miR-222 and -339 may prove beneficial in cancers and cancer immunotherapy.
Materials and Methods
Cell lines, antibodies, peptides, and Flow cytometric analyses (See SI Text).
Patients and Samples.
This study was approved by the local ethical review board of University of Pittsburgh. Frozen GBM tissues were obtained from the brain tumor bank of the Division of Neuropathology, Department of Pathology, University of Pittsburgh School of Medicine, as archived, deidentified samples.
In Vitro Induction of CTL in PBMCs and Cytotoxicity Assay.
CTL induction and 51Cr release cytotoxicity assay were performed as previously described (37) and detailed in SI Text.
Conjugate Assay.
Conjugate assays were performed as described in SI Text.
MiR and mRNA Real-Time Quantitative RT-PCR Analysis.
TRIzol-extracted (Invitrogen), DNase I-treated total RNA (10 ng) was subjected to qRT-PCR analysis using the TaqMan miR Reverse Transcription Kit and miR Assays (Applied Biosystems), and the Real-Time thermocycler iQ5 (Bio-Rad). The small nucleolar RNU43 was used as the housekeeping small RNA reference gene. All reactions were done in triplicate and relative expression of RNAs was calculated using the 2 ΔCT method (38). QRT-PCR analysis of ICAM-1 mRNA was performed as detailed in SI Text.
Dicer RNA Interference, Inhibition of Endogenous miRs, and Ectopic Expression of PremiRs.
We used a synthetic RNA duplex with the sense strand sequences CACUGGUCAGGGAAGACAUU (Dicer siRNA) (20) for Dicer RNA interference and Scrambled II duplex (39) as a control duplex, both of which were purchased from Dharmacon. MiRs-222 or -339 were expressed in cell lines by transfection with a premiR precursor molecule (premiR) (Ambion) and inhibited with miR inhibitor (miR inhibitor) (Ambion). Amaxa Nucleofector technology was used for transfection according to the manufacturer's instructions. Cells were seeded (5 × 105/well) into 6-well plates and were harvested after 20 h, and RNA and protein extractions were performed.
Luciferase Activity Assay.
U87 cells were cotransfected with premiRs or miR inhibitors and 2 μg of renilla/firefly luciferase reporter plasmid as described in SI Text. Cells were harvested 24 h later for analysis of luciferase activity. Luciferase assays were performed using the Dual Luciferase Assay System (Promega) according to the manufacturer's instructions. Relative luciferase activity was obtained by normalizing the renilla luciferase activity to the firefly luciferase activity.
Western Blotting.
Western blot was performed as previously described (40) and detailed in SI Text.
Immunohistochemical Staining and Scoring Staining.
Immunohistochemistry was done as previously described (37) and detailed in SI Text.
Locked Nucleic Acid-Based in Situ Detection of miRs in Primary GBM Tissues.
In situ hybridizations were performed in 12-μm cryosections from primary GBM tissues as previously described (41) and detailed in SI Text.
Statistical Analysis.
Data are presented using means and standard deviations. The 2-sided Student's t test was used for comparison of 2 samples with unequal variances. Spearman's rank correlation and exact test was used to investigate the association of the level of ICAM-1 staining with miR-222, miR-339, and Dicer staining.
Supplementary Material
Acknowledgments.
We thank Ms. Amy Gardner (Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, Pittsburgh, PA) for providing technical assistance. The current work was supported by the National Institutes of Health [1R01NS055140, 2P01 NS40923, and 1P01 CA100327 to H.O. and 1P01 CA101944 to M.T.L.], the Doris Duke Charitable Foundation and James S. McDonnell Foundation [220020036 to H.O.], and the Walter L. Copeland Fund of The Pittsburgh Foundation [D2007–0624 to K.S.].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811817106/DCSupplemental.
References
- 1.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiosea S, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiosea S, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 4.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 5.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 6.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp RA, Ronchese F. Tumor-specific Tc1, but not Tc2, cells deliver protective antitumor immunity. J Immunol. 2001;167:6497–6502. doi: 10.4049/jimmunol.167.11.6497. [DOI] [PubMed] [Google Scholar]
- 11.CBTRUS. The Central Brain Tumor Registry of the United States Statistical Report: Primary Brain Tumors in the United States, 2000–2004. Hinsdale, IL: Central Brain Tumor Registry of the United States; 2008. [Accessed June 5, 2009]. Available at www.cbtrus.org/reports//2007-2008/2007report.pdf. [Google Scholar]
- 12.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 13.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 14.Hatano M, et al. EphA2 as a glioma-associated antigen: a novel target for glioma vaccines. Neoplasia. 2005;7:717–722. doi: 10.1593/neo.05277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eguchi J, et al. Identification of interleukin-13 receptor alpha2 peptide analogues capable of inducing improved antiglioma CTL responses. Cancer Res. 2006;66:5883–5891. doi: 10.1158/0008-5472.CAN-06-0363. [DOI] [PubMed] [Google Scholar]
- 16.Morrison J, et al. Identification of the nonamer peptide from influenza A matrix protein and the role of pockets of HLA-A2 in its recognition by cytotoxic T lymphocytes. Eur J Immunol. 1992;22:903–907. doi: 10.1002/eji.1830220404. [DOI] [PubMed] [Google Scholar]
- 17.Vujanovic L, et al. Regulation of antigen presentation machinery in human dendritic cells by recombinant adenovirus. Cancer Immunol Immunother. 2008;58:121–133. doi: 10.1007/s00262-008-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann MF, et al. Distinct roles for LFA-1 and CD28 during activation of naive T cells: Adhesion versus costimulation. Immunity. 1997;7:549–557. doi: 10.1016/s1074-7613(00)80376-3. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson SR, Williams NA, Morgan DJ. The role of intercellular adhesion molecule-1/LFA-1 interactions in the generation of tumor-specific CD8+ T cell responses. J Immunol. 2005;174:3401–3407. doi: 10.4049/jimmunol.174.6.3401. [DOI] [PubMed] [Google Scholar]
- 20.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 21.Tokumaru S, et al. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- 22.Gaur A, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 23.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visone R, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 26.Felicetti F, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 27.Ciafre SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Medina R, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corsten MF, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 30.Anastassiou G, et al. Expression of the cell adhesion molecules ICAM-1, VCAM-1 and NCAM in uveal melanoma: a clinicopathological study. Oncology (Basel) 2000;58:83–88. doi: 10.1159/000012083. [DOI] [PubMed] [Google Scholar]
- 31.Shirai A, Furukawa M, Yoshizaki T. Expression of intercellular adhesion molecule (ICAM)-1 in adenoid cystic carcinoma of the head and neck. Laryngoscope. 2003;113:1955–1960. doi: 10.1097/00005537-200311000-00019. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa Y, et al. Expression of intercellular adhesion molecule-1 in invasive breast cancer reflects low growth potential, negative lymph node involvement, and good prognosis. Clin Cancer Res. 1998;4:31–36. [PubMed] [Google Scholar]
- 33.Shibata MAK, Amano S, Kurosu Y. Local expression and circulating form of ICAM-1 in colorectal cancer. Ann Cancer Res Ther. 1996;5:29–33. [Google Scholar]
- 34.Arnold JM, Cummings M, Purdie D, Chenevix-Trench G. Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer. 2001;85:1351–1358. doi: 10.1054/bjoc.2001.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poggi A, et al. NKG2D and natural cytotoxicity receptors are involved in natural killer cell interaction with self-antigen presenting cells and stromal cells. Ann NY Acad Sci. 2007;1109:47–57. doi: 10.1196/annals.1398.007. [DOI] [PubMed] [Google Scholar]
- 36.Tang KF, et al. Decreased Dicer expression elicits DNA damage and up-regulation of MICA and MICB. J Cell Biol. 2008;182:233–239. doi: 10.1083/jcb.200801169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda R, et al. Spontaneous immune responses against glioma-associated antigens in a long term survivor with malignant glioma. J Transl Med. 2007;5:68. doi: 10.1186/1479-5876-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (Amsterdam, Neth.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin RM, et al. Protection of glioblastoma cells from cisplatin cytotoxicity via protein kinase Ciota-mediated attenuation of p38 MAP kinase signaling. Oncogene. 2006;25:2909–2919. doi: 10.1038/sj.onc.1209312. [DOI] [PubMed] [Google Scholar]
- 40.Hatano M, et al. Vaccination with EphA2-derived T cell-epitopes promotes immunity against both EphA2-expressing and EphA2-negative tumors. J Transl Med. 2004;2:40. doi: 10.1186/1479-5876-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.