Abstract
Potassium channels are among the core functional elements of life because they underpin essential cellular functions including excitability, homeostasis, and secretion. We present here a series of multivalent calix[4]arene ligands that bind to the surface of voltage-dependent potassium channels (Kv1.x) in a reversible manner. Molecular modeling correctly predicts the best candidates with a conical C4 symmetry for optimal binding, and the effects on channel function are assessed electrophysiologically. Reversible inhibition was observed, without noticeable damage of the oocytes, for tetraacylguanidinium or tetraarginine members of the series with small lower rim O-substituents. Apparent binding constants were in the low micromolar range and had Hill coefficients of 1, consistent with a single site of binding. Suppression of current amplitude was accompanied by a positive shift in the voltage dependence of gating and slowing of both voltage sensor motion and channel opening. These effects are in keeping with expectations for docking in the central pore and interaction with the pore domain “turret.”
Keywords: electrophysiology, Kv1.x channels, ligand–protein surface interactions, molecular recognition, multivalent calix[4]arene ligands
Potassium channels are major targets in biomedical and pharmacological research, because many diseases, mostly nervous disturbances, are linked to these membrane proteins (1). The fact that >90 genes for different K+ channel subunits have been so far identified in the human genome constitutes a major challenge in the development of ligands that specifically bind a certain channel subtype (2). The X-ray 3-dimensional structures of the human voltage dependent potassium channel Kv1.2 (3, 4), and other related channels (5) show that the α-subunits assemble as tetramers to form a pore comprising the turret loop, the P-helix and the selectivity filter, which contains the so-called K+ channel signature, a segment with the highly conserved sequence (XGYG) (6). Although other parts of the channel differ in structure, the pore-forming region is highly homologous among members of the Kv family. In particular, members of the Kv1.x subfamily share >50% identity in the pore-forming region thus rendering these targets particularly challenging to differentiate. Naturally occurring peptides from poisonous animals as well as synthetic compounds have been reported as inhibitors for Kv1.x channels (7). However, despite the high affinity of some of these inhibitors, none makes full use of the 4-fold symmetry of the homotetrameric channels.
Here, we report a family of ligands for Kv1.x channels based on calix[4]arenes. This classical building block of supramolecular chemistry has recently been used as a platform for multivalent ligands to efficiently bind proteins and nucleotides (8), but their application as ligands for protein assemblies of similar symmetry has been described only recently by Gordo et al. (9). Along these design principles, Gradl et al. reported that water-soluble ligands with 4-fold symmetry and endowed with short positively charged peptide arms anchored to a flat, rigid tetraphenylporphyrin core efficiently blocked the Kv1.3 channel in a reversible but nonspecific way (10). Although porphyrins fulfill the symmetry requirement, they are flat scaffolds that do not ideally complement the conical pore entrance of the Kv channels. In fact, solid-state NMR and molecular dynamics have shown that the porphyrins do not sit like a lid on top of the channel, which was the original model, but are oriented in parallel to the channel axis, with only 1 positively charged side arm actually interacting with the selectivity filter (Ader et al.) (11).
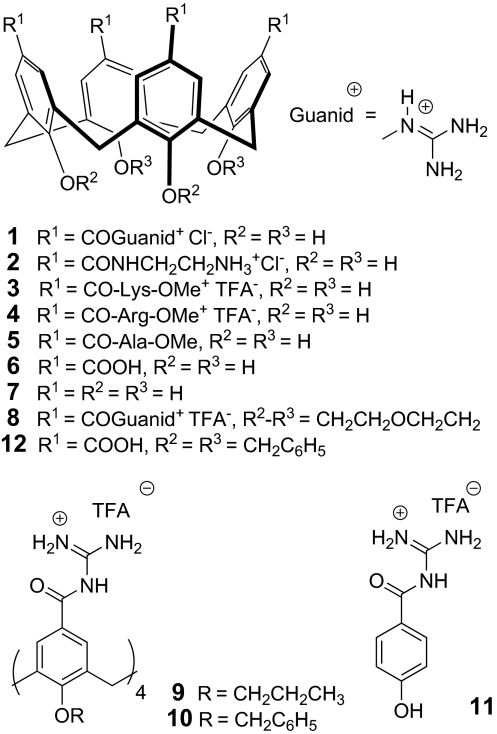
Unlike porphyrins, calix[4]arenes display conical shapes that would likely complement the shape of the outer vestibule of a Kv channel. Thus, ligands 1–8 were synthesized and evaluated (Fig. 1). Structures 1–7 are free-OH calix[4]arenes, to avoid steric hindrance at the entrance of the channel, whereas 8 contains 2 short loops that cannot enter the channel simultaneously but could allow the molecule to adjust in a slightly distorted way, which cannot be possible with longer or less-structured side chains, such as in 9 and 10. Additionally, cationic substituents (guanidines) were attached to the upper rim of the calix[4]arenes to ideally complement the mainly negatively charged extracellular surface groups of the channel at the “turret loop,” specifically residue Asp-379 in Kv1.2, thus improving binding affinities and selectivities by both electrostatic interactions and hydrogen bonding.
Fig. 1.
Ligand structures. Chemical formulae of calix[4]arenes 1–10 and phenol 11 studied as channel blockers.
Results and Discussion
Synthesis.
The synthesis of OH-free calix[4]arenes was based on the common tetracarboxylic acid precursor 12, allowing an easy introduction of the guanidine function, as well as any amino acid derivative or peptide chain by simple amide formation. Thus, compound 12 was guanidilated by following procedures by Schmuck et al. (12), followed by simultaneous debenzylation and guanidine deprotection to afford 1. Cationic ligands 2–4 and the neutral compound 5 were obtained from an activated O-succinimidoyl intermediate, followed by deprotection [supporting information (SI) Text and Fig. S1A]. The tetraanionic negative control 6 resulted from O-debenzylation of 12. O-alkyl calix[4]arenes 8–10 were synthesized by similar procedures from the corresponding carboxylic acids (Fig. S1B). Finally, phenol derivative 11 was selected as a non-calix[4]arene negative control and readily prepared in 2 steps from 4-benzyloxybenzoic acid.
Molecular Dynamics.
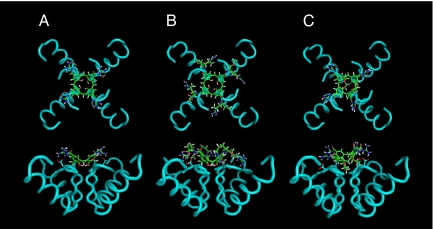
Docking studies employing the coordinates of the crystal structure of the Kv1.2 potassium channel (3) indicated that the conical platform of OH-free calix[4]arenes was best suited to deeply penetrate the extracellular outer vestibule of the channel (Fig. 2). Protein–ligand stabilities were assessed by analyzing the trajectories obtained in 1,500-ps molecular dynamics simulations in vacuo at 300 K with positional restraints. Additional details are provided in Methods. To evaluate protein–ligand binding affinity, we computed both the Xscore empirical scoring function and the variation in the guanidinium–carboxylate (Asp-379) distances versus time. Xscore was used to evaluate, for each trajectory, 128 structures collected every 12 ps. The values obtained for the 3 calixarenes were very similar and only slightly fluctuated during the molecular dynamics simulations. The average Xscore value of all of the complexes formed with 1 was 6.65 (see Fig. S2). The complexes in which 4 and 8 intervened had values of 6.30 and 6.32, respectively. This demonstrated high and similar protein–ligand affinity in the 3 cases. To estimate the variation of the hydrogen bond guanidinium–carboxylate distances we measured all of the NH–OC distances at every step. The 2 values corresponding to a same guanidinium–carboxylate interaction were averaged. Thus, the result depended on the fine orientation of both interactions. Comparison of the plots versus time (see Figs. S3–S5) pointed out a slightly larger fluctuation of the hydrogen bond distances in 1. In this ligand, the 4 interactions oscillated between 1.6 and 2.4 Å during the whole trajectory. In compound 4, the 4 of them were highly fixed at a distance of 1.6–1.7 Å. Three of the interactions in compound 8 were maintained at this distance, but the remaining highly fluctuated between values of 1.6 and >3 Å (Fig. S5). This was due to the tilted disposition of the calixarene on the Kv1.2 surface, which caused one of the guanidinium groups of the calixarene to be not as close to the corresponding aspartate. Thus, 4 was the ligand with most favorable hydrogen bonding distances, followed by 8. In any case, the 3 compounds showed to maintain all of the guanidinium–carboxylate groups at standard hydrogen bonding distances.
Fig. 2.
Molecular dynamics. Top and side wire frame views of calix[4]arenes 1 (A), 4 (B), and 8 (C) docked into the X-ray structure of the human voltage dependent Kv1.2 potassium channel. Only the backbone helices of the α-subunits are shown. The Asp-379 side chains are represented, showing the 4-ion-pair, hydrogen-bonded contacts with the guanidinium residues of the calix[4]arene. Structures were optimized in vacuo at 300 K applying positional restraints to Kv1.2.
In conclusion, the 3 ligands bind the protein with high affinity by establishing 4 hydrogen bonds and by maintaining the lower rim embedded in the channel pore. The 2 crown ether bridges do not avoid the interaction of 8 with Kv1.2 but cause the ligand to be slightly twisted with respect to 1 and 4.
Inhibition of Kv1 Channels by Calix[4]arene Compounds.
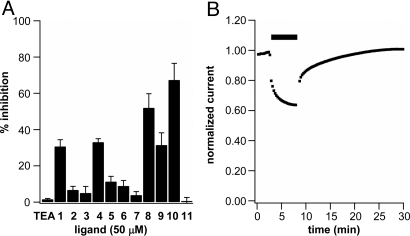
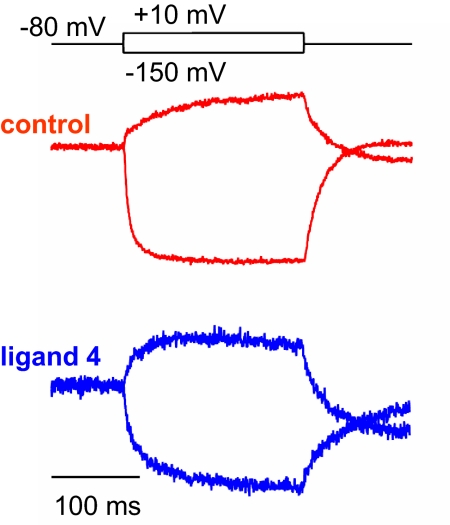
The interaction of the calix[4]arene compounds with potassium channels was investigated by using electrophysiological methods on Xenopus laevis oocytes expressing the Shaker potassium channel, a prototypical member of the Kv1.x subfamily. Ionic currents were measured in cells expressing the Shaker channel in the presence of each calix[4]arene compound. The percentage inhibition elicited by application of 50 μM compounds 1–11 was compared with that induced by the pore blocker tetraethylammonium (TEA) (13) and these are summarized in Fig. 3A. Fig. 3B shows the typical reversible inhibition response by 50 μM compound 4.
Fig. 3.
Electrophysiological screen of calix[4]arene ligands. (A) Percentage of inhibition elicited by application of 50 μM compounds 1–11 and tetraethylammonium cation (TEA) to Xenopus laevis oocytes expressing the Shaker channel. (n ≥ 4 for each condition; error bars indicate SEM). (B) Reversible reduction of Shaker ionic current in response to application of 50 μM ligand 4 (percentage of inhibition vs. time). Black bar indicates time of exposure to ligand 4. Note that inhibition and recovery occur in 2 distinct phases.
At 50 μM, compounds 1–11 caused different levels of reduction in Shaker ionic current, which in most cases was reversible: Compounds functionalized with carboxylates, ammonium, or neutral functional groups at the upper rim did not exert significant reduction in Shaker ionic current. On the contrary, ligands 1 and 4 containing guanidinium groups at the upper rim of a free-OH calix[4]arene scaffold, aimed at interacting with negatively charged residues and hydrogen bonding in the turret loop, proved promising ligands, as anticipated by the modeling studies. When applying O-alkyl derivatives 9 and 10 to oocytes expressing the Shaker channel, decreases in ionic current were also seen, but these effects were not reversible, and cell health was visually compromised. Lower-rim alkylation with hydrophobic residues result in amphiphilic structures that may act as detergents for oocytes' integrity and might cause nonspecific interactions with the membrane lipids or other hydrophobic portions of the Shaker channel that could not be restored by application of the aqueous recording solution used for the washout. Quite remarkably, but once again in agreement with the computer modeling studies, ligand 8, carrying 2 crown ether loops at the lower rim, caused reduction in Shaker ionic currents in a reversible manner and even to a larger extent than the free-OH analogues 1 and 4. It is known that crown ether bridges at vicinal lower-rim positions in a calix[4]arene strongly contribute to keeping the conical shape of the macrocycle, preventing collapse into the so-called pinched conformations (14). The conical shape is also maintained in the free-OH derivatives, because of the cyclic array of hydrogen bonds. At the same time, the crown ether bridges on 8 do not create a severe steric effect, because the ligand can comfortably fit at the top of the channel in a slightly tilted orientation, as indicated by molecular modeling (Fig. 2).
Changes in the counteranions (either chloride or trifluoroacetic acid) had no measurable effect on the channel binding properties of the calix[4]arenes. Furthermore, the presence of <2% dimethyl sulfoxide (DMSO) in the bath solution was not harmful to the cells. Finally, the effect of the multivalency effect present in the calix[4]arene macrocycle was evidenced by the fact that a simple guanidilated phenol such as 11 showed no inhibition at 50 μM. Even at 4 times the concentration used for ligand 1, thus at the same effective concentration, phenol 11 induced a minor increase in current flow (Figs. S6 and S7).
Among the most active ligands (1, 4, and 8), ligand 1 had a limited solubility and ligand 8 with bulkier substituents on the lower rim did dock into the pore in a tilted orientation. Therefore, in the rest of our work, we focused on ligand 4.
Inhibition Includes Gating Modification.
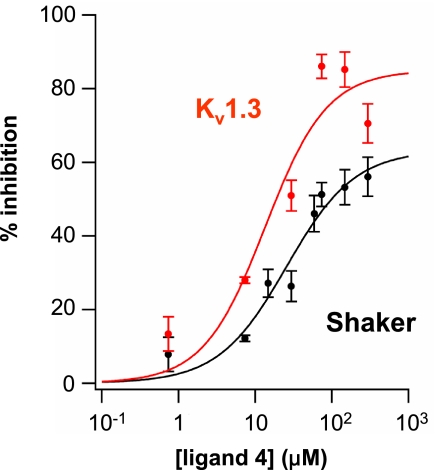
Ligand 4 showed a concentration-dependent inhibition of both Shaker and Kv1.3, 2 related members of the Kv1.x subfamily of ion channels (Fig. 4). Although the inhibition was greater in Kv1.3, the apparent affinity was similar for the 2 channels. The Hill coefficients for Shaker and Kv1.3 (1.0 ± 0.3 and 1.1 ± 0.7, respectively) were close to 1, fitting with the proposed molecular model in which a single calix[4]arene docks in the outer mouth of the pore and interacts with residue Asp-379 (Kv1.2 sequence numbering), which is shared by all Kv1.x subfamily members. Even at saturating concentrations of calix[4]arene 4, Shaker ionic current was not completely inhibited (Fig. 4), suggesting, an incomplete occlusion of the pore.
Fig. 4.
Concentration-dependent effects of ligand 4. Inhibition curve on macroscopic ionic currents in X. laevis oocytes expressing Shaker channels (black circles) or Kv1.3 channels (red circles). (n ≥ 3 for each data point; error bars indicate SEM). Data fit to a single Boltzmann as described in Methods. Shaker: max = 63 ± 10, Kd = 25 ± 11 μM, h = 1.0 ± 0.3. Kv1.3: max = 85 ± 16, Kd = 14 ± 9 μM, h = 1.1 ± 0.7.
The effect of ligand 4 could not be entirely attributed to pore block. The rate of channel opening was slowed (Fig. S8) and the voltage dependence of channel opening was shifted in the positive direction (Fig. 5A) by ligand 4, suggesting that binding relatively stabilizes the resting state or an intermediate state preceding opening in the activation pathway. The onset of current inhibition and of the gating effect occurred in parallel, with reduction in current amplitude tracked by the slowing of channel opening (Fig. 5B and Fig. S9).
Fig. 5.
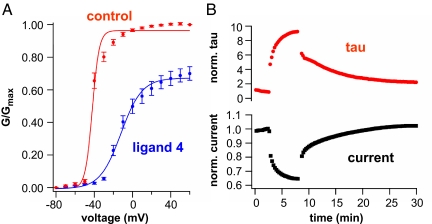
Electrophysiological analysis of kinetic effects of ligand 4. (A) Ligand 4 (50 μM) shifts the voltage dependence of channel opening to the right. Conductance measured from inward tails in 98 mM external K+. (n = 4, error bars indicate SEM). Data fit to a single Boltzmann as described in Methods. Control solution before exposure: max = 0.97 ± 0.04, Vmid = −42 ± 1 mV, m = 3.6 ± 0.9. During ligand 4 application: max = 0.68 ± 0.02, Vmid = −12 ± 1 mV, m = 11 ± 1.2. (B) Time course of reduction of current amplitude parallels slowing of channel opening after application of 50 μM ligand 4.
We wondered whether the gating effect reflects an influence of ligand 4 on the protein motions underlying voltage sensing. To address this we used voltage clamp fluorometry (15) to measure structural rearrangements in the S4 voltage sensor. Recordings were carried out in the W434F mutation, which locks the channel in the nonconducting P-type inactivated state, but permits free motion of S4 (16, 17). A single cysteine mutation was also introduced in the extracellular portion of the Shaker S4 segment (A359C) to allow for site-specific conjugation with tetramethylrhodammine-6-maleimide, an environmentally sensitive fluorophore, and monitor the movement of the voltage-sensing domain (18, 19). Ligand 4 (200 μM) was found to slow the rate of S4 motion at the top of the voltage-dependence curve where channels open (Fig. 6). This result is consistent with the observation that channel opening is slowed by the ligand (Figs. S8 and S9 and Fig. 5B).
Fig. 6.
Fluorescence analysis of kinetic effects of ligand 4. Representative normalized fluorescence traces showing Shaker voltage sensor movement in the absence (Upper) and presence (Lower) of ligand 4. A slowing in kinetics upon hyperpolarization and depolarization steps is seen upon application of ligand 4 (200 μM).
In conclusion, we have shown that guanidilated calix[4]arenes with free phenolic OH groups at the lower rim, as well as bis-crowned calix[4]arenes endowed with 4 acylguanidinium groups at their upper rims, constitute a versatile class of oocyte-benign, reversible ligands for the voltage-dependent K+ channel. Molecular modeling predicts an interaction with the turret loop and docking in the outer mouth of the pore of Kv channels for compounds 1, 4, and 8. A Hill coefficient of 1 for inhibition of the current is consistent with this model. The inhibition is incomplete at saturating concentrations, suggesting that the ligand does not completely block the pore. In addition to current inhibition, the ligand shifts the voltage dependence of gating and slows both the opening of the gate and the voltage-sensing motion of S4. These gating effects are consistent with recent observations of opening rearrangements in the turret (20).
Based on these results, a new generation of ligands remains to be studied, taking advantage of the divergent synthetic strategy for upper-rim functionalization of OH-free, the rigidity of bridged calix[4]arenes, and a study of the footprints of toxins like AgTx (21) and δ-dendrotoxin (22) on the surface of Shaker channel. Increasing the affinity of the channel-calix[4]arene interaction through further functionalization of 4 with suitable peptide chains attached to the upper rim is yet to be completed. Ultimately, the development of this class of compounds might lead to therapeutic agents against various ailments such as autoimmune disorders, diabetes, epilepsy, or cardiac diseases (23).
Methods
Molecular Dynamics.
Simulations were performed with Kv1.2 in the presence of 1, 4, and 8 at 300 K in vacuo. The starting tetramer Kv1.2 was obtained from the X-ray structure (Protein Data Bank ID code 2a79) (3), excluding chain A and part of chain B (32–131 residues). Thus, the integral membrane component of Kv1.2 was considered. To form each of the 3 complexes, the calixarene was docked manually on the surface of Kv1.2, the 4 guanidinium groups in the upper rim interacting through hydrogen bonds with the carboxylates of Asp-379 surrounding the channel pore. The lower rim of the calixarene lay on the middle of the pore. The Maestro interface (Schrodinger Inc.) was used to construct the complexes. Simulations were carried out by using Batchmin, the computational back-end of the Macromodel package (Schrodinger Inc.). Amber* force field was chosen to model the systems. Each complex was relaxed, following a protocol already reported by Magis et al. (24) in the design of Kv1.2 potassium channel blockers. The protocol consisted of an energy minimization, followed by molecular dynamics simulation applying positional restraints. The restraint force constants for residues located at a distance <7 Å from the ligand were set to 5 kcal mol−1Å−2 for the Cα and to 0.005 kcal mol−1Å−2 for nonhydrogen atoms other than the Cα. For residues located at a distance >7 Å from the ligand, the respective values were 50 kcal mol−1Å−2 and 5 kcal mol−1Å−2. No restraints were applied to the ligand. The Polak–Ribiere conjugate-gradient (PRCG) algorithm (25) was used to initially minimize the structures. Next, a molecular dynamics of 1,500 ps was carried out with a time step of 1 fs. The charges applied came from the force field. Van der Waals interactions were truncated at a cutoff distance of 7 Å. Electrostatic interactions were truncated at a distance of 12 Å. The cutoff distance for the hydrogen bonds was 4 Å. Temperature was maintained constant by using a standard velocity Verlet algorithm (26). Maestro and XSCORE (27) programs were used to analyze the trajectories.
Channel Expression.
The Shaker clone Sh H4 IR [Delta] (6–46) (N-type inactivation removed) in the pBluescript2 KS+ vector was used for all experiments unless otherwise noted. Any point mutations were made by performing site-directed mutagenesis by using the QuikChange protocol (Stratagene) from the Sh H4 IR clone.
The rKv1.3 construct was in the pGEM-HE vector. Fluorescence experiments were conducted with the Sh H4 IR W434F (nonconducting channel) A359C (maleimide attachment site) construct that contained 2 additional mutations (C245V and C462A) to prevent background labeling of native cysteines in the Shaker channel.
All DNA was confirmed by DNA sequencing. RNA was transcribed by using mMessage mMachine T7 transcription kit (Ambion). Surgically extracted X. laevis oocytes were injected with 0.5–4 ng of channel RNA (50 nL). The cells were incubated in ND-96 [96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 50 mg mL−1 gentamicin, 2.5 mM sodium pyruvate, and 5 mM Hepes (pH 7.6)] at 12–18 °C for 12–48 h before experiments.
Electrophysiology.
All electrophysiology experiments were conducted at room temperature. Two-electrode voltage-clamp (TEVC) experiments were performed by using a Dagan CA-1B amplifier (Dagan Corporation), Digidata-1440A board, and pClamp10 software. The oocytes were placed in a perfusion dish to allow for prewash, wash, and postwash recordings. All recordings were performed in ND-96 recording solution [96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM Hepes (pH 7.2)] unless otherwise noted. For each experiment, the oocyte membrane voltage was held at −80 mV, followed by a continuous train of 100- or 300-ms pulses to +20 mV every 15 s.
Current–Voltage (I–V) relationships (Fig. S7) were measured by using the same experimental conditions as above, except that the oocyte membrane voltage was held at −80 mV, followed by a depolarizing train of +10-mV steps lasting 300 ms and then returned to −80 mV. Current was measured from −80 mV to +60 mV. The resulting data were normalized to the current measured at +60 mV.
All compounds were dissolved in ND-96 recording solution to make a 500 μM stock solution. When needed, a maximum of 5% (vol/vol) DMSO was added to the stock solution to facilitate solubilization. For inhibition experiments, small quantities of the calixarene stock solutions were manually applied and mixed in a stagnant bath containing the oocyte. For all recordings, DMSO was present at levels <2% (vol/vol). Oocytes were bathed in the calixarene solution until the current measurements taken at +20 mV reached steady state. The compounds were washed away with ND-96 recording solution administered by using a gravity perfusion system at ≈80 mL/h.
All ionic current measurements taken at +20 mV were leak-subtracted by using the following formula:
The percentage of inhibition (B) was determined by using the following formula, using leak subtracted current measured at +20 mV for steady-state levels before and after addition of the calix[4]arene:
For dose–response experiments (Fig. 4), varying concentrations of ligand 4 were applied to oocytes expressing either the Shaker or rKv1.3 channel. Each experiment was repeated with at least 3 different oocytes, and the data were then averaged. Then, the average reduction in ionic current elicited by application of ligand 4 (B) was plotted as a function of the log of ligand 4 concentration. The plotted data were then fit by using Igor Pro software with the modified Hill equation:
where L represents the concentration of calix[4]arene used in the experiment, Kd the equilibrium dissociation constant, and h the Hill Coefficient.
Tail currents (Fig. 5A) were measured by using the I–V protocol described above with cells being incubated in a high-potassium ND-96 recording solution [98 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM Hepes (pH 7.2)]. Measurements taken from 4 different oocytes were averaged and inverted to obtain conductance (G) values and then normalized to the maximum conductance measured before the application of ligand 4 and then plotted as a function of the step voltage. The plotted data were then fit by using Igor Pro software with the modified Boltzmann equation:
where max represents the maximum G/Gmax value, V represents the step voltage, Vmid the midpoint voltage, and m the slope.
Voltage-clamp fluorometry experiments were conducted according to ref. 20. Briefly, oocytes were injected with 4 ng of Sh H4 IR W434F-A359C (C245V, C462A) cRNA. After 1 h, endogenous cysteines on the oocytes surface were treated with tetraglycine-maleimide for 20 min at 18 °C. Cells then were extensively washed with ND-96 and incubated at 18 °C for 3–4 days. On the day of experiments, oocytes were incubated with 12.5 μM tetramethylrhodamine-6-maleimide (Invitrogen) for 30 min at 0 °C and then extensively washed with ND-96. After labeling, cells were kept at 12 °C. Recordings were done in ND-96 recording solution at room temperature. Oocytes were voltage clamped at −80 mV, and fluorescence changes were evoked by a series of +20-mV steps from −150 mV to +50 mV (300-ms step length).
Supplementary Material
Acknowledgments.
Thanks to Dr. Francesco Tombola for assistance with experimental design and data analysis and Drs. Maximilian Ulbrich and Harald Janovjak for help with data analysis. This work was supported by Spanish Ministry of Science and Education (MEC) Project CTQ2005-08948-C02-01/BQU, Consolider Ingenio 2010 Grant CSD2006-0003, the Institute of Chemical Research of Catalonia (ICIQ) Foundation, and National Institutes of Health Grant 5R01NS035549 (to E.Y.I.). V.M. thanks MEC and the ICIQ Foundation for predoctoral fellowships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813396106/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Ashcroft FM. Ion Channels and Disease: Channelopathies. San Diego: Academic; 2000. [Google Scholar]
- 3.Long SB, Campbell EB, MacKinnon R. Crystal structure of mammalian voltage-dependent shaker family K+ channel. Science. 2005;309:897–902. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 4.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like. Nature. 2007;450:376–383. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 5.Jiang YX, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 6.MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 7.Wulff H, Zhorov BS. Channel modulators for the treatment of neurological disorders and autoimmune diseases. Chem Rev. 2008;108:1744–1773. doi: 10.1021/cr078234p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldini L, Casnati A, Sansone F, Ungaro R. Calixarene-based multivalent ligands. Chem Soc Rev. 2007;36:254–266. doi: 10.1039/b603082n. [DOI] [PubMed] [Google Scholar]
- 9.Gordo S, et al. Stability and structural recovery of the tetramerization domain of p53–R337H mutant induced by a designed templating ligand. Proc Natl Acad Sci USA. 2008;105:16426–16431. doi: 10.1073/pnas.0805658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradl SN, Felix JP, Isacoff EY, Garcia ML, Trauner D. Protein surface recognition by rational design: Nanomolar ligands for potassium channels. J Am Chem Soc. 2003;125:12668–12669. doi: 10.1021/ja036155z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ader C, et al. A structural link between inactivation and block of a K+ channel. Nat Struct Mol Biol. 2008;15:605–612. doi: 10.1038/nsmb.1430. [DOI] [PubMed] [Google Scholar]
- 12.Schmuck C, Machon U. Amino acid binding by 2-(guanidiniocarbonyl)pyridines in aqueous solvents: A comparative binding study correlating complex stability with stereolectronic factors. Chem Eur J. 2005;11:1109–1118. doi: 10.1002/chem.200400652. [DOI] [PubMed] [Google Scholar]
- 13.Blaustein RO, Cole PA, Williams C, Miller C. Tethered blockers as molecular ‘tape-measures’ for a voltage-gated K+ channel. Nat Struct Biol. 2000;7:309–311. doi: 10.1038/74076. [DOI] [PubMed] [Google Scholar]
- 14.Arduini A, et al. Calix[4]arenes blocked in a rigid cone conformation by selective functionalization at the lower rim. J Org Chem. 1995;60:1454–1457. [Google Scholar]
- 15.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 16.Perozo E, MacKinnon R, Bezanilla F, Stefani E. Gating currents from a noncondiucting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 1993;11:353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 17.Loots E, Isacoff EY. Protein rearrangements underlying slow inactivation of the Shaker K+ channel. J Gen Physiol. 1998;112:377–389. doi: 10.1085/jgp.112.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 19.Mannuzzu LM, Isacoff EY. Independence and cooperativity in rearrangements of a potassium channel voltage sensor revealed by single subunit fluorescence. J Gen Physiol. 2000;115:257–268. doi: 10.1085/jgp.115.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathak MM, et al. Closing in on the resting state of the Shaker K(+) channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Gross A, McKinnon R. Agitoxin footprinting the Shaker potassium channel pore. Neuron. 1996;16:399–406. doi: 10.1016/s0896-6273(00)80057-4. [DOI] [PubMed] [Google Scholar]
- 22.Imredy J, Chen C, McKinnon R. A snake toxin inhibitor of inward rectifier potassium channel ROMK1. Biochemistry. 1998;37:14867–14874. doi: 10.1021/bi980929k. [DOI] [PubMed] [Google Scholar]
- 23.Coghlan MJ, Carroll WA, Gopalakrishnan M. Recent developments in the biology and medicinal chemistry of potassium channel modulators: Update from a decade of progress. J Med Chem. 2001;44:1627–1653. doi: 10.1021/jm000484+. [DOI] [PubMed] [Google Scholar]
- 24.Magis C, et al. Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers. J Am Chem Soc. 2006;128:16190–16205. doi: 10.1021/ja0646491. [DOI] [PubMed] [Google Scholar]
- 25.Polak E, Ribiere G. Note on convergence of conjugate direction methods. Rev Fr Inf Rech Oper. 1969;16:35–43. [Google Scholar]
- 26.Swope WC, Andersen HC, Berens PH, Wilson KR. A computer-simulation method for the calculation of equilibrium-constants for the formation of physical clusters of molecules–Application to small water clusters. J Chem Phys. 1982;76:637–649. [Google Scholar]
- 27.Wang R, Lu Y, Fang X, Wang S. An extensive test of 14 scoring functions using the PDB bind refined set of 800 protein–ligand complexes. J Chem Inf Comput Sci. 2004;44:2114–2125. doi: 10.1021/ci049733j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.