Abstract
The xeroderma pigmentosum complementation group E (XP-E) gene product damaged-DNA binding protein 2 (DDB2) plays important roles in nucleotide excision repair (NER). Previously, we showed that DDB2 participates in NER by regulating the level of p21Waf1/Cip1. Here we show that the p21Waf1/Cip1 -regulatory function of DDB2 plays a central role in defining the response (apoptosis or arrest) to DNA damage. The DDB2-deficient cells are resistant to apoptosis in response to a variety of DNA-damaging agents, despite activation of p53 and the pro-apoptotic genes. Instead, these cells undergo cell cycle arrest. Also, the DDB2-deficient cells are resistant to E2F1-induced apoptosis. The resistance to apoptosis of the DDB2-deficient cells is caused by an increased accumulation of p21Waf1/Cip1 after DNA damage. We provide evidence that DDB2 targets p21Waf1/Cip1 for proteolysis. The resistance to apoptosis in DDB2-deficient cells also involves Mdm2 in a manner that is distinct from the p53-regulatory activity of Mdm2. Our results provide evidence for a new regulatory loop involving the NER protein DDB2, Mdm2, and p21Waf1/Cip1 that is critical in deciding cell fate (apoptosis or arrest) upon DNA damage.
Keywords: apoptosis, cell cycle, DDB2, Mdm2, p21
Mutation in the gene encoding damaged-DNA binding protein 2 (DDB2) is linked to xeroderma pigmentosum (XP, complementation group E) (1, 2). XP is a human genetic disorder characterized by sun sensitivity, DNA repair deficiency, and high risk of skin tumor development. DDB2 knockout (DDB2−/−) mice exhibit enhanced skin cancer susceptibility in response to chronic ultraviolet (UV) radiation (3–5), providing further evidence of the role of DDB2 in inhibition of UV-induced skin tumorogenesis. Cells from XP patients exhibit deficiency in the nucleotide excision repair (NER), a major DNA repair mechanism that mends UV-induced DNA damage.
Several models for the NER function of DDB2 have been proposed; all of those models are related to the E3 ligase complexes of Cul4A or Cul4B that bind to DDB2 through the adapter protein DDB1. Two groups of researchers have suggested that the DDB2-associated E3 ligase mono-ubiquitinates histones on damaged chromatin to remodel the chromatin structure and to support recruitment of the NER proteins onto damaged chromatin (6, 7). Another model contends that DDB2-containing ligase ubiquitinates the NER factor XP-C to activate NER (8). DDB2 and its proteolysis by Cul4A have been reported to be involved also in the recruitment of XP-C (9). DDB2 is degraded by Cul4A-DDB1 after UV irradiation (10). It has been suggested that the degradation of DDB2 allows recruitment of XP-C.
Recently we showed that, in murine cells, DDB2 could participate in NER indirectly by regulating the cellular levels of p21Waf1/Cip1 (11). In cells exposed to low doses of UV irradiation, DDB2 enhances nuclear accumulation of DDB1, which binds to a modified form of p53 phosphorylated at Ser-18 (p53S18P) and, together with Cul4A, targets it for degradation. In the DDB2−/− cells, the UV-induced accumulation of DDB1 is impaired, causing accumulation of p53S18P and higher expression of p21Waf1/Cip1 (11). P21Waf1/Cip1 was shown to directly interact with proliferation cell nuclear antigen (PCNA) and to inhibit NER, both in vitro and in vivo (12, 13). We showed that depletion of p21Waf1/Cip1 reversed the NER-deficient phenotype observed in the DDB2−/− mouse embryonic fibroblasts (MEFs), providing strong genetic evidence for a link between the NER activity of DDB2 and regulation of p21Waf1/Cip1 expression.
Regulation of p21Waf1/Cip1 by DDB2 is intriguing because, in human cells, both of these genes are transcriptionally induced by p53 (14, 15). The promoter region of the human DDB2 gene contains p53-binding sites; upon DNA damage, p53 directly binds to the DDB2 promoter to stimulate its expression (15). In murine cells, however, DDB2 is not activated by p53, as the promoter region lacks p53-binding site (15, 16).
Several studies have suggested a role of p21Waf1/Cip1 in inhibiting apoptosis (17, 18). The apoptosis-inhibitory function of p21Waf1/Cip1 poses a problem for cells after DNA damage because both pro-apoptotic pathway and p21Waf1/Cip1 are activated by p53. It has been shown that depletion of p21Waf1/Cip1 sensitizes cells to DNA damage induced apoptosis (19, 20). Thus, p21Waf1/Cip1 functions as a barrier and inhibits apoptosis after DNA damage. In this study, we show that the XP-E gene product DDB2 plays a critical role in attenuating the barrier of p21Waf1/Cip1 to allow apoptosis of cells exposed to DNA-damaging agents. Also, we identify a new role of Mdm2 in that process.
Results
DDB2-Deficient Cells Are Resistant to Apoptosis Induced by a Variety of DNA-Damaging Agents.
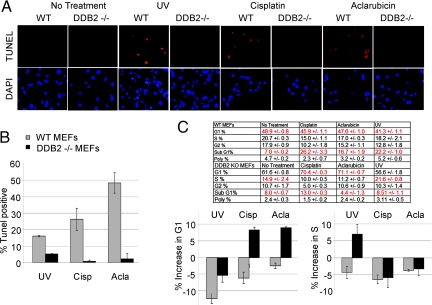
It was shown that the DDB2-deficient cells are defective in the UV-induced apoptosis (3, 21). We investigated whether the deficiency is specific to UV-induced apoptosis. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were used to analyze apoptosis in wild-type and DDB2−/− MEFs. Consistent with the published studies, we observed that our DDB2−/− MEFs are resistant to UV-induced apoptosis (Fig. 1 A and B). In addition to UV irradiation, we analyzed the effects of several other DNA-damaging agents for their ability to induce apoptosis in the wild-type MEFs (not shown). Of those, cisplatin and aclarubicin were found to be potent inducers of apoptosis in the wild-type MEFs. Interestingly, those drugs failed to induce apoptosis in the DDB2−/− MEFs (Fig. 1). The DDB2 −/− MEFs, instead of undergoing apoptosis, exhibited cell cycle arrest (Fig. 1C). Flow-cytometric analyses revealed that cisplatin and aclarubicin increased the G1 population in the DDB2−/− MEFs, whereas UV irradiation increased population of S-phase cells resulting from S-phase delay.
Fig. 1.
DDB2−/− MEFs exhibit cell cycle arrest and apoptosis defect upon DNA damage. (A) Wild-type or DDB2−/− MEFs were treated with UV-C (50 J/m2) or cisplatin (30 μm) or aclarubicin (0.5 μM) for 24 hours. After the treatments, the cells were subjected to TUNEL assay using the ApopTag Red In Situ Apoptosis Detection Kit and procedure provided by the manufacturer (Chemicon International). Average percentages of the TUNEL-positive nuclei from 10 different fields from two independent experiments are plotted (B). (C) After treatment with the DNA-damaging drugs, cells were subjected to flow-cytometric analyses. An average of the cell cycle distribution (including the SubG1 cells) from three different sets is shown.
We investigated whether the function of DDB2 in supporting apoptosis after DNA damage is conserved in human cells. HeLa cells expressing control shRNA or shRNA against DDB2 were compared for their ability to support apoptosis after treatments with DNA-damaging agents (Fig. S1). Flow-cytometric analyses of the control HeLa cells exhibited increase in population of subG1 cells, indicative of apoptosis, after treatments with UV or with aclarubicin or cisplatin (Fig. S1). The DDB2-shRNA expressing cells, on the other hand, did not show any significant increase in the subG1 population of cells. Instead, the DDB2 shRNA expressing cells exhibited S-phase delay (Fig. S1). These results show that DDB2 is required for apoptosis after DNA damage and that, in the absence of DDB2, cells undergo cell cycle arrest.
DDB2-Deficient Cells Support Efficient Activation of the p53-Induced Pro-Apoptotic Genes but Fail to Regulate Accumulation of p21Waf1/Cip1.
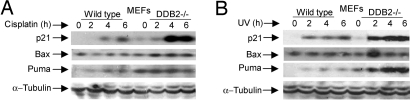
The lack of apoptosis in the DDB2−/− cells did not result from a deficiency in the expression of the pro-apoptotic genes that are induced by p53 upon DNA damage. We compared the wild-type and the DDB2−/− cells treated with UV or cisplatin to analyze expression of the p53-induced pro-apoptotic genes. The pro-apoptotic genes Puma and Bax were expressed at high levels in the DDB2−/− MEFs (Fig. 2 A and B). Moreover, both wild-type and DDB2−/− MEFs exhibited strong induction of p53 after treatments with the DNA-damaging agents (Fig. S2a). It is believed that, after DNA damage, p53 localizes to cytoplasm and mitochondria to activate function of Bax in promoting permeabilization of the outer membrane of mitochondria (22–24). Therefore, we fractionated cells to obtain cytoplasmic, mitochondrial, and nuclear fractions. There was no significant difference in the levels of p53 in those three fractions when wild-type and DDB2−/− cells were compared (Fig. S2a).
Fig. 2.
Expression of pro-apoptotic genes and accumulation of p53 and p21Waf1/Cip1 in DDB2-deficient cells. Wild-type or DDB2−/− MEFs were treated with cisplatin (30 μM) (A) or UV-C (50 J/m2) (B). Cells were harvested 2, 4, and 6 hours after treatment, followed by Western blot analysis for p21, Bax, PUMA, and α-Tubulin (as a loading control).
Previously, we showed that the DDB2−/− MEFs express p21Waf1/Cip1 at a higher level after low-dose UV irradiation because of accumulation of phospho-p53(Ser-18) (11). However, at a high-dose of UV irradiation, a condition used in this study, we did not expect any significant difference in the level of p21Waf1/Cip1 because p53 is equally stabilized in DDB2−/− and wild-type MEFs (Fig. S2a). Surprisingly, when we measured the level of p21Waf1/Cip1, the DDB2−/− MEFs exhibited significantly higher accumulation of p21Waf1/Cip1 (Fig. 2 A and B). The activation of ATR and the accumulation of P-(Ser-18)p53 were comparable (Fig. S2b). In addition, p21-mRNA was induced to a similar extent (Fig. S2c). Therefore, we investigated whether the DDB2-defcient cells are impaired in the decay of p21Waf1/Cip1. We investigated decay rates of p21Waf1/Cip1 in both DDB2−/− MEFs and in HeLa cells expressing DDB2-shRNA. The cells were treated with cycloheximide and the decay-rate of p21Waf1/Cip1 was measured. Clearly, the DDB2−/− MEFs (Fig. S3a), as well as the HeLa cells expressing the DDB2-shRNA (Fig. 3A), exhibited increased stability of p21Waf1/Cip1 compared with the wild-type cells.
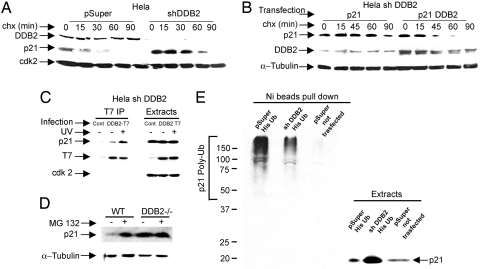
Fig. 3.
DDB2 targets p21Waf1/Cip1 for proteolysis. (A) HeLa cells expressing DDB2-shRNA (shDDB2) or control (pSuper) were treated with cycloheximide (50 μg/ml). At the indicated time points, cells were harvested and the extracts (0.25 mg) were analyzed for the levels of p21 by Western blot. (B) HeLa cells expressing DDB2-shRNA were transfected with plasmids expressing p21 and DDB2 (two plates) or p21 alone (two plates). Twelve hours after transfection, the cells in the two plates were pooled and equally divided into five plates. Twenty-four hours after re-plating, cells were treated with cycloheximide and were harvested at the indicated time points. Extracts were assayed for the levels of p21. (C) HeLa–shDDB2 cells were infected for 16 hours with adenovirus expressing T7-epitope–tagged DDB2. Cells were also treated with or without UV-C. The extracts (2 mg) were subjected to immunoprecipitation with T7-epitope antibody. The immunoprecipitates were analyzed for the presence of p21 by Western blot. (D) Wild-type or DDB2-deficient cells were treated with MG132 for 4 hours, followed by harvesting, extraction, and Western blot analysis for p21. (E) HeLa cells (pSuper) or HeLa cells expressing DDB2-shRNA were transfected with a plasmid expressing His-tagged ubiquitin. The ubiquitinated proteins were isolated following a previously described procedure (11) and then subjected to Western blot assay with p21 antibody.
The DDB2-associated protein DDB1 has been implicated in the proteolysis of p21Waf1/Cip1 (25, 26). To determine whether DDB2 could increase the decay-rate of p21Waf1/Cip1, HeLa cells were transfected with both DDB2 and p21Waf1/Cip1 expression plasmids. The cells were treated with cycloheximide and the decay of p21Waf1/Cip1 was analyzed. We observed that expression of DDB2 reduced the half-life of p21Waf1/Cip1 (Fig. 3B). To investigate an interaction, we expressed T7-epitope tagged DDB2 in the HeLa cells expressing DDB2-shRNA. Immunoprecipitation of DDB2 with T7-antibody co-immunoprecipitated p21 (Fig. 3C). The p21–co-immunoprecipitation was detected also with DNase-treated extracts (Fig. S3b). Moreover, MG132 stabilized p21 in DDB2-containing cells but not in DDB2-deficient cells (Fig. 3D). Next, we compared ubiquitination of p21 in DDB2-containing and DDB2-depleted cells by transfecting a plasmid that expresses His-tagged ubiquitin. The ubiquitinated proteins were isolated by purifying the extracts through Ni-Agarose columns. The bound proteins were analyzed in Western blot assay with p21-atibody to detect polyubiquitinated p21. We observed that, whereas the DDB2-defiient cells accumulated p21, there was a significant reduction in the extent of ubiquitination (Fig. 3E). Together, the observations suggest that DDB2 targets p21Waf1/Cip1 for proteolysis through the ubiquitin–proteasome pathway.
Deletion of p21Waf1/Cip1 Restores Apoptosis in DDB2−/− Cells.
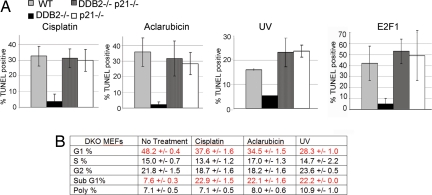
Next, we investigated whether the deficiency in apoptosis in the DDB2−/− cells is related to the high-level accumulation of p21Waf1/Cip1. We observed that the accumulation of p21Waf1/Cip1 in the DDB2−/− cells caused a delay in S-phase entry, and the delay was reversed in the double knockout DDB2−/−p21−/− cells (Fig. S4a). To determine whether the increased level of p21Waf1/Cip1 was the cause of resistance to apoptosis, the DDB2−/−p21−/− double knockout MEFs were compared with DDB2−/− and p21−/− single knockout MEFs for their abilities to undergo apoptosis upon treatments with UV, cisplatin, or aclarubicin. The MEFs were treated with the DNA-damaging agents. Eighteen hours after treatment, the cells were fixed and subjected to apoptotic TUNEL assays (Fig. 4A and Fig. S5). Deletion of p21Waf1/Cip1 restored apoptosis induced by DNA damage in the DDB2−/− cells (Fig. 4; also see PARP cleavage in Fig. S4c). Similar results were obtained with IR treated cells (Fig. S4b). Deletion of p21Waf1/Cip1 also eliminated the cell cycle delay observed in the DDB2−/− MEFs after treatments with the DNA-damaging agents (Fig. 4B). Together these results suggest that DDB2 is required for apoptosis induced by DNA damage, and that DDB2 participates in apoptosis and cell cycle progression by down-regulating the level of p21Waf1/Cip1.
Fig. 4.
Deletion of p21Waf1/Cip1 restores apoptosis in DDB2−/− MEFs. (A) WT DDB2−/− MEFs, p21−/−MEFs, or DDB2−/−p21−/− MEFs were treated with UV-C, cisplatin, or aclarubicin or were infected with adenovirus-expressing E2F1 or virus-expressing lac Z. Twenty-four hours after the treatments, the cells were subjected to TUNEL assay. Averages of the TUNEL positive nuclei from 10 different fields were plotted. (B) The DDB2−/−p21−/− MEFs were subjected to flow-cytometric analysis after treatments with UV-C, cisplatin, or aclarubicin. Averages of the cell cycle distribution from three different sets are shown.
We determined the effect of E2F1, which is a pro-apoptotic gene (27, 28). We used recombinant adenovirus expressing E2F1 to infect the MEFs. Apoptosis was analyzed 24 hours after infection with E2F1-expressing virus. As expected, expression of E2F1 in the wild-type MEFs induced apoptosis. However, E2F1 failed to induce apoptosis in the DDB2−/− MEFs (Fig. 4A). More interestingly, the E2F1-induced apoptosis was restored in the DDB2−/−p21−/− MEFs. Thus, the high level of p21Waf1/Cip1 in the DDB2−/− MEFs also protects the cells from E2F1-induced apoptosis.
Depletion of Mdm2, but Not Inhibition of p53–Mdm2 Interaction, Restores Apoptosis in DDB2-Deficient Cells.
The Cdk-inhibitory function of p21Waf1/Cip1 has been linked to its ability to inhibit apoptosis (29). Interestingly, a recent study indicated that, in cells treated with DNA-damaging agents, efficient inhibition of Cdk2 by p21Waf1/Cip1 requires Mdm2 (30). It was shown that inhibition of the Mdm2-p53 interaction by nutlin-3 and inhibition of Mdm2 levels by Mdm2-siRNA had similar effects on the levels of p53 and p21Waf1/Cip1 expression, but the Cdk2-inhibitory activity of the induced p21Waf1/Cip1 was different. In Mdm2-siRNA treated cells Cdk2-inhibition was inefficient, causing an inefficient arrest of cells after treatment with doxorubicin (30). The nutlin-3–treated cells, on the other hand, exhibited efficient cell cycle arrest in response to doxorubicin. Based on those observations, it was suggested that Mdm2 possesses a Cdk-inhibitory function that is independent of its p53-regulatory activity. If Mdm2 is required for the Cdk2-inhibitory function of p21Waf1/Cip1, we predicted that in the absence of Mdm2 the barrier of apoptosis imposed by p21Waf1/Cip1 would be weak, and the cells would undergo apoptosis without DDB2.
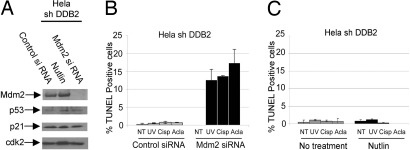
We used HeLa cells deficient in DDB2 expression (stably expressing DDB2-shRNA) because HeLa cells contain very little p53, as it is degraded by the HPV E6 protein. Moreover, we observed that treatment of the HeLa-DDB2shRNA cells with nutlin-3 or Mdm2-siRNA had very little effect on the levels of p53 or its downstream target p21 (Fig. 5A), suggesting that Mdm2 does not regulate p53 in these cells. We investigated whether a depletion of Mdm2 would restore apoptosis. We used Mdm2-siRNA to knockdown Mdm2 in the HeLa cells expressing DDB2-shRNA. Clearly, the knockdown of Mdm2 in DDB2-deficient cells caused significant increase in apoptosis induced by the DNA-damaging agents (Fig. 5B). Mdm2-siRNA had only a slight effect on the apoptosis in the control HeLa cells (Fig. S6b). Nutlin-3 treatment, on the other hand, had very little effect on restoring apoptosis in the DDB2-deficient cells (Fig. 5C). These results identify a new apoptosis-inhibitory activity of Mdm2 that is distinct from its p53-regulatory activity. Thus, both p21Waf1/Cip1 and Mdm2 are required for the inhibition of apoptosis in the DDB2-deficient cells.
Fig. 5.
Mdm2 inhibits apoptosis in DDB2-deficient cells. (A) HeLa cells expressing DDB2-shRNA were transfected with control or Mdm2-siRNA or treated with nutlin-3 (48 hours). Extracts of the cells were assayed for Mdm2, p53, p21, and Cdk2. (B) DDB2-shRNA cells were treated with UV-C (50 J/m2), cisplatin (30 μM), or aclarubicin (0.5 μM), and after 18 hours the cells were subjected to TUNEL assay using the ApopTag Fluorescein In Situ Apoptosis Detection Kit. Averages of TUNEL-positive nuclei from 10 different fields were plotted. (C) HeLa cells expressing DDB2-shRNA were not treated or treated with nutlin-3 for 48 hours, followed by TUNEL assay using the ApopTag Fluorescein In Situ Apoptosis Detection Kit. Averages of TUNEL-positive nuclei from 10 different fields were plotted.
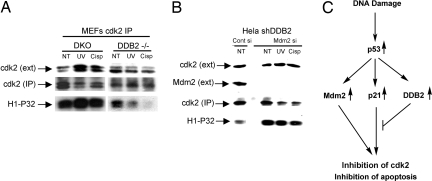
Next, we compared the activity of Cdk2 in DDB2−/− and DDB2−/−p21−/− MEFs after treatments with the DNA-damaging agents. Cdk2 was immunoprecipitated, and the immunoprecipitates were used in histone-H1 phosphorylation assay in the presence of γ-P32 ATP. As expected from p21Waf1/Cip1 accumulation in the DDB2−/− cells, treatments with UV or cisplatin caused severe reduction in the Cdk2-activity, which was not observed with the immunoprecipitates from the DDB2−/−p21−/− cells (Fig. 6A). To further confirm that the apoptosis in the DDB2−/−p21−/− cells resulted from increased Cdk2 activity, we treated those cells with roscovitine, an inhibitor of the Cdk-kinases. Roscovitine inhibited apoptosis in the DDB2−/−p21−/− cells and caused them to undergo cell cycle arrest (Fig. S7). Also, we measured Cdk2 activity after knockdown of Mdm2. Knockdown of Mdm2 caused a significant increase in the Cdk2 activity in cells expressing DDB2-shRNA, and the high activity was maintained after treatments with DNA-damaging agents (Fig. 6B). Together, the results are consistent with the notion that both p21Waf1/Cip1 and Mdm2 are involved in inhibiting Cdk2 after DNA damage, and that DDB2 overcomes the inhibitory effect, at least partly, by down-regulating p21Waf1/Cip1 (Fig. 6C).
Fig. 6.
Stimulation of Cdk2-activity in DDB2-deficient cells by depletion of p21Waf1/Cip1 or Mdm2. (A) Extracts from DDB2−/− MEFs and DDB2−/−p21−/− MEFs were compared for Cdk2 activity after treatments with DNA-damaging agents. (B) DDB2-shRNA expressing HeLa cells were transfected with Mdm2-siRNA. The extracts were compared for Cdk2-activity (B). In both experiments, the Cdk2 activity was measured by assaying for histone-H1 phosphorylation (38). (C) Model for activation of apoptosis by DDB2.
Discussion
The work presented here is significant in several ways. First, we show that the NER factor DDB2 is a key determinant in deciding fate of a cell after DNA damage. In the presence of DDB2 expression cells undergo apoptosis, whereas in the absence of DDB2 expression cells undergo cell cycle arrest. Second, we provide genetic and biochemical evidence that DDB2 supports apoptosis by regulating the levels of p21Waf1/Cip1. Moreover, using a specific inhibitor of p53-Mdm2 interaction, we provide evidence for a role of Mdm2 in inhibiting apoptosis. We suggest a model in which, after DNA damage, Mdm2 and p21Waf1/Cip1, which are induced by p53, act together to inhibit apoptosis. DDB2, which is induced also by p53 in human cells, lowers the level of p21Waf1/Cip1 to overcome the inhibition of apoptosis (Fig. 6C).
Previously we showed that, in low-dose UV–irradiated cells, DDB2 regulates the stability of a modified form of p53 that is phosphorylated at the Ser-15/18 residue (11). In low-dose UV–irradiated cells, a small population of p53 exists in Ser-1–5/18 phopshorylated form, which is specifically targeted for proteolysis by the Cul4-DDB1 E3 ligase (11). In the absence of DDB2, the phoshorylated p53 accumulates leading to higher mRNA-expression of p21Waf1/Cip1. Thus, by lowering the level of p53 that is phosphorylated at Ser-15/18, DDB2 regulates new synthesis of p21Waf1/Cip1 (11). In this study, we used a much higher dose of UV irradiation or other DNA-damaging agents that activate p53 to a very high level irrespective of the presence of DDB2. Under these conditions, the pro-apoptotic genes were turned on in both wild-type and DDB2-deficient cells. The DDB2-deficient cells exhibited a significantly higher accumulation of p21Waf1/Cip1. We provided evidence that the high accumulation of p21Waf1/Cip1 in DDB2-deficient cells is a result of deficiency in p21Waf1/Cip1 proteolysis, suggesting that DDB2 also regulates p21Waf1/Cip1 at the protein level. We observed that DDB2 associates with p21Waf1/Cip1 and that expression of DDB2 reduces the half-life of p21Waf1/Cip1. Although several mechanisms of p21Waf1/Cip1 regulation have been characterized (31–33), our results provide genetic evidence for the DDB2-mediated regulation of p21Waf1/Cip1 in determining the outcome of DNA damage response. That is particularly significant because expression of DDB2 in human cells is induced after DNA damage (15).
We observed similar regulation of p21Waf1/Cip1 by DDB2 in both murine and human cells. However, there is a difference in DDB2 expression between human and murine cells. In murine cells, DDB2 is constitutively expressed, whereas in human cells DDB2 expression is dependent upon p53 (15). It is interesting that, in human cells, p53 induces expression of DDB2 to regulate the level of p21Waf1/Cip1, which is induced also by p53 (14). Therefore, it would appear that in human cells the function of DDB2 is tightly linked o the p53-pathway and that DDB2 is a significant regulator of p21Waf1/Cip1 only after DNA damage or after induction by p53, whereas in murine cells DDB2 is constitutively available for regulation of p21Waf1/Cip1. The reason behind the evolutionary change from murine to human cells with regard to regulation of DDB2 expression is not obvious, because the defects in the DDB2−/− MEFs are mostly reversed by deletion of p21Waf1/Cip1. It is possible that, in murine cells, DDB2-mediated regulation of p21Waf1/Cip1 is important also in the absence of DNA damage.
The mechanisms by which p21Waf1/Cip1 inhibits apoptosis have been studied extensively (17, 18, 29). It was shown that high levels of p21Waf1/Cip1 inhibited activation of caspases, a process believed to depend upon active Cdks (29). A recent study indicated that, in cells treated with DNA-damaging agents, efficient inhibition of Cdk-kinase by p21Waf1/Cip1 involves Mdm2 (30). That function of Mdm2 is independent of its interaction with p53 because Mdm2 could support Cdk-inhibition by p21Waf1/Cip1 in the presence of nutlin-3, a drug that specifically disrupts the interaction between p53 and Mdm2. It is noteworthy that a previous study identified two growth-inhibitory domains in Mdm2 that could inhibit the G1–S transition (35, 36). Nevertheless we predicted that, if Mdm2 is required for p21Waf1/Cip1-mediated inhibition of Cdk and inhibition of the cell cycle progression, depletion of Mdm2 would restore apoptosis in the DDB2-deficient cells after DNA damage. Consistent with that prediction, we observed that siRNA-mediated depletion of Mdm2 could restore apoptosis in the DDB2-deficient HeLa cells. Moreover, nutlin-3 treatment had no effect on restoring apoptosis in the DDB2-deficient cells. Mdm2 is expected to inhibit apoptosis by regulating p53, but that does not explain our observation that nutlin-3 failed to support apoptosis in the DDB2-deficient cells. Therefore, the observations identify a new function of Mdm2 in inhibiting apoptosis that is likely to involve the same pathway by which p21Waf1/Cip1 inhibits apoptosis.
Materials and Methods
MEFs and Cell Line.
MEFs (11) were generated from 13.5-day-old embryos and were grown in Dulbecco modified Eagle's medium containing 10% fetal bovine serum (FBS). Control and DDB2 short hairpin RNA (shRNA) stable cell lines have been described before (11).
Drug Treatment and UV Irradiation.
Cisplatin (Sigma, P4394) was used at a final concentration of 30 mM, and aclarubicin (Sigma, A8959) was used at a final concentration of 0.5 mM. Cells were treated with Nutlin-3 for 48 hours (Cayman Chemical) at a final concentration of 2.5 mM. UV irradiation (50 J/m2) of cells was carried out with a Stratalinker (Fisher Scientific) adjusted to UV-C irradiation.
Western Blot Analysis, Decay Rates, and Immunoprecipitation.
Western blot, decay rate, and immunoprecipitation were performed using previously described procedures (11).
Cell Fractionation.
Cells were washed with phosphate-buffered saline (PBS) and were then treated with cisplatin or aclarubicin or UV irradiated. Eighteen hours after treatment, cells were harvested and fractioned following a previously described procedure (37).
Assays for Apoptois.
Cells were grown on glass cover slips and treated with cisplatin, aclarubicin, or UV, or were infected with recombinant adenovirus expressing E2F1 at 50 PFU/cell. Eighteen hours after treatment or infection, cells were fixed in 1% paraformaldehyde pH 7.4. The ApopTag Red In Situ Apoptosis Detection Kit (S7165) or ApopTag Fluorescein In Situ Apoptosis Detection Kit (S7110) (Chemicon) was used according to the manufacturer's protocol.
Cdk2 Activity Assay.
The activity of Cdk2 was measured following a previously described procedure (38).
Supplementary Material
Acknowledgments.
We thank J.R. Nevins, Duke University Medical Center, for the E2F1-expressing adenovirus construct. We also thank N. Hay, University of Illinois at Chicago, for critically reading the manuscript. The work was supported by grants from the National Cancer Institute (CA77637 to P.R.) and National Institute of Aging (AG 024138 to P.R. and S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812254106/DCSupplemental.
References
- 1.Nichols AF, Itoh T, Graham JA, Liu W, Yamaizumi M, et al. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 2.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 3.Itoh T, Cado D, Kamide R, Linn S. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc Natl Acad Sci USA. 2004;101:2052–2057. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon T, Chakrabortty A, Franks R, Valli T, Kiyokawa H, et al. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene. 2005;24:469–478. doi: 10.1038/sj.onc.1208211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alekseev S, Kool H, Rebel H, Fousteri M, Moser J, et al. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 2005;65:10298–10306. doi: 10.1158/0008-5472.CAN-05-2295. [DOI] [PubMed] [Google Scholar]
- 6.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Zhai L, Xu J, Joo HY, Jackson S, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 8.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 9.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, et al. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 10.Rapic-Otrin V, McLenigan MP, Bisi DC, Gonzalez M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoyanova T, Yoon T, Kopanja D, Mokyr MB, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 activates nucleotide excision repair by regulating the level of p21Waf1/Cip1. Mol Cell Biol. 2008;28:177–187. doi: 10.1128/MCB.00880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper MP, Balajee AS, Bohr VA. The C-terminal domain of p21 inhibits nucleotide excision repair in vitro and in vivo. Mol Biol Cell. 1999;10:2119–2129. doi: 10.1091/mbc.10.7.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan ZQ, Reardon JT, Li L, Flores-Rozas H, Legerski R, et al. Inhibition of nucleotide excision repair by the cyclin-dependent kinase inhibitor p21. J Biol Chem. 1995;270:22008–22016. doi: 10.1074/jbc.270.37.22008. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 15.Tan T, Chu G. p53 Binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol Cell Biol. 2002;22:3247–3254. doi: 10.1128/MCB.22.10.3247-3254.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols AF, Itoh T, Zolezzi F, Hutsell S, Linn S. Basal transcriptional regulation of human damage-specific DNA-binding protein genes DDB1 and DDB2 by Sp1, E2F, N-myc and NF1 elements. Nucleic Acids Res. 2003;31:562–569. doi: 10.1093/nar/gkg152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S, Shu L, Dilling MB, Easton J, Harwood FC, et al. Sustained activation of the JNK cascade and rapamycin-induced apoptosis are suppressed by p53/p21(Cip1) Mol Cell. 2003;11:1491–1501. doi: 10.1016/s1097-2765(03)00180-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhan J, Easton JB, Huang S, Mishra A, Xiao L, et al. Negative regulation of ASK1 by p21Cip1 involves a small domain that includes Serine 98 that is phosphorylated by ASK1 in vivo. Mol Cell Biol. 2007;27:3530–3541. doi: 10.1128/MCB.00086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian H, Wittmack EK, Jorgensen TJ. p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 2000;60:679–684. [PubMed] [Google Scholar]
- 20.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 21.Itoh T, O'Shea C, Linn S. Impaired regulation of tumor suppressor p53 caused by mutations in the xeroderma pigmentosum DDB2 gene: Mutual regulatory interactions between p48(DDB2) and p53. Mol Cell Biol. 2003;23:7540–7553. doi: 10.1128/MCB.23.21.7540-7553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 23.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 24.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, et al. p53 Has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 25.Cang Y, Zhang J, Nicholas SA, Kim AL, Zhou P, et al. DDB1 is essential for genomic stability in developing epidermis. Proc Natl Acad Sci USA. 2007;104:2733–2737. doi: 10.1073/pnas.0611311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sohn D, Essmann F, Schulze-Osthoff K, Janicke RU. p21 Blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase-mediated caspase-9 activation. Cancer Res. 2006;66:11254–11262. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- 30.Giono LE, Manfredi JJ. Mdm2 is required for inhibition of Cdk2 activity by p21, thereby contributing to p53-dependent cell cycle arrest. Mol Cell Biol. 2007;27:4166–4178. doi: 10.1128/MCB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, Mdm2. Mol Cell Biol. 2008;28:1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Amazit L, Long W, Lonard DM, Monaco J J, et al. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 35.Brown DR, Thomas CA, Deb SP. The human oncoprotein Mdm2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998;17:2513–2525. doi: 10.1093/emboj/17.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr C J, et al. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222–1230. [PubMed] [Google Scholar]
- 37.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Major M, Lepe R, Costa R. Forkhead Box M1B transcriptional actvity requires binding of CDK-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.