Abstract
Adhesion pili (fimbriae) play a critical role in initiating the events that lead to intestinal colonization and diarrheal disease by enterotoxigenic Escherichia coli (ETEC), an E. coli pathotype that inflicts an enormous global disease burden. We elucidate atomic structures of an ETEC major pilin subunit, CfaB, from colonization factor antigen I (CFA/I) fimbriae. These data are used to construct models for 2 morphological forms of CFA/I fimbriae that are both observed in vivo: the helical filament into which it is typically assembled, and an extended, unwound conformation. Modeling and corroborative mutational data indicate that proline isomerization is involved in the conversion between these helical and extended forms. Our findings affirm the strong structural similarities seen between class 5 fimbriae (from bacteria primarily causing gastrointestinal disease) and class 1 pili (from bacteria that cause urinary, respiratory, and other infections) in the absence of significant primary sequence similarity. They also suggest that morphological and biochemical differences between fimbrial types, regardless of class, provide structural specialization that facilitates survival of each bacterial pathotype in its preferred host microenvironment. Last, we present structural evidence for bacterial use of antigenic variation to evade host immune responses, in that residues occupying the predicted surface-exposed face of CfaB and related class 5 pilins show much higher genetic sequence variability than the remainder of the pilin protein.
Keywords: crystal structure, pili, diarrheal disease, adhesion, proline isomerization
Enterotoxigenic Escherichia coli (ETEC) use surface fimbriae (also called pili) to attach to host intestinal epithelia, an early, vital step in diarrhea pathogenesis (1). Colonization factor antigen I (CFA/I) fimbriae are prevalent among ETEC strains and represent the archetype of class 5 fimbriae, the largest class of human-specific ETEC colonization factors (2, 3). A 4-gene operon for fimbria-related proteins is shared by all class 5 fimbriae, which are assembled via the “alternate” chaperone pathway, as distinguished from the classic chaperone-usher pathway that guides assembly of class 1 fimbriae, such as P- and type I pili (4). Mature CFA/I fimbriae are a polymer typically consisting of >1,000 copies of the major pilin subunit CfaB, and 1 or a few copies of the tip-residing adhesive minor subunit CfaE (5–7).
Structures of fimbriae-associated components and pilin complexes from bacterial fimbriae have been defined by using X-ray crystallography, electron microscopy (EM), and NMR (8–12). Despite low sequence similarity among Gram-negative bacterial fimbriae, all pilin subunit structures determined to date, except those of type IV, share an immunoglobulin (Ig)-like fold with an exposed hydrophobic groove (10, 13). A β-strand contributed by the adjacent subunit fills this groove via “donor strand exchange” (9, 10), visualized in Fig. 1B. By engineering recombinant proteins in which a subunit is created that fills its own groove, researchers have successfully expressed subunits suitable for structure determination (8, 14). Because fimbriae are capable of inducing protective immune responses against bacterial disease (15), structural insights gained into fimbrial assembly and adhesion will lead to new strategies to prevent and treat such diseases.
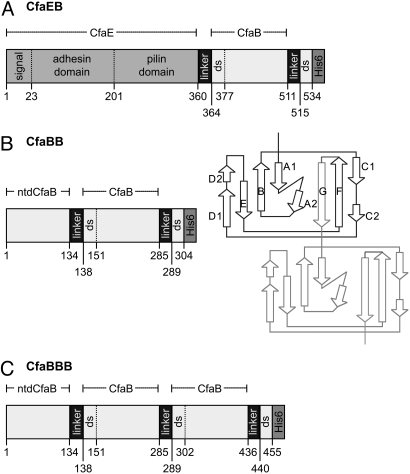
Fig. 1.
Schematic depiction of recombinant fusion proteins containing CfaB. (A) CfaEB comprises (in sequence from N to C terminus) native full-length CfaE, including the N-terminal signal peptide, adhesin domain, and pilin domain; a tetrapeptide linker (designated hereafter as linker); mature CfaB (i.e., without signal peptide); linker; the first 19 aa of a mature CfaB (containing the donor strand, ds); and a hexahistidine affinity tag (His)6. (B) CfaBB comprises mature CfaB with an N-terminal deletion (ntd) of its first 15 residues; linker; a copy of mature CfaB; linker; the first 15 residues from the mature CfaB N terminus (containing the ds); (His)6. The diagram illustrates how this genetic construct replicates the donor-strand exchange mechanism of assembled CFA/I fimbriae. In the native fimbria, 1 CfaB subunit (dark lines) has a hydrophobic groove filled by the N-terminal extension of the next subunit (light lines). In our construct, this connection is enforced by addition of a 4-residue linker between strand F of the first CfaB subunit and strand G of the second subunit. (C) CfaBBB comprises CfaBB with an additional copy of the linker and mature CfaB, as shown. The starting residue number of each segment is denoted along the bottom of each protein depiction, whereas the extent of CfaE, ntdCfaB, and mature CfaB are shown above each depiction.
An atomic-resolution structure for a class 5 fimbrial component was recently reported for the minor adhesive subunit CfaE (16). This advance notwithstanding, a detailed elucidation of the structure and assembly of the CFA/I fimbria has posed a challenge. We have shown that gold-labeled anti-CfaE polyclonal antibodies decorate only the tip region of a mature CFA/I fimbria (17). In addition, EM imaging of CFA/I filaments showed that although most fimbriae are helical filaments, regions of CFA/I fimbriae can undergo unwinding to a thin fibrillum (18). The structural basis for this morphologic heterogeneity has not been determined.
To better understand the structure and assembly of CFA/I fimbria, we have determined the atomic structure of the major pilin subunit. The geometry of adjacent CfaB subunits covalently linked by genetic engineering was examined in the crystal structures, and compared with fibrillar and helical filament models assembled using constraints provided by EM data. We found evidence that isomerization of Pro13 provides a switch for the transition from CFA/I pilins arranged in an extended fibrillar form to the helical filamentous form. Last, we provide a spatial framework for understanding class 5 pilus antigenic variation (19).
Results
Structure of CfaEB.
Because previous characterizations showed that CfaB subunits follow right after the initiating minor adhesive subunit CfaE in a mature pilus (7) and the structure of dscCfaE is known (PDB ID 2HB0) (16), we extended dscCfaE at its C terminus to include CfaB, with the resulting fusion protein designated CfaEB (Fig. 1A). Resulting crystals diffracted X-rays to 2.1-Å resolution (Table S1), yielding data for atomic models of CfaE and CfaB, and insights into intersubunit orientations (Fig. 2 A–C).
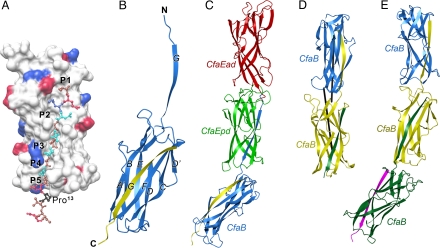
Fig. 2.
Crystal structures of CFA/I fimbrial subunits and their recombinant fusions. (A) The surface structure of the donor-strand exchanged CfaB, with the bound N-terminal extension shown in ball-and-stick format. Positively charged surfaces are shown in blue; negatively charged surfaces, in red. The 5 hydrophobic pockets in the hydrophobic groove are labeled P1 to P5. In the 16 N-terminal residues shown, hydrophobic residues are brown, polar residues cyan, and Pro13, which sits outside the groove, is shown in black. (B) Ribbon diagram of CfaB with native CfaB colored blue and the donor strand G contributed from an associating subunit in yellow. (C) Ribbon diagram of the structure of the CfaEB fusion. The 3 domains are colored in red, green, and blue, for CfaEad, CfaEpd, and CfaB, respectively. The yellow strand associating with the blue CfaB is an engineered donor strand filling the hydrophobic groove of the CfaB subunit. (D) Ribbon representation of the structure of CfaBB. The 2 associating CfaB subunits are colored blue and yellow, respectively. The green strand is an engineered donor strand. The 2 longest inertial vectors for the subunits are drawn as black rods. (E) Ribbon diagram of the structure of CfaBBB. The 3 subunits are colored blue, yellow, and green, respectively. The engineered donor strand filling the final groove is colored magenta.
The CfaB structure has an anticipated 7-stranded β-sandwich fold, belonging to the Ig-like family (Fig. 2B). As in CfaE, CfaB has a barrel-like shape, with the engineered donor strand filling the hydrophobic groove that would be exposed in a native monomeric CfaB. A DALI search (http://ekhidna.biocenter.helsinki.fi/dali_server/) identified CfaE as the best match for structural similarity to CfaB. Indeed, CfaB can be superimposed onto either the adhesin domain (CfaEad) or pilin domain (CfaEpd) of dscCfaE, yielding rms deviations of 2.4 Å and 2.0 Å, respectively.
The structure of the CfaE subunit in CfaEB is nearly identical to that previously reported for dscCfaE (16) (rms deviation, 0.77 Å). Each of the 2 domains of CfaE in CfaEB (i.e., CfaEad and CfaEpd) forms a β-barrel. They are tightly associated with each other, with a joint angle of 173.5° between them (Fig. 2 and Table S2), giving the overall appearance of a rigid cylinder. This is very similar to the composite cylindrical shape and interdomain joint angle of dscCfaE (171.6–173.5°) (16). The twisting angle, defined as a rotation angle that brings superimposition of 1 domain to the other, was calculated in the different crystal forms to range from 169.8° to 171.8° between CfaEad and CfaEpd (Table S2). Thus, both solved structures of CfaE show highly conserved interactions at the interface between the adhesin and pilin domains of CfaE, despite being crystallized in different forms (this study and ref. 16).
The CfaEB structure elucidates the structural relationship between CfaE and the adjoining CfaB subunit. Chemical interactions between CfaEpd and CfaB in CfaEB are very limited, featuring only 4 hydrogen bonds and no salt bridges. The total buried surface area at the CfaEpd–CfaB interface is 505 Å2 (Table S3), indicative of minimal interactions between the 2 domains. Last, there is a significant bend (joint angle, 138°; twisting angle, 138.4°) between CfaEpd and CfaB, such that the entire CfaEB fusion is J-shaped (Fig. 2C and Table S2).
Structures of Multiple CfaB Subunit Fusions.
As characterized biochemically, a CFA/I fimbria is composed of about 1,000 copies of the major fimbrial subunit arranged in an ordered assembly governed by the donor-strand exchange mechanism. To elucidate the geometric relationship between 2 neighboring CfaB subunits, we followed the same strategy as that used to design the CfaEB fusion, genetically engineering a CfaBB fusion protein that emulates 2 adjacent, noncovalently linked CfaB subunits (see Methods and Fig. 1B). Crystals of CfaBB diffracted X-rays to 2.3-Å resolution.
Molecular replacement methods were used to identify 2 CfaBB molecules, called “A” and “B,” in a crystallographic asymmetric unit cell, corresponding to 4 pilin subunits for which atomic models were refined (Table S1). The 2 CfaB subunits in each CfaBB are identically folded (Fig. 2D). Although both subunits in molecule A are tightly packed, 1 subunit of molecule B is flexible, giving rise to poor density. These differences between molecules A and B suggest a degree of flexibility in the CfaB intersubunit spatial orientation. This significant degree of freedom is corroborated by the relatively large variations in joint (8°) and twisting (18.4°) angles between adjacent major subunits (Table S2). In addition, contacts between 2 CfaB subunits are weak, with only 3 hydrogen bonds (2 water-mediated), and no salt bridges (Table S3).
We genetically engineered a trimeric, donor-strand-exchanged CfaB fusion, designated CfaBBB (Fig. 1C), to determine whether the relationship derived from the CfaBB fusion is generally applicable to all polymerized CfaB subunits. Crystals of CfaBBB diffracted X-rays to 2.3-Å resolution (Table S1). In the structure of a single CfaBBB molecule (Fig. 2E), where the 3 subunits are covalently linked in tandem, the first 2 CfaB subunits are well ordered, whereas the last subunit displays considerable disorder. This is most likely due to lack of sufficient contacts with neighboring molecules. Assessment of the intersubunit geometry revealed significant flexibility in the interactions between neighboring CfaB molecules (Table S2), providing more information for the modeling of CFA/I fimbriae.
Conservation of the Donor Strand and Its Binding Groove.
The dscCfaE donor strand, derived from the N terminus of CfaB, had well-ordered electron density, suggesting that binding between the donor strand and CfaEpd has stringent specificity and high affinity, and the artificial linker does not interfere with this binding (16). The same binding mode is found in all of the structures described here (Fig. 1B and 2), which permits a detailed structural definition for the donor-strand exchange mechanism of class 5 fimbriae. The sequence alignment of 8 ETEC major pilins (Fig. S1) shows that the highly conserved donor-strand sequence V1EKNITVTASVD12 exhibits an alternating pattern of hydrophobicity (Fig. 2A). Evenly spaced hydrophobic depressions of the binding groove (Fig. 2A, labeled P1 to P5) precisely complement the contours of the side chains of hydrophobic residues in the donor strand. Two highly conserved charged residues, Glu2 and Lys3, at the beginning of the donor strand interact with the equally conserved CfaB residues Lys61 and Asp16, respectively, on the groove side. Pro13 is positioned outside the groove (Fig. 2A and Fig. S2).
Modeling the Native and Unwound States of CFA/I Fimbriae.
CFA/I fimbriae are visualized as both helical fimbrial filaments and thin fibrillar structures (18, 20, 21). The latter appear to be an unwound (extended) fibrillar form of the helical conformation (Fig. 3A). A fibrillar model was generated by extending the CfaBB dimer with multiple CfaB subunits, repeating the same protein–protein interactions defined by the dimeric crystal structure. The resultant fibril has a periodicity consistent with measurements of fibrillae in EM images, and the model visualized as a projection image strongly resembles the EM data (Fig. 3A). Specifically, the fibrillar model has a diameter of 45 Å, ≈4.2 subunits per turn, and a 49-Å rise per subunit. This contrasts with the 3-dimensional reconstruction of EM data for the helical form of CFA/I fimbria (Fig. 4A), which exhibits a diameter of 74 Å, 3.17 subunits per turn, and a rise per subunit of 8.3 Å along the helical axis (18). Thus, the crystal structures of CfaBB and CfaBBB conform more closely to unwound CFA/I rather than to the helical filamentous structure.
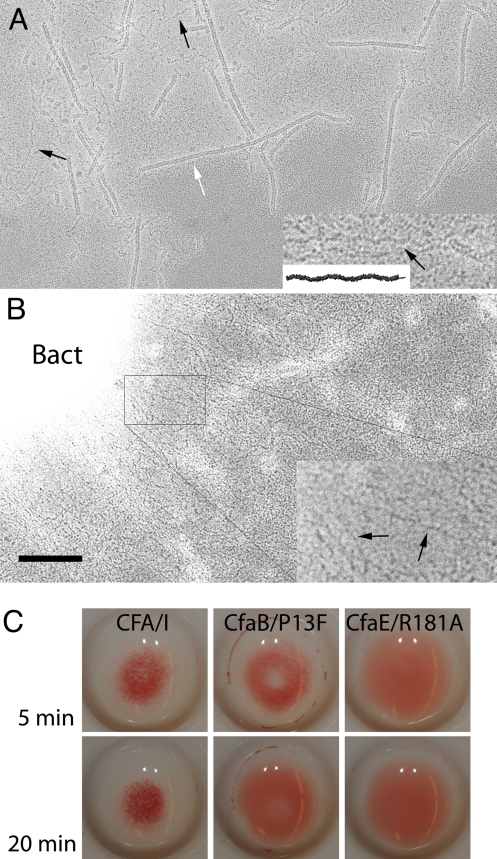
Fig. 3.
Structure and function of CFA/I fimbriae. (A) Wild-type CFA/I fimbriae can be seen as helical filaments (white arrow) and in an extended conformation (black arrows). The extended fibrillar form (Inset, black arrow) is indistinguishable, at this resolution, from a model built from repetition of the subunit–subunit interaction visible in the CfaBB crystal structure (Inset, black fibrillum). In the crystal, structure Pro13 is in the trans conformation, producing the fibrillar conformation of CfaB multimers. (Magnification bar corresponds to 1000 Å—400 Å in the Insets—of A and B.) (B) Mutation of Pro13 (CfaB with the point mutation P13F) yields bacteria (labeled Bact) that express fimbriae unable to coil into a helical form (see Inset, black arrows, for examples of the extended conformation), suggesting that Pro13 is important for the transition of the thin fibrillar structure to its helix form. (C) Mannose-resistant hemagglutination (MRHA) of recombinant bacteria expressing native CFA/I, CFA/I with the CfaB/P13F mutation, and CFA/I with the CfaE/R181A mutation, shown after incubation with bovine erythrocytes at 5 and 20 min, rocking on ice. Native CFA/I mediates 2+ hemagglutination detectable by 5 min and remaining positive throughout the observation period. This contrasts with early detection of 1+ hemagglutination with the mutant CFA/I-CfaB/P13F fimbriae at 5 min, which thereafter dissolves to become negative after 8–10 min (see 20-min time point). The CfaE/R181A mutation served as a negative control (16). Results were the same when the bacterial preparations were induced for 3 h before performing the MRHA assay.
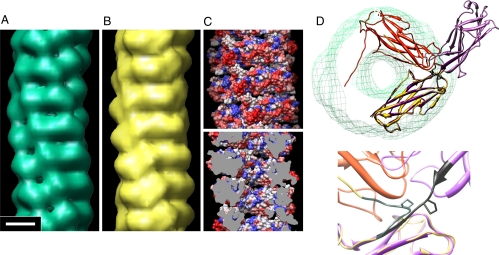
Fig. 4.
Model of CFA/I fimbria, visualization of the high genetic sequence variability of the outer fimbrial surface, and possible mechanism for conversion between extended and helical filament forms. (A) EM reconstruction of CFA/I fimbriae (18). (Magnification bar corresponds to 25 Å for A–C.) (B) CfaB subunits placed in a filament by using the helical symmetry determined by EM and the structure of CfaB determined by X-ray crystallography and reduced to 20-Å resolution. (C) Homology map of CFA/I, in which a surface view of the CFA/I pilus model is colored by using the color scheme blue-to-white-to-red for highly conserved to highly variable residues, respectively. The outside surface and surface in the inner channel are shown in Upper and Lower, respectively. To view the inner channel (Lower), the filament is sliced down the center of the helix axis; cutting plane colored gray. (D) Illustration to show the change in assembled structures when Pro-13 is in the trans conformation or in the cis conformation. The conversion of Pro13 from trans to cis converts an extended fibrillar structure (purple) to a helical filament structure (yellow/orange) of CfaB multimers. When Pro13 is in the trans conformation, as in the solved X-ray crystal structure of CfaBB (Upper, light purple and purple subunits), a thin extended fibril is assembled (see also Fig. 3A Inset). In contrast, computational isomerization of Pro13 into the cis conformation (Upper orange and yellow subunits) acts as a hinge at the intersubunit junction, so that the polypeptide backbone swings into position to allow coiling of the subunits into a helical filament that fits into the EM reconstruction (green mesh). Lower, the change in backbone direction is highlighted by coloring residues 10–16 in black when Pro13 is in trans, and gray when Pro13 is in cis. In both diagrams, Pro13 is the only side chain shown.
To build a helical superstructure from CfaB subunits, an electron density map of CfaB was calculated at 20-Å resolution, and subunits were placed on the helical positions from the EM map (Fig. 4 B–D). Thousands of possible orientations of CfaB were tested by cross-correlation, and the top 5 models were retained for further analysis. The best model (Fig. 4 B and C) has a cross-correlation with the EM map of 0.92, and an ω angle of 11° for Pro13 (Fig. 4D), whereas the next best model has values of 0.81 and 66°, respectively. Because prolines are expected to be in either the cis (idealized ω = 0°) or trans (ω = 180°) conformation, an ω angle of 66° is unacceptable, and this model was rejected.
All peptide bonds are synthesized in the trans isomer, with some later isomerized to cis by an isomerase (see, e.g., ref. 22) or in concert with conformational changes of a macromolecular assembly (23). The crystal structures of CfaBB and CfaBBB show that Pro13 is located just beyond the donor β-strand that fills the hydrophobic groove of the preceding subunit (Fig. 2A and Fig. S2), where it may provide a hinge between the barrel-like core and the N-terminal extension of CfaB. It is part of a highly conserved residue pair in all ETEC class 5 fimbriae, Asp12-Pro13 (Fig. S1). This proline is in trans in our crystallographic data, and in cis in the helical filament model, as seen by the change in backbone direction in Fig. 4D. To test whether Pro isomerization is important for helical filament assembly, recombinant CFA/I was expressed with a point mutation that encoded CfaB/P13F. Phenylalanine was chosen because it is nonpolar, has a pKa similar to that of proline, and very rarely isomerizes to the cis conformation (24). Surface expression of CFA/I was confirmed in both wild-type and mutant CfaB bacteria by using dot blot analysis, showing labeling of bacteria by antibodies against CfaE and whole fimbriae (Fig. S3). EM data showed that bacteria primarily assembled fibrillar (unwound) structures when expressing CFA/I with the mutant pilin (Fig. 3B). Short segments of helical filaments were extremely rare (<0.1%), and no long filaments (>25 nm) were seen. Notably, this structural derangement of CFA/I filaments is associated with mannose-resistant hemagglutination of bovine erythrocytes during incubation on ice with agitation for short intervals, but loss of hemagglutination after 8–10 min (Fig. 3C).
Clustering of Highly Variable Residues on the Exterior Surface of the Helical Filament Model.
ETEC class 5 major fimbrial subunits exhibit significant polypeptide sequence variability, which inversely correlates with serological cross-reactivity of these fimbriae (25). We surmised that this would be explicable by greater sequence variability in residues positioned on the exterior surface of the helical fimbrial shaft. By using sequences of the major pilins from 8 class 5 pilus types from representative clinical ETEC isolates (19), we scored each position on a scale from highly similar to highly variable. Overall, 31% of all subunit residues were highly variable. Based on our model for CFA/I helical fimbriae, 48% of predicted surface-exposed residues were highly variable, as compared with 20% of internal residues. The homology map (Fig. 4C), shows the clustering of highly variable residues on the outer fimbrial surface.
Discussion
The CfaB major pilin subunit from class 5 CFA/I fimbriae displays the Ig-like fold adopted by all known class 1 pilins, subunits from Caf1 (26), and Dr adhesins (8). The hydrophobicity of the pilin groove and the alternating hydrophobicity of the pilin N-terminal extension are highly conserved, facilitating subunit–subunit interactions of high specificity and affinity. To this point, CfaB from CFA/I fimbriae, its close homolog CooA from CS1 fimbriae (27), and PapA from P-pili (12) all exhibit similar sequence patterns. Thus, there is a single structural theme for the formation of fimbrial filaments from class 1 and class 5 fimbriae that includes structural similarity and donor-strand exchange, belying the separate classification of class 5 fimbriae into an alternate chaperone pathway. The structural differences between fimbriae appear correlated with the host niche environment rather than class designation, as P-pili (class 1) and CFA/I (class 5) fimbriae appear more structurally similar to each other than to Hib pili (class 1). Unwinding is observed in both P-pili and CFA/I fimbriae, whereas this fibrillar conformation is not seen in Hib pili that survive in an environment of even higher shear forces (28).
The CfaEB structure provides information on how the adhesive subunit is attached to the major pilin stalk, and it describes extensive contacts between the adhesin and pilin domains of CfaE, strongly supporting the notion that rigidity rather than flexibility is an intrinsic feature of the minor adhesive subunit under static conditions. In contrast, CfaB subunits can rotate with respect to each other, as demonstrated by the large and variable twisting angle between CfaB subunits and between CfaB and CfaE. Our model for CFA/I fimbriae includes the adhesin protruding from the tip at the edge of the filament, parallel to the helical axis, maintained in this position by the rigidity of the interaction between the 2 domains, CfaEad and CfaEpd. This is consistent with immunolabeling of the CfaE adhesin occurring at the tip of the fimbria, but offset from the helical axis (17).
Several lines of evidence indicate that Pro13 of CfaB functions as a hinge in CFA/I organelle morphogenesis. First, multimers of CfaB with Pro13 in the trans conformation form an extended thin fibrillum. Second, computational trans-to-cis isomerization of Pro13 in the CfaBB crystal structure is key for constructing the best model for the helical CFA/I fimbria filament. Third, polymers of CfaB forced to retain a trans conformation at position 13 by substitution of Phe for Pro retain the extended conformation in vivo and are unable to coil into the CFA/I fimbrial helix form. We therefore suggest that Pro13 isomerization may be essential for the transition from its slender fibrillar form, which must be maintained to transit the outer membrane usher assembly, to its helical fimbrial form, which is the dominant form after exiting the usher assembly. Notably, this specific mechanism may be operative for all ETEC class 5 fimbriae, which all feature a proline at position 13 (Fig. S1). The helical filament is clearly a stable structure, as shown by thousands of EM images, yet there are always thin fibrillae visible, suggesting the possibility that metamorphic transition occurs in vivo via isomerization back to trans. Thus, the CFA/I fimbria appears to be a filament primed for unwinding. Although the current data do not address the mechanism by which the proline isomerization occurs, the requirement for a cis peptide in the helical pilus filament suggests the possibility of a new role for CfaC, whereby this outer membrane usher protein either facilitates or induces trans-to-cis isomerization at Pro13 of CfaB during extrusion through the usher.
The CfaB pilin surface exposed to the environment (vs. internal residues or surfaces that are involved in subunit–subunit interactions) includes the majority of highly variable residues of the subunit. This antigenic variation is presumably driven by immune pressure from exposure to humans. By presenting varying antigenic surfaces, ETEC can limit the immune response of the host upon repeated infections with similar fimbrial types. Our model presents a structural framework for this evasive bacterial strategy of genetic diversification of class 5 fimbriae associated with human diarrheal disease.
The structural findings reported bear on research to define effective fimbria-related approaches to prevention of ETEC diarrhea, a few aspects of which are mentioned here. First, a consensus peptide comprising the N-terminal 36 residues of class 5 pilin subunits has been promoted as a possible class 5-wide vaccine antigen based on the high amino acid conservation along this stretch (29). Our CFA/I structural model indicates that this stretch is buried within the helical stalk, detracting from the appeal of this strategy. Second, the flexibility of the CFA/I structure afforded by the weak interactions between turns of the helix and the Pro13 cis-trans hinge suggest a mechanism for the protective effects of anti-fimbrial antibodies. If such antibodies cross-link repeating epitopes on the same or different fimbriae and thereby impede localized filament unwinding and winding, this could disrupt adhesive function. Last, in this work we have defined the native-like structure of the CfaEB fusion protein, which emulates the tip of the CFA/I fimbria. This fusion protein presents the 2 structural subunits, CfaE and CfaB, in a 1:1 ratio rather than the >1,000:1 ratio found in native fimbriae. Further studies are needed to determine whether this antigen might be superior to either recombinant dscCfaE adhesin or native CFA/I fimbriae in eliciting a combination of neutralizing antibodies that provide the protective effects of anti-fimbrial antibodies, as previously observed, and in addition directly block bacterial adhesion.
Methods
Strains and Chemicals.
Plasmids and bacterial strains used in expression and purification of CfaEB, CfaBB, and CfaBBB are listed in Table S4. Various chemicals, reagents, and enzymes for cloning, expression, protein purification, and crystallization are of the highest grade possible and were purchased commercially.
Protein Expression, Purification, and Crystallization.
We genetically engineered fusion proteins comprising covalently linked, donor-strand exchanged CfaE (adhesin) and CfaB (major pilin) proteins in various combinations. For example, to produce the dimeric covalently linked CfaBB fusion protein, a genetic construct was made to encode the mature CfaB polypeptide from which the first 13 residues of the N terminus had been removed (N-terminal-deleted CfaB, designated “ntdCfaB”); at its C terminus a tetrapeptide linker was added, followed by a second copy of this donor-strand exchanged variant of CfaB. Methods for construction of the expression vectors for CfaEB, CfaBB, and CfaBBB are provided in SI Materials and Methods, Table S5, and ref. 30. The purification procedure for CfaEB is very similar to that for CfaE (31); see SI Materials and Methods. The purification procedure for CfaBB and CfaBBB followed a different protocol: see ref. 30 and SI Materials and Methods. After purification, all proteins were crystallized by the standard vapor diffusion technique, and conditions for their crystallization (30) are detailed in SI Materials and Methods.
X-Ray Diffraction Data Collection, Structure Determination, Model Building, and Refinement.
Crystals were tested for diffraction quality and for cryoprotection conditions before data collection at the SER-CAT beam line of Advanced Photon Source (APS), Argonne National Laboratory. Crystals of CfaEB display monoclinic space group P21 symmetry. Those for CfaBB and CfaBBB belong to the space groups of P21212 and C2, respectively. The atomic structure of the CfaEB fusion was solved by molecular replacement using the dscCfaE model (PDB ID 2HB0) (16) as a phasing template. Difference Fourier calculations revealed clear electron density for the covalently linked, donor-strand-complemented CfaB subunit. Diffraction data sets for CfaBB and CfaBBB crystals were phased similarly by using the donor-strand-exchanged CfaB domain from the CfaEB structure as the phasing model.
Imaging and Atomic Modeling of Native and Unwound CFA/I Fimbriae.
CFA/I fimbriae were isolated as described previously (18). Details of EM sample preparation, imaging, and digitization are included in SI Materials and Methods. CfaB subunits were placed in the CFA/I pilus filament based on electron density from the EM map, maximizing the cross-correlation between the electron density maps of the 3D-reconstruction from EM data and the CfaB subunit from crystallographic data. The subunit used for modeling is shown in Fig. 1B, residues 151–303, but with the linker (286–289) removed: CfaB with its hydrophobic groove self-filled. In the native filament, residue 14 follows residue 13, whereas in the model the groove of a subunit is self-filled and does not connect directly to the adjacent subunit. Minimization of the distance between residues 13 from the self-filled groove of 1 subunit to residue 14 from the adjacent subunit was used to refine the 5 best models. Constraints for a planar peptide bond at the end of the N-terminal extension (Asp12-Pro13 in a native filament, corresponding to residues 151 and 303 in the CfaBB construct), distances, and energy minimization were done with the programs Coot (32) and Chimera (33). The fibrillar filament was modeled by using the CfaBB crystal structure, superimposing 1 subunit of a second copy of the dimer onto the end subunit, and repeating this process to produce an extended fiber.
Homology Between Class 5 Major Pilin Subunits.
The Scorecons server (34) was used to score residue conservation among major pilins representing each of the 8 known ETEC class 5 pilus types (19). Surface residues were defined as those with >10% of their area exposed on the subunit surface, and internal residues were defined as those with ≤10% surface exposure. Results of this analysis are displayed with Chimera (33).
Construction and Analysis of Recombinant Bacteria Expressing CFA/I Fimbriae with CfaB/P13F Mutation.
Site-directed mutagenesis was used to introduce a point mutation in cfaB that directed expression of CFA/I fimbriae containing CfaB/P13F. Along with appropriate controls, this construct, BL21-SI(pMAM2-CfaB/P13F), was examined by EM to determine the morphology of its CFA/I filaments, and by mannose-resistant hemagglutination (MRHA) to assess its impact on adhesive function. Detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank the staff at Southeast Regional Collaborative Access Team for assistance in data collection; L. Esser for discussions; N. Pattabiraman for programming; and P. Guerry, A. Fasano, and D. Isaac for critical reading of the manuscript. This research was supported by the National Cancer Institute Intramural Research Program (D.X.), the Trans National Institutes of Health/Food and Drug Administration Intramural Biodefense Program (D.X.), the U.S. Army Military Infectious Disease Research Program Work Unit A0307 (S.J.S.), the Henry M. Jackson Foundation (S.J.S.), and National Institutes of Health Grant GM055722 (to E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3F83 for CfaEB, 3F84 for CfaBB, and 3F85 for CfaBBB).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812843106/DCSupplemental.
References
- 1.Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: Epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Low D, et al. Fimbriae. In: Neidhart FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol 2. Washington, DC: Am Soc Microbiol; 1996. pp. 146–157. [Google Scholar]
- 3.Wolf MK. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic E. coli. Clin Microbiol Rev. 1997;10:569–584. doi: 10.1128/cmr.10.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soto GE, Hultgren SJ. Bacterial adhesins: Common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froehlich BJ, Karakashian A, Melsen LR, Wakefield JC, Scott JR. CooC and CooD are required for assembly of CS1 pili. Mol Microbiol. 1994;12:387–401. doi: 10.1111/j.1365-2958.1994.tb01028.x. [DOI] [PubMed] [Google Scholar]
- 6.Sakellaris H, Scott JR. New tools in an old trade: CS1 pilus morphogenesis. Mol Microbiol. 1998;30:681–687. doi: 10.1046/j.1365-2958.1998.01088.x. [DOI] [PubMed] [Google Scholar]
- 7.Sakellaris H, Balding DP, Scott JR. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 8.Anderson KL, et al. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol Cell. 2004;15:647–657. doi: 10.1016/j.molcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Sauer FG, et al. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 10.Choudhury D, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 11.Zavialov AV, et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: Preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 12.Verger D, Bullitt E, Hultgren SJ, Waksman G. Crystal structure of the P pilus rod subunit PapA. PLoS Pathogens. 2007;3:e73. doi: 10.1371/journal.ppat.0030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerlund-Wikstrom B, Korhonen TK. Molecular structure of adhesin domains in Escherichia coli fimbriae. Int J Med Microbiol. 2005;295:479–486. doi: 10.1016/j.ijmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Barnhart MM, et al. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc Natl Acad Sci USA. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freedman DJ, et al. Milk immunoglobulin with specific activity against purified colonization factor antigens can protect against oral challenge with enterotoxigenic Escherichia coli. J Infect Dis. 1998;177:662–667. doi: 10.1086/514227. [DOI] [PubMed] [Google Scholar]
- 16.Li YF, et al. A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J Biol Chem. 2007;282:23970–23980. doi: 10.1074/jbc.M700921200. [DOI] [PubMed] [Google Scholar]
- 17.Poole ST, et al. Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol Microbiol. 2007;63:1372–1384. doi: 10.1111/j.1365-2958.2007.05612.x. [DOI] [PubMed] [Google Scholar]
- 18.Mu XQ, Savarino SJ, Bullitt E. The three-dimensional structure of CFA/I adhesion pili: traveler's diarrhea bacteria hang on by a spring. J Mol Biol. 2008;376:614–620. doi: 10.1016/j.jmb.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anantha RP, et al. Evolutionary and functional relationships of colonization factor antigen I and other Class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect Immun. 2004;72:7190–7201. doi: 10.1128/IAI.72.12.7190-7201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutton S, Lloyd DR, Candy DC, McNeish AS. Ultrastructural study of adhesion of enterotoxigenic Escherichia coli to erythrocytes and human intestinal epithelial cells. Infect Immun. 1984;44:519–527. doi: 10.1128/iai.44.2.519-527.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat Struct Biol. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 24.Pal D, Chakrabarti P. Cis peptide bonds in proteins: residues involved, their conformations, interactions and locations. J Mol Biol. 1999;294:271–288. doi: 10.1006/jmbi.1999.3217. [DOI] [PubMed] [Google Scholar]
- 25.Gaastra W, et al. Antigenic variation within the subunit protein of members of the colonization factor antigen I group of fimbrial proteins in human enterotoxigenic Escherichia coli. Int J Med Microbiol. 2002;292:43–50. doi: 10.1078/1438-4221-00189. [DOI] [PubMed] [Google Scholar]
- 26.Zavialov AV, et al. Donor strand complementation mechanism in the biogenesis of non-pilus systems. Mol Microbiol. 2002;45:983–995. doi: 10.1046/j.1365-2958.2002.03066.x. [DOI] [PubMed] [Google Scholar]
- 27.Starks AM, Froehlich BJ, Tamara F, Jones TN, Scott JR. Assembly of CS1 pili: The role of specific residues of the major pilin, CooA. J Bacteriol. 2006;188:231–239. doi: 10.1128/JB.188.1.231-239.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu XQ, Egelman EH, Bullitt E. Structure and function of Hib pili from Haemophilus influenzae type b. J Bacteriol. 2002;184:4868–4874. doi: 10.1128/JB.184.17.4868-4874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassels FJ, Jarboe DL, Reid RH, Lees A, Deal CD. Linear epitopes of colonization factor antigen I and peptide vaccine approach to enterotoxigenic Escherichia coli. J Ind Microbiol Biotechnol. 1997;19:66–70. doi: 10.1038/sj.jim.2900416. [DOI] [PubMed] [Google Scholar]
- 30.Li YF, et al. Crystallization and preliminary x-ray diffraction analyses of several forms of the CfaB major subunit of enterotoxigenic Escherichia coli CFA/I fimbriae. Acta Crystallogr F. 2009;65:242–247. doi: 10.1107/S1744309109001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YF, et al. Crystallization and preliminary x-ray diffraction analysis of CfaE, the adhesive subunit of the CFA/I fimbriae from human enterotoxigenic Escherichia coli. Acta Crystallogr F. 2006;62:121–124. doi: 10.1107/S1744309105043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.