Abstract
Small regulatory RNAs (sRNAs) in eukaryotes and bacteria play an important role in the regulation of gene expression either by binding to regulatory proteins or directly to target mRNAs. Two of the best-characterized bacterial sRNAs, Spot42 and RyhB, form a complementary pair with the ribosome binding region of their target mRNAs, thereby inhibiting translation or promoting mRNA degradation. To investigate the steady-state and dynamic potential of such sRNAs, we examine the 2 key parameters characterizing sRNA regulation: the capacity to overexpress the sRNA relative to its target mRNA and the speed at which the target mRNA is irreversibly inactivated. We demonstrate different methods to determine these 2 key parameters, for Spot42 and RyhB, which combine biochemical and genetic experiments with computational analysis. We have developed a mathematical model that describes the functional properties of sRNAs with various characteristic parameters. We observed that Spot42 and RyhB function in distinctive parameter regimes, which result in divergent mechanisms.
Keywords: gene regulation, mRNA silencing, RyhB, small RNA, Spot42
Small regulatory RNAs (sRNAs) play numerous roles in stress adaptation and development in eubacteria, archeabacteria, and eukaryotes (1, 2). These molecules are generally noncoding and <300 nt. Today, ≈70 sRNA genes have been identified in Escherichia coli (3–5). Those characterized are involved in diverse cellular functions such as initiation of DNA replication (6), regulation of transcription (7), regulation of translation (8–12), and mRNA stability (13, 14). Such sRNAs are usually expressed in response to a specific stimulus that signals the cell about a possible threat to survival.
A critical stimulus to which growing cells must respond rapidly is the level of glucose, which regulates genes involved in carbon metabolism such as those belonging to the galactose operon (galETKM). In the absence of glucose, d-galactose can induce transcription of all 4 genes of the galETKM operon in equimolar fashion, which is required for catabolism of d-galactose. In contrast, when cells grow in the presence of glucose, mainly the first 2 genes of the operon are expressed, which are necessary for providing substrates for biosynthetic glycosylation reactions (15–17). Spot42, a 109-nt-long sRNA, is involved in this glucose-dependent regulation. When glucose is used as a carbon source, Spot42 represses the translation of galK, the third gene of the galactose operon that is unnecessary when glucose is present (11, 18). However, when cells grow in the absence of glucose, Spot42 levels decrease significantly, allowing translation of galK. It has been suggested that the sRNA only acts at the translation level, because Spot42 expression does not inhibit GalE and GalT synthesis (11).
Another important cellular response mediated by a sRNA is the adaptation to iron (Fe) starvation (19). The sRNA RyhB, normally repressed by the transcriptional regulator Fur (ferric uptake regulator) in presence of sufficient Fe, is expressed in the bacterium E. coli specifically during Fe depletion (20). Thus, in conditions of low Fe, RyhB induces the rapid degradation of at least 18 mRNAs, all of which encode nonessential Fe-using proteins, to redirect the metal into essential parts of the cell (19, 21). The rapidity of this sRNA-mediated mRNA degradation (within 2–3 min) suggests that the target mRNAs have to be silenced as soon as possible under low Fe conditions. We recently confirmed this hypothesis by demonstrating an inhibitory growth phenotype of a ryhB mutant during Fe starvation (22). Furthermore, data from our group indicates that RyhB plays a role in the regulation of intracellular Fe homeostasis (22, 23). Because Fe is an essential factor that is carefully distributed among the Fe-using proteins, it is likely why many organisms share a RyhB-like mechanism of Fe regulation (21, 24).
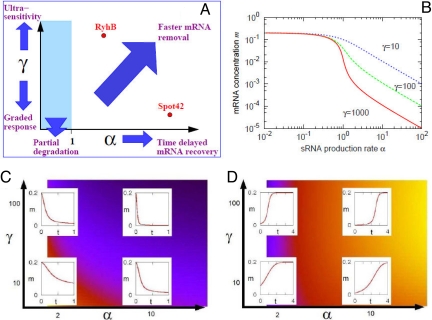
Despite the importance of sRNAs, their regulatory features are not yet fully understood. Here, we analyze quantitative properties of regulation where a sRNA binds to target mRNAs, which is the mechanism of action of a large family of sRNAs in bacteria. The pairing between the sRNA and a target mRNA facilitates irreversible inactivation of both RNAs in the complex (in the case of RyhB both the sRNA and mRNA are actively degraded), as illustrated in Fig. 1. sRNA regulation can exhibit ultrasensitivity (25), hierarchy of action (26), cross-talk between downstream targets (19, 23, 27), prioritization of targets (28), fast response against sudden environmental changes (13, 28, 29), and discoordination of translation in a polycistronic mRNA (11, 30). Most of these abilities, except discoordination, are constrained by 2 key parameters: the system's capacity to overexpress the sRNA relative to its target (α) and the speed at which the target is irreversibly inactivated (γ). By performing a computational analysis of the potential functional properties of sRNA regulation, covering all parameters, we show which functions can be achieved for which parameter values. Using 2 well-known sRNA systems, Spot42 and RyhB, we demonstrate different methods for the determination of the above 2 key parameters by combining biochemical and genetic experiments with computational analysis.
Fig. 1.
Schematic mechanism of sRNA regulation. The formation of the sRNA–mRNA complex irreversibly inactivates both RNAs. The behavior is entirely determined by 3 parameters (see SI Text): the ratio of the sRNA production rate to mRNA production rate, α = αs/αm; the dimensionless rate of inactivation of the mRNA via sRNA–mRNA complex formation, γ = δαmτs2; and the ratio of the mRNA lifetime to the sRNA lifetime, τ = τm/τs.
Results
Ultrasensitivity and Graded Regulation.
Fig. 2B shows the steady-state concentration of the target mRNA, m, as a function of α for different values of the degradation rate γ (the units were chosen as described in SI Text). For α < 1 there is insufficient sRNA to inactivate much mRNA, whereas when α > 1 the mRNA levels are significantly reduced. This reduction is much sharper for a larger γ value, i.e., a small change of α results in a large difference in mRNA concentration m. Such switch-like regulation has been termed “ultrasensitivity” (25). At lower γ values, m declines in a more “graded” manner.
Fig. 2.
Capabilities of sRNA regulation. (A) Schematic diagram showing functional features observed in the model behavior at different values of the parameters, α and γ. These are shown more quantitatively in B–D. (B) The steady-state level of sRNA as a function of α for γ = 10 (blue dotted line), γ = 100 (green dashed line), and γ = 1,000 (red solid line). The value of mRNA concentration m for α > 1 is scaled with 1/γ, which results in a ≈10-fold difference between lines. m changes gradually for small γ, and more abruptly when γ is large (termed “ultrasensitivity”). (C) Dynamics of m (red solid lines) when the value of α is increased, at time 0, from 0.01 to the value shown on the horizontal axis. The background color indicates the time to reach the new steady state, with darker colors indicating shorter times. (D) Dynamics of m when the value of α is reduced, at time 0, from the value shown on the horizontal axis to 0.01. The background color indicates the time to reach the new steady state; darker colors indicate shorter times.
Speed of Response.
In addition to steady-state regulation, the speed of the response is also important. Perturbations in the external stimuli sensed by a sRNA will result in changes in the value of α. When α is abruptly increased from 0.01 to a higher value (which we vary from α = 2 to 10), then the mRNA level decreases rapidly in response. Fig. 2C shows that the time required for m to reach its new steady state is smaller for larger α and larger γ, because less time is needed to produce more sRNA for large α, whereas a large γ results in quick degradation of m. Because increasing α can compensate for lower γ, and vice versa, the behavior of m with α = 10 and γ = 10 looks very similar to the case of α = 2 and γ = 100. Note, however, that the former has a much higher sRNA level in the new steady state.
When α is abruptly decreased from a starting value (which we vary from α = 2 to 10) to 0.01, the response is slower than in the case of increasing α, as shown in Fig. 2D (note the larger span on the x axis relative to Fig. 2C). This difference is because recovery of m level begins only after all of the sRNAs (which are not being produced any more) present are degraded. The larger α is, the larger this initial pool of sRNA, and therefore the longer the time delay before recovery of mRNA level.
In both scenarios (increasing or decreasing α), however, a larger value of γ results in relatively sharper regulation and faster response.
Determination of α and Calculation of γ.
The parameter α can be measured directly, both in vivo and in vitro by measuring the promoter strengths for the sRNA and target mRNA in steady-state conditions, and computing their ratio. However, direct measurement of γ is difficult: not much is known about the mechanism of Hfq-mediated sRNA–mRNA coupling. Once α is determined, however, the value of γ can be calculated by measuring either (i) the timing of sRNA–mRNA degradation or (ii) the steady-state level of mRNA and sRNA.
Measurement of α and Estimation of γ for RyhB and Target mRNA Using Degradation Kinetics.
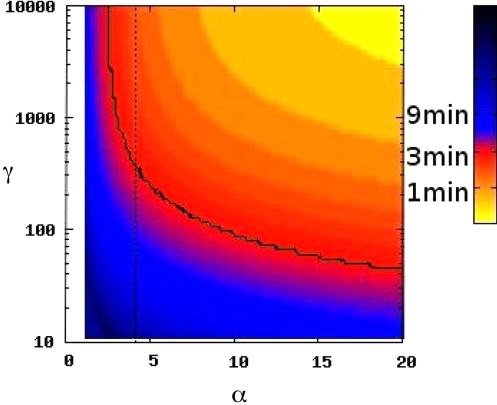
For the Fe–Fur system in E. coli, it has been observed that 1 of the RyhB targets, sodB, is depleted ≈5-fold within 3 min after full induction of the PryhB promoter (13), which constrains the value of α and γ. We numerically calculated the time required to deplete mRNA 5-fold, T, by changing the value of α from ≈0 to higher values (see Fig. 3). T strongly depends on α and γ. The contour line of T = 3 min is the solid green line in Fig. 3. Further, we measured the value of promoter activity of RyhB and major target mRNAs (see Fig. 4) from which we calculate α, the ratio of sRNA production to sum of the production of all its target mRNAs, to be ≈4. For α = 4, the T = 3-min contour in Fig. 3 leads to an estimate of ≈400 for the γ value for sodB. Therefore, this system represents the low-α, high-γ case, which is consistent with our previous predictions (23). Note that we use one single α, as if all target mRNAs have similar γ values. The effect of this assumption in determination of γ for sodB is discussed in SI Text. More careful analysis requires detailed knowledge of the variation of γ across the target mRNAs, as outlined in ref. 28.
Fig. 3.
Measurement of γ from dynamics of target degradation. The color indicates the time T required, in model simulations, to deplete mRNA levels by 5-fold in respond to a sudden increase of α from near 0 to the value on the x axis. The solid line shows the contour corresponding to 3 min, the time observed in RyhB degradation of sodB mRNA (13). In the simulation, the lifetimes of sodB τm = 5/ln2 min and RyhB τs = 25/ln2 min were used. The dotted line indicates α = 4, the value obtained from our measurements of promoter activities. To calculate α, we assume to have measured all of the target mRNAs of RyhB in Fig. 4. If there are more targets, the computed α value will be less, hence γ will be higher. That is, 4 is an upper bound for α, and 400 is a lower bound for γ. Therefore, our conclusion that RyhB is characterized by low α and high γ is true for sodB.
Fig. 4.
Specific β-galactosidase activities of promoter fusions of the Fe–Fur system without Fe (in the presence of 200 μM the Fe chelator 2,2′-dipyridyl). In these conditions, the promoter of ryhB is fully active. Promoter activities of 4 target mRNAs are indicated by the gray bars, and their sum gives αm ≈ 20 + 19 + 12 + 2 = 53. The promoter activity of PryhB is αs ≈217(left black bar, ryhB+), therefore α = αs/αm ≈ 4. See SI Text and Table S1 for experimental details.
Measurement of α and Estimation of γ for Spot42 and Target mRNA Using in Vitro Transcription Assays.
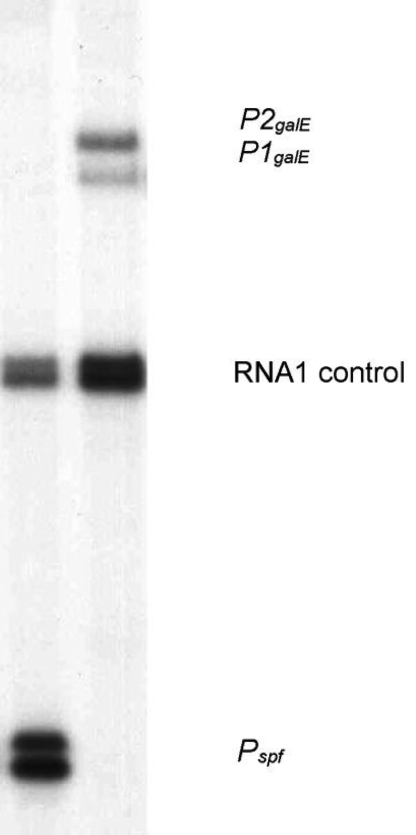
The Spot42 sRNA specifically binds the galK Shine-Dalgarno region of the galETKM mRNA and inhibits production of GalK by blocking ribosome binding. Because translation of the upstream regions is not affected, Spot42 action results in discoordinated translation of the gal operon genes (11). Transcription of the spf gene encoding the Spot42 sRNA is reduced 3- to 5-fold in the presence of cAMP-CRP (31). The galETKM operon is transcribed from 2 promoters, P1galE and P2galE. Both promoters are repressed in the absence of galactose by GalR. d-galactose inhibits GalR-mediated repression of both promoters. When d-galactose is present, cAMP-CRP can activate P1galE and repress P2galE (32). To determine the relative intensities of sRNA and mRNA transcription we measured the intrinsic activities of the Pspf (for sRNA) and P1galE and P2galE (for mRNAs) promoters in the absence of regulators (Fig. 5). Quantitation of band intensities (see SI Text) resulted in α = 18. The promoter activities measured approximate the conditions in ref. 11 when Spot42 sRNA is expressed at full strength (not repressed by cAMP–CRP) and the gal operon is transcribed at the maximum rate possible in the absence of cAMP–CRP and GalR. In cells grown in glucose-rich media the unrepressed Pspf promoter activity, approximated in the in vitro transcription assay, results in ≈200 copies of Spot42 sRNA per cell with a half-life of 12–13 min (18). Therefore, by our calculations, the sRNA is produced at a rate of 11 per min and the mRNA, which has a half-life of 4–5 min (33), is produced at a rate of 0.6 per min. This level of Spot42 sRNA reduces the amount of the GalK protein to ≈30% of the level observed in Δspf or glycerol-grown (high cAMP–CRP) cells (11). Because the GalK level is not affected by the 3- to 5-fold reduced Spot42 level in the presence of cAMP–CRP (11), we assume that mRNA–sRNA pairing is negligible when the Spot42 sRNA is present at <50 copies per cell.
Fig. 5.
In vitro transcription assays were performed on pSA850spf (Pspf, Left) and pSA850 (P1galE and P2galE, Right) supercoiled plasmid DNA templates in the presence of σ70 RNA polymerase. The RNA1 transcript was used as an internal control between lanes. See SI Text for experimental details.
In this case, the degradation kinetics is not known, but the amount of sRNA for different values of α and the according fold change of mRNA level has been measured. As can be seen in the plot of the steady state in Fig. 2B, for a given value of α, the mRNA level (and also the sRNA level) varies depending on γ, thus once the fold change of mRNA by changing α is known, γ can be calculated. Let m1 (s1) denote the concentration of mRNA (sRNA) with Pspf unrepressed and m2 (s2) denote the concentration of mRNA (sRNA) with Pspf repressed. Assuming steady state in both cases, we find, from the equations of Fig. 1, that
from which γ can be determined:
Inserting (m1/m2) = 0.3, s1 = 200 copies per cell, and s2 = 50 copies per cell, we get γ ≈ 2. This system thus represents a high-α, low-γ case.
Summary and Discussion
Many features of sRNA-mediated mRNA regulation are determined by 2 key parameters: the system's capacity to overexpress the sRNA relatively to its target mRNA (α) and the speed at which the target mRNA is irreversibly inactivated (γ). In this article, we used a mathematical model to analyze how the regulatory behavior changes depending on these parameters. We then determined α experimentally in 2 sRNA systems, RyhB and Spot42, using 2 different approaches to estimate γ. For the sRNA RyhB, we used the time course of depletion of target mRNA upon sudden derepression of the sRNA promoter, from ref. 13. Using our model simulations of the depletion process and our measurement of α to be 4 or less, we estimate that γ is at least 400 for the target sodB. For Spot42, we measured α to be ≈18. From the relation between concentrations and model parameters at 2 different steady states, we estimated γ to be ≈2. The results indicate that sRNA–mRNA systems can function in different parameter regimes.
We suggest that evolution of sRNA–mRNA systems functioning at different parameter regimes has a physiological significance. In our description of sRNA–mRNA systems, RyhB represents sRNAs that recognize target mRNAs very efficiently (high γ) but where sRNA excess (α) is low. According to our model predictions, this parameter regime optimizes the system for efficient and fast inactivation of target mRNAs in case of stress and, at the same time, a slightly time-delayed recovery of target mRNA levels after the sRNA synthesis is stopped (when stress is removed). Indeed, the RyhB system is responsible for the quick rearrangement of the intracellular channeling of available Fe. RyhB inhibits translation of nonessential Fe-using proteins, allowing a larger fraction of the limited Fe to be available for essential Fe proteins (22). To achieve recovery in the absence of stress, the sRNA expression is rapidly stopped, making the sRNA level decline, which after some time results in production of target genes, coding for nonessential Fe-using proteins. The length of the associated delay depends on how much α exceeds 1 and could be seen as buffering time to confirm that stress is persistently absent. However, because of high γ, the mRNA level quickly recovers after the time-delay period is over. Our results are in agreement with the previously observed in vivo expression of RyhB (13). This type of regulation demonstrates a “dual” response where the cell adapts rapidly to substrate deprivation and more slowly when the substrate becomes newly available.
As opposed to RyhB, the sRNA Spot42 is expressed at higher levels relative to the target mRNA (α large). Nevertheless, the pairing between Spot42 and the gal mRNA is inefficient (γ small). Modeling predicts that the activation of Spot42 gives similar fast decline of its target gene. However, turning off of the Spot42 system should give a more graded response with a long time delay. In a recent article by Mehta et al. (34) in which the stochastic response of sRNA regulation was characterized, they suggest that graded response by sRNAs may be limited by transcriptional noise (transcriptional burst). Nevertheless in the case of Spot42, our data indicate that having a low γ would allow filtering of the noise. Thus, even though transcriptional bursts may occur for Spot42, the effect is reduced by the low binding capacity of Spot42 on the target mRNA. Unlike RyhB, Spot42 modulates the amphibolic utilization of the sugar d-galactose by discoordinating the synthesis of the enzymes involved in galactose metabolism (11, 35). Theoretically, partial regulation can be achieved by having low α (even <1) and high γ or having high α and low γ. In the case of Spot42, we observed the latter case presumably implemented by weak pairing between Spot42 and galK mRNA. The possible advantage of this parameter regime is that the mRNA level is less sensitive to any noise in the sRNA level. If Spot42 were expressed at a lower abundance than the galETKM message (≈0.6 per min), however, the noise level would be quite significant and might interfere with efficient functioning.
In principle γ can be regulated by global changes in the cellular physiology (e.g., affecting the level or activity of the RNA chaperone Hfq). However, because those changes are difficult to quantify we did not consider them in our equations. Nevertheless, our analysis does indicate ways in which global changes can affect the system, especially through the parameter α, which can be specifically regulated by changes in gene expression of the sRNA and/or the target mRNA. For example, in the galactose utilization network of E. coli, transcription of both the spf gene and the gal operon is simultaneously regulated by cAMP–CRP (11, 35). When cAMP–CRP level is increased, spf transcription is inhibited, while gal transcription is activated, therefore removal of Spot42 sRNA can be facilitated, resulting in a faster but still graded recovery of GalK.
In this work although we focused on 2 particular sRNAs, we assume that our findings can be extended to most, if not all, sRNAs using antisense pairing to regulate their target mRNAs. As demonstrated with RyhB and Spot42, the determination of both α and γ parameters in other sRNAs would indicate the type of physiological response corresponding best to a specific signal. One could imagine a sRNA that acts with a high α and γ, both parameters being necessary high for a rapid and efficient inactivation of a highly expressed target mRNA. Another example is low α and low γ, which would only slightly modulate the target mRNA expression, thus allowing a more graded response. In such a case however, a protein regulator, which allows greater quantitative adjustments than sRNAs, may be more appropriate (34). Indeed unlike protein regulators, the regulation by sRNAs was shown to work best in a situation where a large change in signals is observed (34).
In addition to silencing target mRNAs, some sRNAs activate their target mRNAs by making a complex that can be readily translated into proteins. For example, the sRNAs RyhB, DsrA, and GlmY bind to the 5′ UTR of cis-repressed mRNAs such as shiA, rpoS, and glmS, respectively, to activate translation initiation (10, 26, 36–38). In such cases, the sRNA–mRNA complex formation is also considered irreversible, and the response speed will depend on the lifetime of this complex and α and γ. The model can be easily extended to such an activation scenario, which in addition opens for regulation with α substantially smaller than 1.
Our findings are also applicable to synthetic RNA molecules that are engineered to regulate gene expression in vivo (reviewed in ref. 39). Indeed, the rational design of synthetic base-pairing RNAs with specific α and γ will help to achieve tunable and programmable effects on the cell. For example, a single RNA molecule combining multiple binding domains, each with a different γ, could allow a concerted and complex response. This type of molecule could also be used as a tool to disrupt several genes at once to study a complex genetic network.
The combined use of genetic experiments and mathematical calculation allowed us to determine fundamental differences in the regulatory mechanisms of 2 well-characterized sRNAs. We showed that the RyhB system has an ultrasensitive response, whereas the response in the Spot42 system is more graded. It would be of interest to investigate similar parameters for other sRNAs, which may open up additional perspectives on these sophisticated regulatory molecules.
Materials and Methods
Mathematical Model of sRNA and mRNA Dynamics.
We model the coupling between an sRNA (s) and its target mRNA (m) using the equations in Fig. 1 (23, 25, 27–29). In SI Text we describe how the number of parameters can be decreased by appropriately choosing units of time and concentrations. Three dimensionless parameters then determine the model behavior. The 2 most important are: the sRNA production rate relative to the mRNA production rate, α = αs/αm, and the mRNA inactivation rate γ = δαmτs2. Each individual sRNA–mRNA pair would have a specific value of γ; in general, one would expect that more basepair matching between the sRNA and its target would correlate with a higher value of γ. The third parameter, τ, is the passive degradation time of the mRNA relative to the sRNA. Typically, sRNAs have longer natural half-lives than mRNAs, i.e., τ < 1, partly because Hfq can protect the sRNAs (11, 13, 40). For sodB, a target of RyhB, τ = 0.2 (20), and we use this as a default value, unless otherwise specified.
Supplementary Material
Acknowledgments.
We thank Karine Prévost and Takácsné Botond Judit for excellent technical assistance. This work was supported by Canadian Institute for Health Research Grant MOP69005, the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, the Danish National Research Foundation, and Hungarian Scientific Research Fund Grant PD75496. E.M. is a Canadian Institute for Health Research New Investigator Scholar. S.S. is supported by a János Bolyai fellowship from the Hungarian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901466106/DCSupplemental.
References
- 1.Gottesman S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman S. Micros for microbes: Noncoding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Argaman L, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 4.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eguchi Y, Itoh T, Tomizawa J. Antisense RNA. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 7.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 8.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 10.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an antiantisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udekwu KI, et al. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullmann A, Joseph E, Danchin A. Cyclic AMP as a modulator of polarity in polycistronic transcriptional units. Proc Natl Acad Sci USA. 1979;76:3194–3197. doi: 10.1073/pnas.76.7.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semsey S, Virnik K, Adhya S. Three-stage regulation of the amphibolic gal operon: From repressosome to GalR-free DNA. J Mol Biol. 2006;358:355–363. doi: 10.1016/j.jmb.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Lee HJ, Jeon HJ, Ji SC, Yun SH, Lim HM. Establishment of an mRNA gradient depends on the promoter: An investigation of polarity in gene expression. J Mol Biol. 2008;378:318–327. doi: 10.1016/j.jmb.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 18.Sahagan BG, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. II. Accumulation and distribution. J Mol Biol. 1979;131:593–605. doi: 10.1016/0022-2836(79)90009-3. [DOI] [PubMed] [Google Scholar]
- 19.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Jacques JF, et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 23.Semsey S, et al. Genetic regulation of fluxes: Iron homeostasis of Escherichia coli. Nucleic acids Res. 2006;34:4960–4967. doi: 10.1093/nar/gkl627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massé E, Arguin M. Ironing out the problem: New mechanisms of iron homeostasis. Trends Biochem Sci. 2005;30:462–468. doi: 10.1016/j.tibs.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitarai N, Andersson AM, Krishna S, Semsey S, Sneppen K. Efficient degradation and expression prioritization with small RNAs. Phys Biol. 2007;4:164–171. doi: 10.1088/1478-3975/4/3/003. [DOI] [PubMed] [Google Scholar]
- 29.Shimoni Y, et al. Regulation of gene expression by small noncoding RNAs: A quantitative view. Mol Syst Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhya S. Suboperonic regulatory signals. Sci STKE. 2003;2003:pe22. doi: 10.1126/stke.2003.185.pe22. [DOI] [PubMed] [Google Scholar]
- 31.Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol. 1988;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weickert MJ, Adhya S. The galactose regulon of Escherichia coli. Mol Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta P, Goyal S, Wingreen NS. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol. 2008;4:221. doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semsey S, Krishna S, Sneppen K, Adhya S. Signal integration in the galactose network of Escherichia coli. Mol Microbiol. 2007;65:465–476. doi: 10.1111/j.1365-2958.2007.05798.x. [DOI] [PubMed] [Google Scholar]
- 36.Kalamorz F, Reichenbach B, Marz W, Rak B, Gorke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 37.Prévost K, et al. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 38.Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 40.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.