Abstract
InfoMax and FastICA are the independent component analysis algorithms most used and apparently most effective for brain fMRI. We show that this is linked to their ability to handle effectively sparse components rather than independent components as such. The mathematical design of better analysis tools for brain fMRI should thus emphasize other mathematical characteristics than independence.
Independent component analysis (ICA), a framework for separating a mixture of different components into its constituents, has been proposed for many applications, including functional magnetic resonance imaging (fMRI) (1–3). Separating a signal mixture into its components is impossible in general; however, many cases of interest allow for special underlying assumptions that make the problem tractable. ICA algorithms decompose into a sum of signals that “optimize independence.” Several algorithms and software packages are available for ICA (4, 5).
The first blind source separation in fMRI via ICA used InfoMax (1); other ICA algorithms for fMRI followed, such as FastICA (2, 5). These algorithms work well if the components have “generalized Gaussian” distributions of the form p(x) = Cexp(−α|x|γ), with γ≠2. More general ICA algorithms that assume less about the components can separate into independent components mixtures for which Infomax and FastICA fail. Nevertheless, these 2 are the most used ICA algorithms for brain fMRI.
Stochastic processes are independent if the distribution of either remains the same if the other is conditioned to any subregion of their range. Detecting deviations from independence requires large samples. In fMRI experiments, brain activity is measured in small volumetric regions or voxels v ∈ V, at times tn, n = 1,…,N. (fMRI measures brain function via the associated increase of oxygen-enriched blood flow. The hemodynamic response function is the flow's time profile for 1 pulse in brain activity; from a signal analysis point of view, it blurs the signal in time.) Often the voxels outnumber N by far. Thus, one often prefers to view the voxel-index v as labeling the samples over which independence is sought (spatial ICA, or SICA), rather than the tn (temporal ICA, or TICA).
In the linear model for brain activation (6), the total brain activity X(t,v) is assumed to be a linear superposition of the different ongoing brain activity patterns: X(t,v) = ∑ℓ = 1L Mℓ(t)Cℓ(v), where the Cℓ correspond to the brain activity patterns, and the “mixing matrix” M gives the corresponding time courses. At high signal amplitudes, saturation effects “spoil” linearity; nevertheless, the linear model is remarkably effective. We shall stick to it here.
Typically, the brain function under study is turned “off” and “on” by having subjects perform a task during defined periods, punctuated by either resting states or other tasks. The activation map of interest Cact(v) associated with a time course Mact(t) related to the task paradigm, is then identified via a statistical analysis. When a strict paradigm is not possible, or to capture more complex task-related time dependence, “blind” decomposition techniques are of interest; they decompose X(t,v) without reference to the task paradigm. The time paradigm used (convolved with the hemodynamic response), is used only to identify, among the Cℓ(v) found by the algorithm, the one with the closest resembling time course.
ICA (or another source separation algorithm) is thus used to identify the different components Cℓ by decomposing X(t,v). In brain fMRI, there is no physical reason for the spatial samples to correspond to different activity patterns Cℓ(v) with independent distributions. Several of the seminal papers suggest that ICA decomposition is particularly effective when the brain patterns one seeks are spatially sparse, with negligible or no overlap (1). Such components are “near” to independence in the following sense. Consider 2 binary-valued components C1 and C2. Define V1 (V2) as the collection of all v where C1(v) [C2(v)] equals 1, and V12 as V1 ∩ V2. Then the random processes that consist in evaluating C1(v) and C2(v) for a voxel v, picked randomly (uniformly distributed) in V, are independent if and only if or, equivalently, , regardless of the value of these fractions, where the notation #V stands for “number of voxels in V.” If the components are sparse (i.e., #V1/#V and #V2/#V are much smaller than 1) and well separated (V12 is tiny), then these equations always hold after, at most, a small change in #V12; in this (loose) sense, sparse and well-separated components are always “nearly independent.” Similar arguments hold for multivalued components. We want to understand better, beyond this heuristic, when an ICA algorithm gives a valid decomposition, or the reasons why some types of ICA algorithm work better for brain fMRI. This article summarizes the findings of several years of interaction between applied mathematicians and neuroscientists, expert in fMRI, concentrating on probing ICA methods for brain fMRI. It raises questions, informed by mathematical considerations, that are investigated by using numerical simulations and specially designed fMRI experiments. Although other authors have investigated to what extent ICA decomposition techniques for brain fMRI data can be validated (1, 7), our emphasis and conclusions are of a different nature. In particular, we conclude that independence is not the right mathematical framework for blind source separation in fMRI; representations in which the fMRI signal is sparse are more promising. A similar observation was made about ICA for image processing (8).
Spatial Variation Captured by ICA Algorithms
We first investigate the ability of InfoMax and FastICA to extract fine-grained spatially varying brain function maps. To this end, we carried out an ICA analysis of the data described in ref. 9, which demonstrated distributed patterns of activation, to find out to what extent ICA analysis could reproduce these findings. We first briefly recall the setup and results of this experiment (9).
Overlapping Representations of Faces and Objects in Ventral Temporal Cortex.
The subjects in the original experiment were shown visual stimuli during the “on” intervals of a simple on/off block time paradigm. The stimuli were images that, depending on their subject, belonged to 1 of 8 categories: Faces, Houses, Cats, Bottles, Scissors, Shoes, Chairs, or a control category of Scrambles (random noise patches). Images viewed in each 24-s stimulus block belonged to the same category. Each run consisted of 8 stimulus blocks (1 for each category), separated by 12-s rest intervals. The blocks visited the categories in a different random order for each run. To keep their attention on the images, the subjects were assigned a 1-back memory task. The object of the study was, however, not linked to this task but to the representation in the ventral temporal cortex of the different broad classes of objects.
For each subject, 12 fMRI time series were recorded. The object-selective cortex was determined (for each subject separately) by selecting the voxels whose responses differed significantly across categories, including voxels that showed no or weak activation for 1 or several categories. The ventral temporal object-selective cortex S was defined as the intersection of the anatomically defined ventral temporal cortex and the functionally defined object-selective cortex. The data, reduced to 12 time series for each voxel in S, were then split into training and test sets (even- vs. odd-numbered time series).
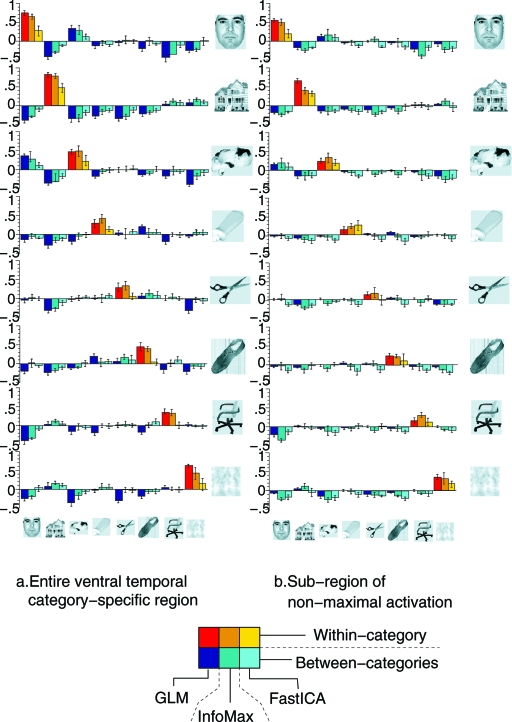
Based on the training dataset, S was partitioned into 8 subsets Sc by determining for each voxel v ∈ S the category c for which the response deviated most from the average response (over categories) of that voxel. This partition was used in the second, most important, step of the study. In the first step, the activation patterns (restricted to S), extracted for each stimulus time block in the test dataset were correlated with each of the category-specific activation patterns on S observed in the training dataset. The correlation scores were systematically and significantly higher “within category” than “between category,” so that the activition pattern in S for any particular stimulus block in the test set allows one to identify, with high confidence, the category the subject viewed during that time block. (Detailed data are in ref. 9; see also Fig. 2 below.)
Fig. 2.
Correlation scores (averaged over subjects, standard error indicated by error bar) between pairs of patterns from the training and test datasets for all pairs of categories. (Left) Correlations computed by using all the voxels in S. (Right) Correlations for each pair (c,c′) computed by using only the voxels in S\(Sc ∪ Sc′). In each block, triplets of columns give the results for every pair (c,c′) with 3 different methods: left = the GLM analysis in ref. 9, middle = InfoMax, and right = FastICA. For each algorithm (GLM, InfoMax, or FastICA), the sets Sc are defined by using a first-step analysis with the same algorithm.
In the second step, similar pairwise correlations were computed, except that, for every pair c,c′, the computation of the correlation between the 2 activation patterns was carried out summing over only voxels in S\(Sc ∪ Sc′), i.e., excluding the voxels that were most active for either c or c′. Although the correlation scores for these amputated object-selective zones were smaller than before, they still permitted correct identification of the categories viewed during the test time blocks; with high confidence. The information about the identity of the categories was thus stored not just in a small specialized zone but also, almost as importantly, in a more distributed spatially varying pattern, with lower amplitudes.
ICA Analysis of This Dataset.
Because N was too large for the ICA algorithms, we first performed a dimensionality reduction via PCA, retaining only the L PCA components with largest singular value, with L < N. Choosing L appropriately is nontrivial (10); we used an automatic estimation method for each dataset, maximizing the evidence of the model (11), as implemented in the FSL package (12) [the FSL (and its subtools BET, FLIRT, FEAT, MELODIC) can be downloaded from www.fmrib.ox.ac.uk/fsl]. (We also checked that augmenting L beyond our cutoff dimension did not impact the contrast of the components isolated by the ICA algorithm.) The output of this PCA was used as input for spatial ICA, to obtain L “spatially independent” components. We used 2 algorithms: InfoMax, as implemented in the NIS package (nisica, available at http://kraepelin.wpic.pitt.edu/nis), and FastICA, as implemented in the FSL package (12); in both cases the standard nonlinearities were used, maximizing the non-Gaussianity of the spatial sources. (Note: the FSL package outputs Z-score maps for the components isolated by the FastICA algorithm, whereas the NIS package provides the ICA maps themselves. To make the results comparable, the raw ICA components computed within FSL were used, not the corresponding Z-score maps.)
As shown by the original data analysis in ref. 9, the patterns of activation associated with the different separate categories are highly overlapping; InfoMax and FastICA lacked sufficient sensitivity to distinguish the responses across categories. When either ICA algorithm was applied to 1 of the original 8-block functional time series (containing 1 block of trials for each of the 8 categories), the time series of the resulting ICA components did not correlate strongly with any of the category-specific reference functions (which consisted of single 24-s “on” blocks). Instead, a single consistently task-related (CTR) component was produced systematically, with a time series that correlated strongly with the full 8-block time paradigm. This result persisted when several original time series were concatenated (“creating” a signal in which the category-specific paradigms had several active blocks).
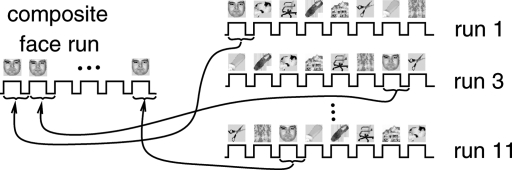
To identify category-specific activation maps by ICA, we re-organized the data. Only voxels in the ventral temporal cortex were studied, as in the original data analysis in ref. 9. For these voxels, new time series were constructed by concatenating blocks (consisting of 24 s of task plus the following 12 s of rest) of images corresponding to the same category, adjusting the mean of each time series, and high-pass filtering to avoid baseline drifts. This was done separately for training and test datasets, thus creating 16 new composite time series (2 for each of the 8 categories), each containing 6 blocks of stimulus of a unique category (see Fig. 1). For each of these, we identified the component of interest generated by ICA; because the analysis used a dataset with trials corresponding to 1 category only, we expected these components to contain category-specific information. From the resulting 16 maps, we computed the correlations between Cc,training and Cc′,test within and between category, i.e., for c = c′ and c ≠ c′. The results are given in Fig. 2 Left, for both InfoMax and FastICA; the within-category correlations are in most cases significantly higher than the between-category scores, leading to an identification accuracy of 82% for FastICA and 89% for Infomax, significantly better than chance level, albeit not as high as the 96% identification accuracy obtained in ref. 9. This high identification accuracy confirmed that category-specific information was indeed present in the CTR components estimated from the composite runs; it also confirmed that the preliminary dimension-reducing PCA step had not removed the correlations observed in ref. 9.
Fig. 1.
Concatenation to create “face run” from the odd-numbered runs.
Next, we concentrated on the “off-peak” information in these ICA components. Following the lead of ref. 9, we partitioned S into 8 subsets c; each c was composed of the voxels that responded more strongly to the category c stimulus than the other stimuli. For each pair c,c′, we defined the c,c′-off-peak region c,c′OFF := \(c ∪ c′), and we computed the corresponding correlations between the components c,testOFF and c′,trainingOFF. The results are shown in Fig. 2 Right: Though within-category scores are smaller over these off-peak regions, they remain significantly higher than between-category correlations in most cases, still producing high identification accuracy (73% for FastICA and 80% for InfoMax, both significantly higher than chance level, albeit lower than the 94% accuracy for the GLM-based analysis). In this analysis, InfoMax systematically outperformed FastICA.
It is standard practice, in the use of ICA for fMRI, to threshold the CTR component obtained. If a binary ground truth of activation is available, this can be done by means of ROC curves. Typically such “ground truth” maps are not available, and one thresholds based on the deviation from the mean in the distribution of the voxel amplitudes (see, e.g., refs. 13 and 14). The thresholds determined by standard practice are fairly high, retaining only the voxels with the very highest response. A post hoc analysis of the ICA CTR maps in our case shows that this would have eliminated, on average, >60% of the voxels in the off-peak regions; restricting the correlation computations to only the remaining voxels reduced the identification accuracy to much smaller values, for both FastICA and InfoMax. This shows the importance of voxels that experience lower amplitude activation, and of the spatial variation of this lower-amplitude activation, in encoding category-specific information; it also shows that the ICA algorithms were sufficiently sensitive to pick up the lower-amplitude spatial variation in the distributed category-specific components that enabled good identification accuracy from the off-peak regions. (As stated above, the ICA algorithms did not pick out the category-specific activation patterns from the original data; we refer here to their effectiveness in capturing the correct spatial variation in amplitude of the activation patterns, even among amplitude levels that are normally discounted.)
This ICA-analysis of the data of ref. 9 show that Info Max and FastICA “work:” they capture fine-grained spatially varying information, even though the independence hypothesis is surely violated. We next examine “independence” more closely.
Mathematical Independence: Generalities
Two stochastic variables X and Y are independent if their joint distribution is a product of their marginal distributions. More precisely, X and Y are independent if, for all possible choices of the 4 numbers a ≤ b and c ≤ d, we have Prob(a ≤ X ≤ b|c ≤ Y ≤ d) = Prob(a ≤ X ≤ b), i.e., the probability of observing X in an interval is the same whether we condition the values of Y to a subrange or not. For X and Y to be independent, it is necessary and sufficient that their mutual information equal zero: ∑k,ℓPk,lX,Y log(Pk,lX,Y) − ∑kPkX log(PkX) − ∑ℓPℓY log(PℓY = 0), where P1A,…,PℓA,… are the probabilities with which a random variable A can take its different values a1,…,aℓ,…. Requiring independence for 2 random variables is a very strong condition. Independent random variables are uncorrelated; the converse is not true.
Given linear mixtures Zr = ∑Mr,sXs,r = 1,… R of independent random variables Xs,s = 1,…S (with R ≥ S), one can recover the Xs by identifying the S × R matrix W for which the combinations ∑r = 1RWs,rZr are independent. Often this matrix W is identified via an iterative algorithm that seeks to minimize the mutual information
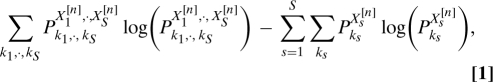 |
where , and “n” numbers the iteration steps. This nonconvex minimization problem is nontrivial. Moreover, when SNR values are low, mutual information is hard to evaluate.
Many ICA algorithms minimize a proxy functional instead of Eq. 1 or simplify the optimization by making assumptions about the distributions of the components. This is the case for InfoMax and FastICA: Both algorithms perform better if the components have distributions of type p(x) = Cexp(−α|x|γ), with γ ≠ 2 (1, 2, 5).
Mathematical Independence: Some Simulations
In this numerical study of InfoMax and FastICA (15), we study the respective roles of independence, sparseness, and separatedness in the success of the algorithms. We experiment on simple models where it is easy to change each of these characteristics separately.
In the InfoMax and FastICA algorithms, as adapted to brain-fMRI studies, the data are first prewhitened via a PCA analysis, leading to XWH.(t,v) = ∑ℓ = 1kMℓWH.(t)CℓWH.(v). Next, the algorithms identify an (orthogonal) L × L matrix W such that the Cℓ(v) = ∑k = 1LWℓ,kCkWH.(v) are as “independent” as possible. FastICA determines W such that the values (Cℓ(v))v ∈ V are distributed as un-Gaussian-like as possible, as measured by the kurtosis or the negentropy. In InfoMax, the different components are fed into a neural network optimizing the mutual information of the “output components.” For details, see refs. 5 and 16. These ICA algorithms fail when the components to be separated have Gaussian distributions.
We construct input mixtures (of independent or dependent components) for InfoMax and FastICA, and run the algorithms to produce their estimate of the “unmixed” components.
Both algorithms come in several forms; we selected those to correspond with the implementations on real fMRI data, described above. The selected InfoMax algorithm is adapted to heavy-tailed components; the nonlinear function characterizing its neural network is 1/(1 + e−x). For the FastICA algorithm the nonlinear function approximating the negentropy is (in the notation of ref. 5) g(y) = y3; the iteration follows the “symmetric approach”.
Our examples are deliberately chosen simple, with few parameters. We consider only 2 components C1 and C2, and 2 “observations,” at times t1, t2. Each component is a realization of a stochastic process that is itself a composite of 2 processes: one “activation process,” restricted to a subset of V, and a “background process” on its complement; the components are both of the type Ci(v) = χVi(v)xvi + [1 − χVi(v)]yvi,i = 1,2, where the Vi,i = 1,2 are different subsets of V (and can be picked differently for different examples), where χA stands for the indicator function of A ⊂ V (i.e., χA(v) = 1 if v ∈ A, χA(v) = 0 otherwise), and where x1, x2, y1 and y2 are 4 independent random variables, of which, as v ranges over V, the xvi and yvi are independent realizations. The “background” random variables y1 and y2 have the same cumulative density function (cdf) Φy(u) = [1 + e−1−u]−1; the “activation” random variables x1 and x2 also have identical cdf, equal to either Φx(u) = [1 + e2−u]−1 or Φx(u) = [1 + e2(2−u)]−1, depending on the example. (For the first choice, the parameters of our ICA implementations provide optimal “detectability” in the sense that the nonlinear function defined by the parameter setting of the algorithm coincides with the cdf of the signal source; for the second, there is a slight mismatch, as can be expected in realistic applications.) Finally, the mixtures of C1 and C2 given as input to the algorithms are X(t1,v) = 0.5C1(v) + 0.5C2(v) and X(t2,v) = 0.3C1(v) + 0.7C2(v).
The joint distribution function of the components C1 and C2 is easy to compute. The probability density functions (pdf) of x and y are, respectively, φx(u) = Φx′(u) and φy(u) = Φy′(u) = (cosh[(u + 1)/2])−2. The pdfs ψ1 and ψ2 of C1 and C2, respectively, are then given by ψ1(u) = (#V1)/(#V)φx(u) + [1 − (#V1)/(#V)]φy(u), ψ2(u) = (#V2)/(#V)φx(u) + [1 − (#V2)/(#V)]φy(u). Likewise, the joint pdf ψ{1,2} of C1 and C2 is . Thus ψ{1,2}(u,v) = ψ1(u)ψ2(v) (i.e. C1 and C2 are independent) if
This condition does not involve the pdfs φx, φy. The unrealistic rectangular shapes of V, V1, and V2 have no bearing on the outcome of the simulations; by spatial rearrangement V, V1, V2 could be shaped closer to physiological reality.
In each example below, the algorithms start from the 2 mixtures and are asked to “unmix” them; in all examples, a simple visual inspection of the mixtures already clearly indicates that there are several different components; the success of the algorithms is judged by the extent to which the “unmixed” output components they provide are close to the original 2 components, i.e., show a contrast boundary at only the edges of V1 for one of the components and only at the edges of V2 for the other.
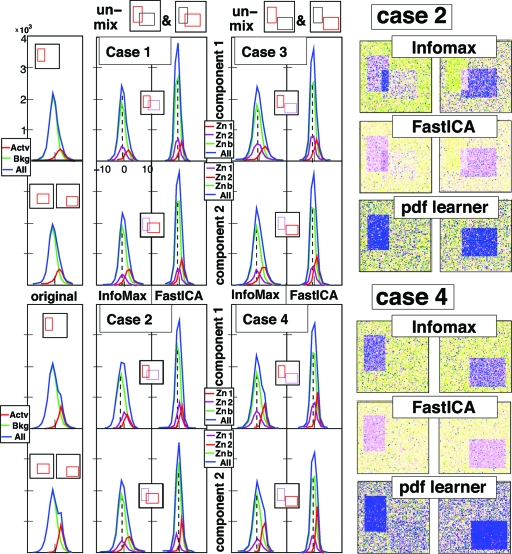
Consider now the following 4 examples, each specified by the choices of V1, V2, and the cdf Φx. These examples are illustrated in Fig. 3, with V = {1,…,100}×{1,…,100}.
Fig. 3.
Unmixing the 4 mixtures of rectangular components described below. (Left) pdfs of original 2 components, of components as identified by InfoMax and by FastICA. Color coding: the whole (blue), the active region (red), the “background” (green); In the ICA outputs, the purple pdf corresponds to the zone associated to the other component; the background pdf is then for the area outside V1 and V2. Separation is completely successful only when the purple and green pdf line up, i.e., in cases 1, 3 and 4, but not in case 2. (Right) false-color rendition of unmixed components for Cases 2 and 4, as obtained by InfoMax, FastICA, and a more sophisticated ICA algorithm that learns pdf distributions. InfoMax and FastICA do better in Case 4 (separated components) than in Case 2 (independent compoenents); for the pdf-learner it is the converse.
Example 1. In this example, V1 = {11,…,40}×{21,…,70}, and V2 = {31,…,80}×{41,…,80}. By Eq. 2, C1 and C2 are independent. For the cdf Φx we choose . Fig. 3, Case 1 shows (Left) the 2 components C1 (Upper) and C2 (Lower), and the 2 components computed by InfoMax (Center) and FastICA (Right). The plots of the pdfs of the components: for the ℓth component (ℓ = 1 or 2) the pdf for the pixels in its own “zone” Zoneℓ = Vℓ\V3−ℓ (red) and in the “rival zone” Zone3−ℓ = V3−ℓ\Vℓ (purple) are shown as well as the pdf for the background (green) and for the whole picture (blue). For both components, and for both algorithms, the rival-zone pdfs align perfectly with the background pdf, showing that the decomposition is effective.
Example 2. All choices are identical to Example 1, except that the cdf Φx is picked differently: . The components C1 and C2 are still independent. Fig. 3, Case 2 (with the same organization as Fig. 3 Example 1) shows that neither Infomax nor FastICA separate the 2 components, even though they are independent. Both components exhibit “ghosting” (see the difference in shape between the ghosting-Zone pdf and the background pdf).
Example 3. A different variant of Example 1: All choices are identical, except the location of V2, now shifted to {43,…,92}×{53,…,92}. The pdfs of C1 and C2 are unaffected. The new V2 and V1 do not intersect, i.e., C1 and C2 are spatially separated, not independent. Fig. 3, Case 3 shows that both InfoMax and FastICA successfully identify the 2 original components C1 and C2.
Example 4. The sets V1 and V2 are as in Example 3, but Φx is as in Example 2; C1, C2 are not independent. Fig. 3, Case 4 shows that both algorithms both successfully separate C1 and C2.
Examples 3 and 4 show that InfoMax and FastICA can identify different but not truly independent components; this is not surprising, because these components, even if not independent, may give the “least dependent” decomposition.
In Examples 2 and 4, the components are very similar—they differ only by a shift of V2, resulting in truly independent C1, C2 in Example 2, but some dependence in Example 4. Yet the 2 algorithms fail to identify the correct components in Example 2 (the independent case) and succeed in Example 4. The failure for both algorithms to identify these truly independent components is due to the fairly strong assumptions about the pdfs of the individual components. For comparison purposes, we also analyzed the mixtures from Example 2 with a more sophisticated ICA algorithm [see supporting information (SI)] that has fewer underlying assumptions on the pdfs of the constituent components, at the price of taking longer to converge. This alternative ICA algorithm not only “learns” the components but also their pdfs, modeling them as a mixture of Gaussians, of which the parameters are “learned.” The Fig. 3 Right shows that this more complex algorithm identifies the correct independent components in Example 2, and does less well in the separated case (Example 4), more consistent with expectation for an ICA. ICA algorithms of this type do not seem to be used in fMRI studies; in our trials, they did not perform well on fMRI data.
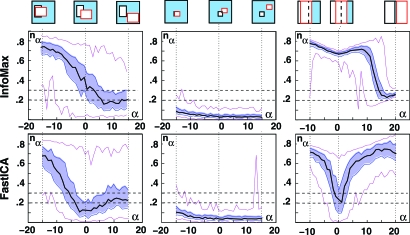
To further investigate the relative influence of separation and independence of both InfoMax and FastICA, we repeated the experiment for more gradual shifts of V2: for α ranging from −15 to 15, we pick V2α = {31+α,80+α}×{41+α,80+α}, leaving all the other settings the same as in Examples 2 and 4. In this family, α = 0 corresponds to mathematical independence (= Example 2), whereas we have complete separation (no overlap of V1, V2) for α ≥ 10. For each α, we find the unmixing matrix Wα, with both FastICA and InfoMax, and quantify the quality of the decomposition by the norm nα = ∥Id − Wα · M∥ of the difference between the 2 × 2 identity matrix and the product of the unmixing matrix Wα and the mixing matrix M. For an accurate decomposition, this product is close to the identity, and nα is close to 0. The less accurate the decomposition, the larger nα. The value of nα depends on the particular realizations of C1, C2; we computed (for each α) 99 different realizations, and we show the median, 1st and 3rd quartiles, and the highest and lowest values of nα in Fig. 4 (leftmost images). “Success of separation” can also be judged by visual inspection; by this crietrium, the components were visually perfectly separated whenever nα < 0.2; they were never separated when nα > 0.3.
Fig. 4.
The dependence on α of the norm nα = ∥Id − Wα · A∥. Ninety-nine realizations were generated for each α, and used for both algorithms. Black curve: medians; shaded blue region: bounded by the 1st and 3rd quartiles; magenta curves: lowest and highest values. When nα ≤ 0.2, the components are separated; when nα ≥ 0.3, they typically are not. (Left) “Medium” boxes (also used for Fig. 3). (Center) “Small.” (Right) “Large boxes.” In each case, α = 0 marks the independent case, where #(V1 ∩ V2,α) = #(V1) × #(V2)/(#V).
In addition to this “medium boxes” family of examples, where the components occupy, respectively, 15% and 20% of the 100 × 100 voxels in V, we repeated the numerical experiment with both sparser and less-sparse choices for V1, V2. In the “small boxes” family, V1 = {41,…,60}×{31,…,50}, V2,α = {57+α,…,81+α}×{46+α,…,65+α}, occupying 4% and 5%, respectively, of V; in the “large boxes” family, V1 = {1,…,48}×{1,…,100}, V2,α = {25+α,…,74+α}×{1,…,100}, occupying 48% and 50% (see top of Fig. 4).
Within each family, Fig. 4 shows the relative importance of independence. For medium and large boxes, the dip at α = 0 shows that FastICA is most successful when the components are indeed independent; InfoMax becomes successful only for larger α, when there is (almost) no overlap. For small boxes, both algorithms identify the components for all α, even when the overlap is large. Comparison across the 3 families shows that sparsity of the components (measured by #V2/#V, #V1/#V) affects the success rate of either algorithm much more than (in)dependence (measured by the deviation).
Mathematical Independence: fMRI Experiments
The fMRI experiments discussed here parallel the simulations above; they are inspried by ref. 7. We analyze* the results of experiments with 2 components, depending on 1 parameter, 1 value of which gives 2 independent components; for other values there is either more or less overlap. We analyze the results with InfoMax and FastICA and discuss their rate of success for different parameter values (17).
Experimental Design.
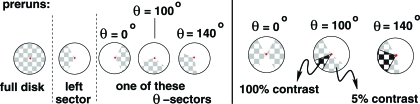
The experimental paradigm consisted of stimulating the right and left visual hemifields using a pair of 8-Hz flashing checkerboard wedges. The subjects were asked to focus on a bright dot at the center of a circular field; the visual stimulus consisted of flashing checkerboards in large wedges of this field. Two parameters, θ and α, characterized each run; quantifying the spatial and temporal overlaps in the paradigm.
The value of θ determined the positions of the 2 flashing checkerboard wedges. Both wedges spanned an angle of 120°. One wedge, S1, remained in the same position; S2(θ) was either completely separated from or overlapped with S1. In the overlap, the contrast of the flashing checkerboard was more pronounced than in regions covered by only 1 wedge. (See Fig. 5.) The contrast levels of the photic stimulations (5% for nonoverlapping wedges, 100% for overlapping wedges) ensured the linearity of the model.
Fig. 5.
(Left) Full disk and “left” wedge used in 2 of the preruns in each session, as explained in Experimental Design. A third prerun featured 1 of the θ -sector wedges. (Right) Superpositions used for the visual stimuli in the runs themselves.
The values of θ and the spanning angles were selected so that the experiment mirrored the setup of the numerical experiments above: for θ(= 100), the overlap of the wedges gives
Within the full disk, the indicator functions of the wedges S1 and S2(100) are thus independent. For θ = 140, the overlap is larger; for θ = 0, the wedges are completely separated. Because retinotopic mapping preserves the relative importance of spatial areas, spatial independence of the visual stimuli translates (in first approximation) into spatial independence of the corresponding retinotopic activity patterns. Within brain areas that preserve retinotopic organization, we thus have the same (or similar) spatial relationships between the activity patterns for the wedges as for the wedge stimuli themselves.
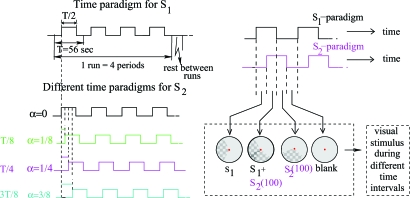
A second parameter, α, indexes shifts in the time paradigms for the 2 wedges. (Even though we studied only SICA, we wanted to test dependence on shifts in the time paradigm seen in ref. 7.) For each α, both wedges are “on” (“off”) half the time, and follow identical time patterns; the only difference is a time lag for S2. The 3 values of α ( and ) represent the fraction of temporal overlap between the “on” time intervals for S1 and S2 (see Fig. 6). Each experimental run had 4 blocks of visual stimulation for a fixed couple (α,θ), with each block corresponding to a 56-s period. Each session had 12 of these 224-s runs (different α, but same θ per session), with 10 s of rest between consecutive runs. To minimize subject fatigue and obtain better fixation of the paradigms, subjects were scanned at 3 different sessions, each dedicated to a specific value for θ. To perform a clear identification of “ground truth,” each session included 3 preruns, corresponding to the individual visualization of the wedges S1, S2(θ) separately (1 prerun for each), as well as a full flashing checkerboard (the third prerun).
Fig. 6.
(Left) Temporal paradigm. (Upper) Time paradigm for visual stimulus S1. (Lower) Time paradigms for S2, for different α. For α = 0 (not used in experimental runs), the paradigm is just that of S1; for α = 1/8,1/4,3/8, respectively, it is shifted by αT. (T = 56 sec. = period of the time paradigm.) (Right) Combined paradigm for (α,θ) = (1/4,100): wedge S2(100) is turned on and off according to the time paradigm shifted by T/4.
Image Acquisition and Preprocessing Stages.
Whole brain images were acquired for 3 healthy subjects (2 males, 1 female) with a 3T Siemens Allegra scanner. A T1-weighted structural image was acquired to localize the anatomy of the activated areas. The functional images were acquired by using a gradient-echo EPI sequence, (TR = 1,000 ms; flip angle = 60°; field of view 192 mm × 192 mm; matrix 64 × 64; slice thickness 7 mm; distance factor of 13%); each run contained 244 volumes of 15 axial slices acquired in the anterior-to-posterior phase encode direction. Preprocessing stages included the application of the FSL FLIRT motion correction algorithm (12) on all EPI images after discarding the first 6 volumes of each run; in order to include only brain voxels in the analysis, we applied a brain extraction algorithm [FSL BET (12)] on motion-corrected data.
Data Analysis of the Preruns.
We performed a general linear model (GLM) regression (6) on each of the preruns using the FSL FEAT package (12), to establish a “ground truth” activation map for each of the individual components. Voxels for which the time series, restricted to stimulus periods, showed a departure of at least 2 standard deviations from their average rest behavior were designated as true positives. The activation map resulting from the stimulation of the full flashing checkerboard was used for a second analysis (see below). The β -maps B1, B2,θ from the GLM analysis on the preruns were used to define ground truth maps for the brain patterns created by visualization of the individual wedges S1, S2(θ).
Using standard GLM statistical analysis, we also defined (separately, for each of the 3 preruns, for each session) binary ground truth maps G(v). We first extracted from (F(v,tn))n=1,…,N the component that most distinguished between “on” and “off” paradigms, and evaluated its significance by comparing it with the remainder of the demeaned (F(v,tn))n=1,…,N; when the distinguishing component was significant at the P = 0.01 level for both v and a contiguous neighbor v′ ≠ v we set G(v) = 1, otherwise G(v) = 0.
Application of SICA.
We carried out both InfoMax and FastICA analyses on the experimental runs for all (α,θ) pairs, using the same methods and software package as the ICA analysis of the Haxby data (see above); this included, in particular, a dimensional reduction via PCA, with a dimensional cutoff chosen so that the output component strength did not change when the dimension was increased. For each of the separated components produced by the ICA algorithm, we computed correlation scores r1(ℓ) and r2,α(ℓ) between its associated time course Mℓ(t) and the 2 time-paradigm functions φ1(t) and φ2,α(t). The component with the highest value of r1 (respectively r2,α) was identified as the CTR component map C1(v) [C2,θ(v)] corresponding to S1 (S2,θ). (This classification is equivalent to classification based on the GLM threshold τ.) To assess and compare the “quality” of the activity maps produced by the ICA algorithm for the different experimental designs, receiver operating characteristics (ROC) curves were used.
Results for the Different Experimental Conditions.
For each α, i.e., for each of the 3 time paradigms, we compare the quality of the component separation by ICA for the 3 values of θ = 0,100, and 140. The spatial paradigm (and thus also the retinotopic brain activity) has 2 independent components when θ = 100°, because
where D is the whole disk, which translates to a similar relation for the corresponding activation patterns,
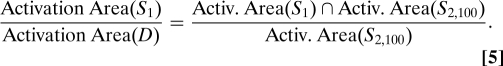 |
In the numerical simulations leading up to Fig. 4, we explored, for each of the small, medium, and large boxes families, the deviation from “true” independence in a fine-grained manner (varying the overlap in small increments) that is not possible in our experimental setting. The distinction among the 3 families can be mimicked, however. Eq. 4 explicitly refers to the disk D as the reference with respect to which independence of S1 and S2,100 are assessed (i.e. D plays here the role of the square V in the numerical study). This means that the corresponding components C1 and C2,100 can be expected to be independent if we compare them within the region := Activ.(D) = {v;Gdisk = 1}, i.e. if we carry out our ICA analysis after restricting ourselves to these voxels only. If we consider a larger collection of voxels as the region within which we carry out the ICA analysis, then Eq. 5 is not satisfied for this larger , and the corresponding components 1; and 2,100; are no longer independent. The deviation from independence increases with the ratio of the areas of regions and .
We carried out 3 different ICA for each (α,θ)-pair, varying the “horizon region” within which the ICA is carried out: = (the region of retinoscopic response to the disk), = V (the whole brain), and = , an intermediate region, ⊂⊂, for which [#]/[#] = [#]/[#], but that was otherwise chosen randomly. To compare these 3 cases via their ROC curves, the ground truth had to be the same for all, with identical regions 1 := {v;G1(v) = 1} and 1 := {v;G1(v) = 0}, and similarly for G2,θ. This “common denominator” was achieved by reducing everything to the intermediate region ; in the case = , we extended the components obtained by the ICA algorithm to V = I by setting them equal to 0 on \; in the case = V, we restricted the components to . Note that this introduces a bias favoring the -components: because 1 and 2,θ are concentrated almost exclusively within , the extension from to automatically (and artificially) ensures that the identification will be overwhelmingly correct on \ for the extended components.
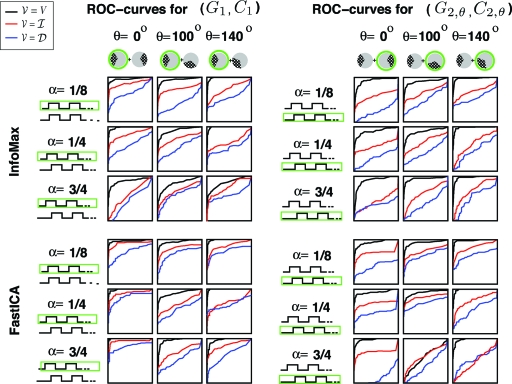
Fig. 7 shows the results of our experiments, for both InfoMax and FastICA, for the 9 different (α,θ)-pairs and the 3 different choices V = , or , for 1 of the subjects (AW).
Fig. 7.
The ROC curves obtained for the ICA-computed component, for the choices = V (black), (red) or (blue); the data are those for 1 subject (AW) in the experiment. This is done for 36 cases: 2 components, 2 ICA algorithms (InfoMax or FastICA), 3 different time paradigms, 3 different overlap angles (θ = 100 is the independent case when = ). The comparison is with the ground truth activation patterns G1 (1st component), G2,θ (2nd component).
For both algorithms we observe that: (i) for the choice = , success in identifying the components is not noticeably higher in the independent case (θ= 100) than for the other values of θ; (ii) in most cases, success in component identification increases as the size of increases, and this despite the bias in our comparison method in favor of the smallest region .
The ROC curves in Fig. 7 were for the data from 1 subject only. The results for the other 2 subjects were similar. Fig. S10 gives average AUCs over all 3 subjects; because the region 0 ≤ false positive rate ≤ 0.05 is the one of most interest, we restricted the AUC computation to this region only.
The averaged ROC powers for the different cases illustrate the relative importance of independence/separation/sparsity.
Role of independence.
When the reference region is , the experimental design mimics the numerical simulations discussed earlier. For InfoMax, the AUC for = turns out to be, at best, only marginally higher for θ = 100 (the “independent” choice) than for the other θ; for FastICA the effect is slightly more pronounced; in neither case is it very convincing. (Note that the ROC power is significantly smaller for = than for the other 2 choices, arguing against any special role of “independence.”)
Role of separation.
For the parameter choices for which the error bars in Fig. S10 are not too wide (i.e., the cases where the mean AUC is most meaningful), there is virtually no difference between the cases θ = 0 (separated activity patterns) and θ = 140 (substantial overlap). The influence of spatial separation of the components, very noticeable in the numerical simulations in the previous section, is thus not so apparent here. However, the θ = 140 case shows substantial differences in effectiveness over the 3 test subjects, so it is not clear how much we can rely on it.
Role of sparsity.
For most (θ,α) [including all (θ,α) with small error bars], AUCV (black) > AUCI (red) > AUCD (blue), for both InfoMax and FastICA. Switching from = to = and then to = V, makes the activation regions more and more sparse with respect to the reference region .
In this experiment, the factor that most influences the success rate of the ICA algorithms is thus sparsity of the components; moreover, this is slightly more marked for InfoMax than for FastICA. Independence of the components plays a marginal role, at best.
Remark. The mention of sparsity in the introduction was within the framework of justifying the use of Independent Component Analysis in brain fMRI analysis: even if components were not truly independent, the heuristic argument went, they were “close to independent” if very sparse. The experiment described above is important in that it teases apart independence and sparsity; it points to sparsity (in its own right, not as promoting independence) as the crucial factor.
Independence Versus Sparsity: Discussion
Summary of our Findings so Far.
InfoMax and FastICA can capture, with considerable accuracy, fine-grained spatial variability in activity patterns.
In our fMRI brain experiments, InfoMax systematically gave better results than FastICA.
In the purely numerical simulations with the small, medium, and large boxes, InfoMax is barely selective for independence; FastICA shows more selectivity towards independence. For both algorithms, sparsity of the components is more important.
Our second fMRI experiment confirms that sparsity affects the success of InfoMax and FastICA more than independence.
A more sophisticated ICA algorithm that performs better than InfoMax and FastICA on independent medium boxes, and is more sensitive to independence, performs less well on the fMRI experiments.
This strongly suggests that when InfoMax and FastICA are effective in brain fMRI, the underlying reason may be other than their striving for independence. Let us now examine sparsity more closely.
Sparsity.
We call a vector v = (v1,v2,…,vN) B-sparse (B ≫ N) if at most B of its entries differ from 0. This notion is basis-dependent; the basis with respect to which the B-sparseness is defined, must be specified. One can also define sparse random vectors, i.e., sparse vectors of which the components are random variables (r.v.). Fig. 8 Left shows realizations of noisy 2-dimensional 1-sparse random vectors, generated as follows: r := γr1 + (1−γ)r2, where γ is an r.v. that takes values 1 and 0 only; r1,r2 are the 2-dimensional random vectors rℓ := αℓa + βℓb, where α1,β2 have the same pdf p(t), and α2,β1 have the rescaled pdf p′(t) = 10 × p(10t). It follows that r is mostly aligned either with a, or with b.
Fig. 8.
Mixtures M1, M2 of 2 2-dimensional processes that are 1-sparse as defined in. Each component has a Gaussian distribution, yet both InfoMax and FastICA succeed in separating them: The directions found with InfoMax are marked by fat red lines, with FastICA by thin green lines.
Very similar figures are often used as “promotional material” for ICA: In this example it is clearly desirable to identify (from the data) the 2 vectors a and b; because these are not orthogonal, they cannot possibly be the outcome of a PCA. Hence (the argument goes) the need for ICA. Yet Fig. 8 depicts processes with 2 sparse rather than independent components: If one conditions r on having a large inner product with b, the distribution of its component along a will be affected. The 2 images of Fig. 8 differ only in that γ equals 1 in 50% for M1, but only 30% for M2. When given this input, InfoMax or FastICA identify the 2 special directions a and b correctly as the components. However, in the example given here, p(t) is gaussian; because ICA methods cannot separate mixtures of independent gaussian processes, the successful separation of components by InfoMax and FastICA underscores again their ability to identify sparse components.
Algorithms for Brain fMRI Adapted to Sparsity.
We conclude that, rather than decomposition methods that search for independent components, one should develop alternate decomposition methods, still striving to write X(t,v) = ∑ℓ = 1L Mℓ(t)Cℓ(v), but targeting decompositions into components that are optimally sparse (only a small number of voxels play a role in each Cℓ) and/or separated (the number of voxels playing a role in more than one Cℓ is small).
It is a good time to look for algorithms involving sparsity. Important progress has been made recently on the problem of recovering a sparse vector from underdetermined linear information. More precisely, if A is a L × N matrix, with L ≫ N, then it is typically impossible to recover an N-dimensional vector u from its image w = Au, or to recover a close approximation to u from w if [∑ℓ = 1L[wℓ − (Au)ℓ]2]1/2 ≤ ɛ. Often, one introduces constraints or penalizations to this type of problem to make it well-defined and well-posed. The knowledge that u is B-sparse, or is close to a B -sparse vector (with B < L), is now known (18–21) to be a sufficient constraint to achieve this, even if the identity of the nonvanishing entries of u is unknown. (There are, of course, technical conditions on A, involving L, N, and B, and satisfied by large classes of matrices, that we shall not discuss here.) The mathematical study for this problem uses insights from computer science, statistics, nonlinear approximation and the geometry of finite-dimensional Banach spaces. Several approaches have been proposed; of special interest are the ultrafast (sublinear in N) methods developed, in e.g., ref. 22, and the ℓ1-optimization methods in refs. 18–21, 23–25, equivalent to ℓ1-penalization methods (26). ℓ1 penalization can be connected with InfoMax and FastICA. Both implicitly assume that the independent components have pdfs of the form p(u) = Ce−α|u|γ. Using this same assumption as a prior for independently distributed components in a Bayesian framework, in which X(v,t) is viewed as ∑ℓMℓ(t)Cℓ(v) corrupted by Gaussian noise, leads to the search for Cℓ that maximize
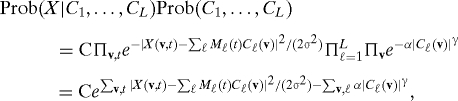 |
where C contains all the appropriate normalization constants, and we assume the Mℓ are normalized. Maximizing this amounts to maximizing the exponent; for γ = 1, this is an ℓ1-penalized functional, also used to obtain sparse decompositions. This may explain why InfoMax and FastICA are good at identifying sparse components.
In the quest for algorithms that are effective for fMRI, based on sparsity of either the components or their intersections, it will be important, to determine the basis with respect to which one expects/hopes for sparsity. In fMRI experiments subjects have as few distractions from the task at hand as possible, in the hope of minimizing the brain activity, so that functional activation maps are less hard to isolate. If this means the signal of interest is confined to a small region only, with little overlap with other activation maps, then sparsity in the voxel domain is likely. For an experiment like the one in ref. 9, where 8 different components all live in the same region of the ventral temporal cortex, sparsity in the voxel domain may be elusive. The original experimental data were not registered so as to make reduction to a common cortical surface model possible for all the subjects. Recently, the experiment was repeated, with such extra registration;† the corresponding new 2-dimensional data on the cortical surface remains to be analyzed, aiming for sparsity not in the purely spatial domain (on the cortical surface), but in, e.g., a corresponding wavelet domain, appropriate for components that have strong peaks in some small areas but behave more smoothly elsewhere.
Acknowledgments.
We thank C. Beckman, D. Donoho, and R. Nowak for their comments. This work was supported in part by National Science Foundation Grants DMS-0421608 and DMS-0245566, National Institute of Mental Health (NIMH) Grant MH067204, and NIMH Conte Center Grant MH62196.
Footnotes
The authors declare no conflict of interest.
Benharrosh M, Takerkart S, Cohen JD, Daubechies I, Richter W, Annual Meeting of the Organization for Human Brain Mapping, June 18–22, 2003, New York.
Sabuncu MR, Singer BD, Bruan RE, Ramadge PJ, Haxby JV, Annual Meeting of theOrganization of Human Brain Mapping, June 11–15, 2006, Florence, Italy.
References
- 1.McKeown MJ, et al. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans Neural Netw. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 3.Bartels A, Zeki S. The chronoarchitecture of the human brain, natural viewing conditions reveal a time-based anatomy of the brain. NeuroImage. 2004;22:419–433. doi: 10.1016/j.neuroimage.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Lee T-W, Ziehe A, Orglmeister R, Sejnowski T. Combining time-delayed decorrelation and ICA: Towards solving the cocktail party problem. Proceedings of the International Conference on Acoustics, Speech, and Signal Processing. Piscataway, NJ: IEEE; 1998. pp. 1249–1252. [Google Scholar]
- 5.Hyvärinen A, Karhunen J, Oja E. Independent Component Analysis. New York: Wiley; 2001. [Google Scholar]
- 6.Worsley KJ, Friston KJ. Analysis of fMRI time series revisited—again. NeuroImage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- 7.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. Spatial and temporal independent component analysis of functional MRI data containing a pair of task related waveforms. Hum Brain Mapp. 2001;13:43–53. doi: 10.1002/hbm.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donoho DL, Flesia AG. Can recent advances in harmonic analysis explain recent findings in natural scene statistics? Netw Comput Neural Syst. 2001;12:371–393. [PubMed] [Google Scholar]
- 9.Haxby JV, et al. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 10.Hansen LK, et al. Generalizable patterns in neuroimaging: How many principal components? NeuroImage. 1999;9:534–544. doi: 10.1006/nimg.1998.0425. [DOI] [PubMed] [Google Scholar]
- 11.Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imag. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 12.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 13.Esposito F, et al. Spatial independent component analysis of functional MRI time series: To what extent do results depend on the algorithm used? Hum Brain Mapp. 2002;16:146–157. doi: 10.1002/hbm.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calhoun VD, Adali T, Pekar JJ, Pearlson GD. Latency (in)sensitive ICA: Group independent component analysis of fMRI data in the temporal frequency domain. NeuroImage. 2003;20:1661–1669. doi: 10.1016/s1053-8119(03)00411-7. [DOI] [PubMed] [Google Scholar]
- 15.Golden C. Spatio-temporal methods in the analysis of fmri data in neuroscience. PhD thesis. Princeton: Princeton Univ; 2005. [Google Scholar]
- 16.Bell AJ, Sejnowski TJ. An information-maximisation approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1003–1034. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- 17.Takerkart S, Benharrosh M, Haxby J, Daubechies I. Independent component analysis of spatially distributed patterns of brain activation measured by fMRI. Proceedings of the 19th GRETSI Symposium, 8–11 September 2003, Paris, France. 2003 [Google Scholar]
- 18.Candès E, Romberg J, Tao T. Stable signal recovery from incomplete and inaccurate measurements. Commun Pure Appl Math. 2006;59:1207–1223. [Google Scholar]
- 19.Candès E. Compressive sampling. Proceedings of the International Congress of Mathematicians, Madrid, Spain, 2006. Madrid: Asociación International Congress of Mathematicians; 2006. [Google Scholar]
- 20.Donoho D. For most large systems of underdetermined equations, the minimum ℓ1 -norm solution is the sparsest solution. Commun Pure Appl Math. 2006;59:797–829. [Google Scholar]
- 21.Donoho D. For most large underdetermined systems of equations, the minimal ℓ-norm near-solution approximates the sparsest near-solution. Commun Pure Appl Math. 2006;59:907–934. [Google Scholar]
- 22.Gilbert A, et al. Near-optimal sparse Fourier representation via sampling. Proceedings of the ACM Symposium on the Theory of Computing, May 19–21, 2002, Montreal, QC, Canada. New York: Assoc for Comput Machinery; 2002. pp. 152–161. [Google Scholar]
- 23.Donoho D, Elad M, Temlyakov V. Stable recovery of sparse overcomplete representations in the presence of noise. IEEE Trans Inf Theory. 2006;52:6–18. [Google Scholar]
- 24.Gribonval R, Nielsen M. Beyond sparsity: Recovering structured representations by ℓ1 -minimization and greedy algorithms—Application to the analysis of sparse underdetermined ICA. J Adv Comput Math. 2008;28:23–41. [Google Scholar]
- 25.Cohen A, Dahmen W, DeVore R. Compressed sensing and best k-term approximation. J Am Math Soc. 2009;22:211–231. [Google Scholar]
- 26.Zhu C. Stable recovery of sparse signals via regularized minimization. IEEE Trans Inf Theory. 2008;54:3364–3367. [Google Scholar]