A 21 kDa Kunitz-type proteinase inhibitor was purified from tamarind (T. indica) seeds, crystallized and characterized by X-ray diffraction.
Keywords: Kunitz-type proteinase inhibitors, Tamarindus indica
Abstract
A Kunitz-type proteinase inhibitor has been purified from tamarind (Tamarindus indica) seeds. SDS–PAGE analysis of a purified sample showed a homogeneous band corresponding to a molecular weight of 21 kDa. The protein was identified as a Kunitz-type proteinase inhibitor based on N-terminal amino-acid sequence analysis. It was crystallized by the vapour-diffusion method using PEG 6000. The crystals belonged to the orthorhombic space group C2221, with unit-cell parameters a = 37.2, b = 77.1, c = 129.1 Å. Diffraction data were collected to a resolution of 2.7 Å. Preliminary crystallographic analysis indicated the presence of one proteinase inhibitor molecule in the asymmetric unit, with a solvent content of 44%.
1. Introduction
Proteinases are proteolytic enzymes that catalyze the hydrolysis of peptide bonds present in proteins. These enzymes are essential for nonspecific digestion of intracellular and extracellular proteins and are specifically required for proteolytic cleavage of inactive precursors, activation of zymogens, processing of hormones and neuropeptides, activation of receptors, protein translocation through membranes etc. (Neurath, 1984 ▶). If the proteolytic activity of these enzymes is not restrained then it can be hazardous for the cells and tissues. Therefore, the activity of proteases is tightly controlled by various regulatory mechanisms such as the regulation of enzyme expression or secretion, the specific degradation of the active form of the enzyme and the inhibition of proteolytic activity by the formation of enzyme complexes with proteinase inhibitors (PIs). Many cells, tissues and organisms produce PIs that are proteins for regulating the enzymatic activity of endogenous proteases (Laskowski & Kato, 1980 ▶; Bode & Huber, 2000 ▶; Krowarsch et al., 2003 ▶).
In plants, PIs consist of a large and assorted group of proteins that have the ability to inhibit proteases by making a reversible inhibitory complex with the enzyme (Laskowski & Kato, 1980 ▶; Richardson, 1977 ▶). Plant proteinase inhibitors (PPIs) play a major role in the defensive mechanism against phytophagous insect infestation and microorganisms. The defensive capability of PPIs is dependent on their ability to inhibit peptidases which are secreted in the gut of insects or by infectious microbes. PPIs isolated from legume seeds have mainly been grouped into Kunitz, Bowman–Birk, potato I and II, squash, cereal superfamily and thaumatin-like proteinacous inhibitors. Proteinase inhibitors of the Kunitz-type family are proteins with a molecular weight of approximately 18–24 kDa and usually contain four Cys residues forming two intramolecular disulfide bridges (Richardson, 1991 ▶; Pandya et al., 1996 ▶). The tamarind tree (Tamarindus indica), a member of the Leguminosae family, grows naturally in many tropical and subtropical regions of the world. Tamarind-fruit pulp is an important food ingredient and tamarind-kernel powder (TKP) is used as a stabilizer and an emulsifier in the food industry (Kumar & Bhattacharya, 2008 ▶).
In this study, we have successfully purified and crystallized a Kunitz-type proteinase inhibitor from the seeds of T. indica. The molecular weight of the purified proteinase inhibitor is about 21 kDa (Araújo et al., 2005 ▶; Rao & Gowda, 2008 ▶). The purified protein is active towards trypsin. Here, we report the isolation, purification, crystallization and preliminary X-ray diffraction analysis of the Kunitz-type proteinase inhibitor from T. indica.
2. Material and methods
2.1. Protein purification
Seeds (15 g) were collected from a tamarind tree growing locally. The fruit pods were soaked overnight at room temperature in 100 mM Tris pH 7.4 (buffer A). The fruit pulp and seed coat were removed manually to obtain the seed kernels. A crude extract was prepared by homogenizing tamarind-seed kernels (TSKs) in buffer A using an electric blender. The extract was centrifuged at 14 000g for 30 min at 277 K and a clear supernatant was obtained. The supernatant was loaded onto Affi-Gel Blue matrix (Bio-Rad Laboratories, Hercules, California, USA) which had been pre-equilibrated with buffer A. After loading the sample, ten column volumes (CV) of buffer A were used for washing and the bound proteins were eluted with a step gradient of NaCl from 0–1.5 M (0.1, 0.2, 0.3, 0.5, 1.0 and 1.5 M) in buffer A. The fractions eluted with 0.5 M NaCl containing trypsin-inhibitory activity were pooled, concentrated using Amicon Ultra 15 (10 kDa cutoff, Millipore) and dialyzed against buffer A. The dialyzed sample was loaded onto a weak cation-exchange matrix [MacroPrep CM (carboxymethyl) support, Bio-Rad] pre-equilibrated with buffer A. The CM flowthrough containing trypsin-inhibitory activity was pooled and loaded onto a weak anion-exchange matrix [MacroPrep DEAE (diethylaminoethyl), Bio-Rad] pre-equilibrated with buffer A. Trypsin-inhibitory activity was detected in the DEAE flowthrough, which was collected and concentrated to about 9 mg ml−1. The homogeneity of the concentrated protein sample was determined by 15% SDS–PAGE stained with Coomassie Brilliant Blue. The protein concentration was determined with the Bio-Rad protein assay kit using BSA as standard and the yield of tamarind-seed proteinase inhibitor was subsequently estimated. The N-terminal amino-acid sequence was obtained by the Edman degradation method on a Shimadzu Automated Protein Sequencer (PPSQ-20).
2.2. Trypsin-inhibition assay
The trypsin-inhibitory activity was determined by measuring the residual enzymatic activity towards the substrate BAPNA–HCl (N-benzoyl-arginine-p-nitroanilide–HCl) predissolved in 20% dimethylsulfoxide (DMSO; Erlanger et al., 1961 ▶). The total reaction-mixture volume was 1.5 ml. 20 µl trypsin at 1 mg ml−1 in 50 mM Tris–HCl pH 7.5 containing 20 mM CaCl2 was incubated with 20 µl protein sample in 160 µl 50 mM Tris–HCl pH 7.5 for 10 min at 310 K. The reaction was initiated by the addition of 1 ml 1.5 mM BAPNA solution. The reaction mixture was incubated at 310 K and 0.3 ml 30% acetic acid solution was added after 10 min to terminate the reaction. The enzymatic hydrolysis of the BAPNA was determined spectrophotometrically by measuring the release of p-nitroaniline at 410 nm.
2.3. Crystallization
For crystallization, the purified protein was used at a concentration of 9 mg ml−1 in 100 mM Tris–HCl pH 7.4. Crystallization was performed by the hanging-drop vapour-diffusion method in 24-well VDX plates (Hampton Research). The drops were prepared by mixing 4 µl protein solution with the same volume of reservoir solution and were equilibrated against 1000 µl reservoir solution at 293 K. The initial crystallization conditions were obtained using Crystal Screen I and Crystal Screen II reagent kits (Hampton Research, USA). Crystals of tamarind-seed proteinase inhibitor were obtained in the presence of 100 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (HEPES) pH 7.0, 100 mM zinc acetate and 10–15%(w/v) polyethylene glycol (PEG) 6000 as precipitant.
2.4. Data collection and analysis
Crystals were mounted using 0.1–0.2 mm cryoloops (Hampton Research) and were flash-cooled in a liquid-nitrogen stream at 100 K generated by an Oxford Cryosystem (Oxford, England). Data were collected with a MAR 345 imaging-plate system using Cu Kα radiation generated by a Bruker Microstar-H rotating-anode generator operated at 45 kV and 60 mA and equipped with Helios optics. Data were collected as 120 images with a crystal-to-detector distance of 200 mm and 1° oscillation per image. The time of exposure per frame was 10 min. The crystal diffracted to 2.7 Å resolution. The diffraction data were processed with the AUTOMAR program (Bartels & Klein, 2003 ▶) and were scaled using the XDS suite of programs (Kabsch, 1993 ▶).
3. Results and discussion
A proteinase inhibitor was isolated and purified to homogeneity from T. indica seeds. Three steps of chromatography involving affinity chromatography and ion-exchange chromatography were used for purification of the tamarind-seed proteinase inhibitor. A trypsin-inhibitory activity assay was performed at each step of purification. The supernatant obtained after centrifugation of the crude extract was applied onto an Affi-Gel Blue column and the bound proteins were eluted with a step gradient of 0–1.5 M NaCl. Trypsin-inhibitory activity was present in the fractions eluted with 0.5 M NaCl, but the fractions were heterogeneous when analyzed on SDS–PAGE. Therefore, fractions containing trypsin-inhibitory activity were pooled, concentrated and dialyzed against buffer A. The protein sample was then loaded onto a CM column and the CM flowthrough containing trypsin-inhibitory activity was collected. It was then loaded onto a DEAE column and the flowthrough was collected. The collected DEAE flowthrough exhibiting trypsin-inhibitory activity was concentrated and assessed for homogeneity by SDS–PAGE; a single protein band appeared. The estimated yield of purified protein was about 0.6 mg per gram of seeds. Molecular-weight estimation of tamarind-seed proteinase inhibitor was performed using protein molecular-weight markers and reducing SDS–PAGE. The estimated molecular weight of the purified protein was about 21 kDa (Fig. 1 ▶; Araújo et al., 2005 ▶; Rao & Gowda, 2008 ▶). The first 15 amino-acid residues of the purified protein were determined by automated N-terminal amino-acid sequencing and the obtained sequence was identical to that of the recently reported proteinase inhibitor from tamarind belonging to the Kunitz inhibitor family (Rao & Gowda, 2008 ▶). The purified protein was concentrated to about 9 mg ml−1 and used for crystallization.
Figure 1.
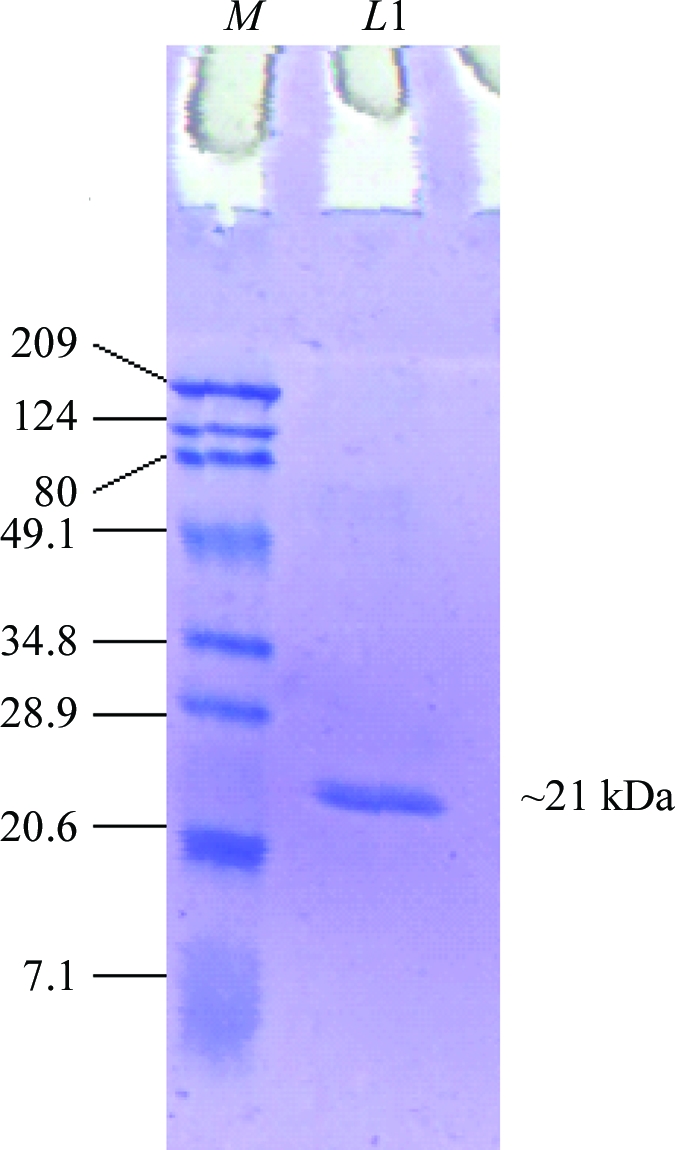
15% SDS–PAGE of tamarind-seed proteinase inhibitor under reducing conditions. Lane M, protein molecular-weight markers (kDa); lane L1, purified protein.
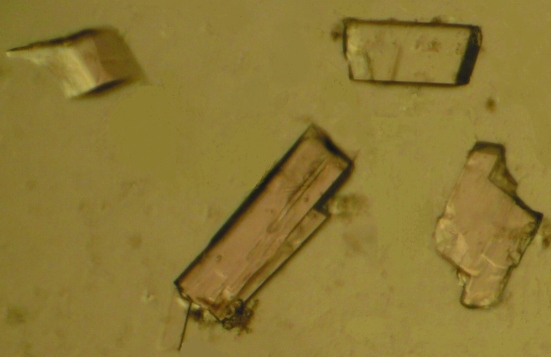
Crystals of the Kunitz-type proteinase inhibitor from T. indica were obtained in 5–7 d using 100 mM HEPES pH 7.0, 10%(w/v) PEG 6000 and 100 mM zinc acetate (Fig. 2 ▶). The crystals belonged to the orthorhombic space group C2221 and diffracted to 2.7 Å resolution in-house. The unit-cell parameters were found to be a = 37.2, b = 77.1, c = 129.1 Å, with one molecule per asymmetric unit. This corresponds to a crystal volume per unit molecular weight (V M) of 2.2 Å3 Da−1, given the molecular weight of 21 kDa for the protein, with a solvent content of 44% (Matthews, 1968 ▶). The data-collection statistics are summarized in Table 1 ▶.
Figure 2.
Crystals of tamarind-seed proteinase inhibitor. The longest dimension of a typical crystal is between 100 and 150 µm.
Table 1. Data-collection statistics for tamarind-seed proteinase inhibitor.
Values in parentheses are for the highest resolution shell (2.8–2.7 Å).
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 37.2, b = 77.1, c = 129.1 |
| Resolution range (Å) | 50–2.7 (2.8–2.7) |
| Completeness (%) | 99.1 (99.8) |
| Rmerge† (%) | 6.7 (22.3) |
| Mean I/σ(I) | 6.4 (2.1) |
| No. of observed reflections | 27781 |
| No. of unique reflections | 5789 (527) |
| Molecules per ASU | 1 |
| Mathews coefficient (Å3 Da−1) | 2.2 |
| Solvent content (%) | 44 |
| Redundancy | 3.5 (2.3) |
| Mosaicity (°) | 0.75 |
R
merge = 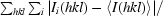
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl) is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl) is the weighted average intensity for all observations i of reflection hkl.
Acknowledgments
The authors are grateful to and thank the macromolecular crystallographic facility (MCU) at the Institute Instrumentation Centre (IIC), Indian Institute of Technology (IIT), Roorkee for data collection. DNP thanks MHRD, Government of India and Preeti thanks CSIR for financial support.
References
- Araújo, C. L., Bezerra, I. W., Oliveira, A. S., Moura, F. T., Macedo, L. L., Gomes, C. E., Barbosa, A. E., Macedo, F. P., Souza, T. M., Franco, O. L., Bloch, C. Jr & Sales, M. P. (2005). J. Agric. Food Chem.53, 4381–4387. [DOI] [PubMed]
- Bartels, K. S. & Klein, C. (2003). The AUTOMAR Manual, v.1.4. Norderstedt, Germany: MAR Research GmbH.
- Bode, W. & Huber, R. (2000). Biochim. Biophys. Acta, 1477, 241–252. [DOI] [PubMed]
- Erlanger, B., Kokowsky, N. & Cohen, W. (1961). Arch. Biochem. Biophys.95, 271–278. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Krowarsch, D., Cierpicki, T., Jelen, F. & Otlewski, J. (2003). Cell. Mol. Life Sci.60, 2427–2444. [DOI] [PMC free article] [PubMed]
- Kumar, C. S. & Bhattacharya, S. (2008). Crit. Rev. Food Sci. Nutr.48, 1–20. [DOI] [PubMed]
- Laskowski, M. Jr & Kato, I. (1980). Annu. Rev. Biochem.49, 593–626. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Neurath, H. (1984). Science, 224, 350–357. [DOI] [PubMed]
- Pandya, M. J., Smith, D. A., Yarwood, A., Gilroy, J. & Richardson, M. (1996). Phytochemistry, 43, 327–331. [DOI] [PubMed]
- Rao, D. H. & Gowda, L. R. (2008). J. Agric. Food Chem.56, 2175–2182. [DOI] [PubMed]
- Richardson, M. (1977). Phytochemistry, 16, 159–169.
- Richardson, M. (1991). Methods in Plant Biochemistry, Vol. 5, edited by P. M. Dey & J. B. Harborne, pp. 259–305. New York: Academic Press.