Abstract
Background
GATA3 activates transcription of the TH2 cytokines, including IL13, an important step in the allergic inflammatory pathway.
Objective
We sought to identify associations of single nucleotide polymorphisms of the genes GATA3 and IL13 and their interactions with rhinitis and allergic sensitization during childhood.
Methods
We performed genetic association studies in a cohort of children (n = 923) who have been evaluated for the development of rhinitis and allergic sensitization by means of skin prick tests (SPTs) at age 10 years. Pyrosequencing was used to genotype 7 polymorphisms from GATA3 and 5 from IL13. A novel model-selection procedure combining logistic regression models and classification was used to study the contributions of the polymorphisms and their interactions.
Results
Combinations of polymorphisms and their interactions increase the risk for rhinitis and allergic sensitization at age 10 years. A model with rs1058240, rs379568, and rs4143094 (GATA3) and rs1800925 (IL13) and their interactions was selected to predict rhinitis and positive SPT responses. rs1058240 was associated with rhinitis and allergic rhinitis (P < .05), and the gene-gene interaction rs1058240:rs1800925 was associated with rhinitis (P = .043). The odds ratios for 4 genotype combinations were significant for rhinitis or SPTs (P < .044).
Conclusion
Gene-gene interaction between GATA3 and IL13 polymorphisms can influence the risk of childhood rhinitis. Our study suggests that set associations of polymorphisms are important in studying genetic associations for complex phenotypes, such as rhinitis and atopy.
Keywords: Rhinitis, allergic rhinitis, atopy, GATA3, IL13, genetic association, gene-gene interactions
Rhinitis is a disease of high prevalence, especially in industrialized countries.1 This high prevalence translates into high cost to society in terms of overall health care use and quality of life of those with moderate-to-severe disease. Recent studies showed that the upward trend in prevalence seen during the last few decades might not yet have reached a peak.2 The reasons for these trends remain unclear but probably reflect environmental influence on genetic predisposition. Noninfectious rhinitis is classified into allergic and nonallergic rhinitis, depending on the presence or absence of atopic sensitization. Familial predisposition for rhinitis, both allergic and nonallergic, is well established.3,4 Atopy is characterized by production of specific IgE after exposure to allergens. In our birth cohort maternal and paternal rhinitis and atopy were shown to be independent risk factors for investigator-diagnosed rhinitis at age 10 years.5 Both atopy and rhinitis have multifactorial inheritance, and it is likely that different combinations of genes increase the risk of phenotypic expression, resulting in allergic and non-allergic rhinitis.
GATA3 was selected as a candidate gene for rhinitis based on its action in driving the TH2 cytokine response. It has been associated with human asthma6 and is located within a quantitative trait locus for allergic asthma in a murine model.7 IL13 is an important cytokine involved in the IgE pathway, and IL13 is one of the genes most consistently associated with asthma and IgE-related phenotypes in association studies.8–14 An association of the IL13 gene with serum IgE levels was shown in patients with allergic rhinitis but not with allergic rhinitis itself.15 Because of the critical role of GATA3 in TH2 cell development and its role in regulating expression of IL4, IL5, and IL13, we believed it is important to examine GATA3 along with IL13. We tested for the association of human GATA3 and IL13 gene polymorphisms with rhinitis and for positive skin prick test (SPT) responses at age 10 years. The combinations of single nucleotide polymorphisms (SNPs) and possible interactions that were statistically significant for rhinitis were then further tested for allergic rhinitis or nonallergic rhinitis and compared with results in the control group of children without rhinitis and with negative SPT responses. We used statistical variable selection procedures and cross-validation to investigate the influence of SNP patterns in the IL13 and GATA3 genes.
METHODS
Study population
A whole population birth cohort (n = 1456) was established on the Isle of Wight, United Kingdom, in 1989 to study the natural history of asthma and allergic disorders and identify genetic and environmental risk factors important in their development. Approval was given by the local research ethics committee, and parental informed consent was obtained. The population is largely white (99%) and living in a semirural environment with no heavy industry. At birth, information was collected on the family historyof atopy and potential environmental risk factors. These children have been followed at the ages of 1, 2, 4, and 10 years.16 At every follow-up, detailed questionnaires were completed with the parents for each child regarding the prevalence of asthma, rhinitis, and atopic dermatitis. Information was also collected and updated on environmental risk factors. At 4 and 10 years, SPTs were performed to 14 common food allergens and aeroallergens (ALK-Abelló, Hørsholm, Denmark) on 981 and 1036 children, respectively.17 Anticoagulated blood samples were collected at age 10 years and stored frozen for subsequent DNA analysis (n = 923).
Genotyping
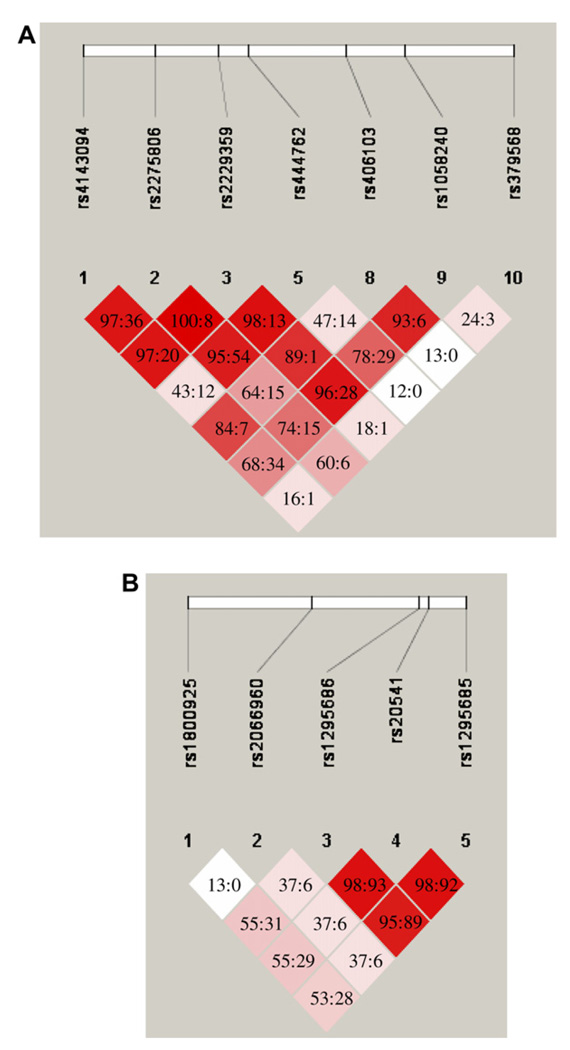
Genomic DNA was isolated from blood samples by using QIAamp DNA Blood Kits (Qiagen, Valencia, Calif) or the ABI PRISM 6100 Nucleic Acid PrepStation (Applied Biosystems, Foster City, Calif). Polymorphisms in the GATA3 and IL13 genes were examined with the SNPper (http://snpper.chip.org/) and Applied Biosystems (http://www.appliedbiosystems.com/) databases. Genotyping was conducted by using biotin-streptavidin–based pyrosequencing performed on PSQ-96 instrumentation (Biotage AB, Uppsala, Sweden) or by using fluorogenic 5′ nuclease chemistry PCR with Assays on Demands kits cycled on a 7900HT Sequence Detection System (Applied Biosystems). Haploview 3.32 software (Broad Institute, Cambridge, Mass) was used to conduct marker quality checks and generate linkage disequilibrium plots for each gene (Fig 1).
FIG 1.
Linkage disequilibrium plot using the Haploview program for the genes IL13 and GATA3. D′ and r2 are pairwise linkage disequilibrium determinants. The standard D′/LOD color scheme for Haploview is used as follows: D′ = 1 and LOD ≥ 2 is bright red, D′ < 1 and LOD ≥ 2 is shades of red/pink, and D′ < 1 and LOD < 2 is white. A, GATA3 linkage disequilibrium plot. D′:r2 values are displayed. B, IL13 linkage disequilibrium plot. D′:r2 values are displayed.
The SNPs were selected to provide data across the GATA3 and IL13 genes. IL13 is a small gene, approximately 3 kb in size, that has been genetically evaluated in a number of studies.8–14,18 The IL13 SNPs genotyped in the present study span from the promoter to the 3′ untranslated region (UTR) and identify 1 linkage disequilibrium block. The GATA3 gene is approximately 20 kb in length, including introns. Initially, 10 validated SNPs identified by using the SNPper database were screened in a subset of our population. Based on minor allele frequencies and the linkage disequilibrium structure of the gene obtained in these preliminary studies,19 7 SNPs located in the promoter, introns, exon, and 3′ UTR were genotyped in the entire population. These SNPs provide information on the entire GATA3 gene. Seven SNPs from the GATA3 gene and 5 from IL13 were used in this study (Table I).
TABLE I.
Polymorphism assay details
| SNP | Pyrosequencing F primer | Pyrosequencing R primer | Pyrosequencing probe |
|---|---|---|---|
| GATA3 | |||
| rs4143094 | ACGCAGCACTGGGGCTATTCT | AGGCACTCTGTGTGACC* | GTAGGGGTTTT |
| rs2275806 | CCCCAAATTAAGCTCTCAGC | CGGGAAAAGAAGACTCCAAA* | GGGGCGAGCAGAG |
| rs2229359 | AGCATGAAGCTGGAGTCGTC* | GAGCTGTACTCGGGCACGTA | GAGGCTCCACCCAG |
| rs444762 | AGCATGAAGCTGGAGTCGTC* | GAGCTGTACTCGGGCACGTA | GAGGCTCCACCCAG |
| rs406103 | AGTGGACTGGGATCAGCAAG* | GAGCGATTCACTTGGAGAGG | GAGGGTGGTGGTGAC |
| rs1058240 | CCAGTTCTGGGCAATCAG* | ATTCTTGTTCACAAAGCATGTAGGC | TGAAACCCTCA |
| rs379568 | ATAAGAGTCAGCTCACTCCTC | CAGCACACTTCTGTCTGG* | AGTTTGGGGATT |
| IL13 | |||
| rs1800925 | NA† | ||
| Rs2066960 | GCATTTGCCAACTGGATTTT* | GGCAAGGAGCGGACTCTAC | AAGGGCGGGCCTAT |
| rs1295686 | NA† | ||
| rs20541 | AAAGAAACTTTTTCGCGAGGG | ATGATGCTTTCGAAGTTTCAGTTG* | ACTTTTTCGCGAGGG |
| rs1295685 | N/A† |
Indicates biotinylated primer.
Applied Biosystems TaqMan SNP genotyping assays used: C_8932056_10 for rs1800925, C_8932053_10 for rs1295686, and C_8932052_10 for rs1295685.
Outcomes
Outcomes of rhinitis and SPTs were evaluated at age 10 years. The relevant question for the period prevalence of rhinitis was as follows: Has your child, in the last 12 months, had a problem with sneezing or a runny or blocked nose when he or she did not have a cold or the flu? A positive response was taken as indicating rhinitis. Children with rhinitis were further classified into allergic and nonallergic rhinitis. A child with rhinitis and a positive reaction on an SPT to 1 or more allergens was said to have allergic rhinitis. Nonallergic rhinitis was defined as the presence of rhinitis in the absence of a positive SPT response. We focused on rhinitis at age 10 years because rhinitis is a clinical diagnosis, and in early childhood responses to standardized questions are not specific to make a firm diagnosis. Furthermore, upper respiratory tract infections are frequent in early childhood, blurring the distinction between infectious and noninfectious rhinitis. The allergic rhinitis and nonallergic rhinitis subgroups were compared with the control group of children without rhinitis and with a negative SPT response. Furthermore, SPT responses at age 10 years were examined as a separate outcome for genetic association analysis because the mechanisms of allergic sensitization, although related, might be different from those specific for nasal symptoms.
Statistical analysis
In this article a 2-step procedure with logistic regression and classification of selecting SNP patterns among the 12 SNPs was used. We performed an exhaustive search of logistic regression models for 12 SNPs in 2 genes involved in the IgE pathway. The model search for the best subset of variables was based on the Akaike Information Criterion (AIC).20 The AIC is a measure of goodness of fit with a trade-off for complexity versus how well the model fits the data. Among the models with up to 4 main effects and all possible 2-way SNP-SNP interactions, we selected those with the smallest AIC, such that the SNPs included in the model have highest rank, as measured in the classification procedure random forests.21 This novel approach allowed the search for unique SNP patterns in combination with selected SNP-SNP interactions. All statistical analyses were performed with R v2.4.1 (http://cran.r-project.org) to identify genetic associations and SNP-SNP interactions with rhinitis, allergic rhinitis, and nonallergic rhinitis at age 10 years. For the model comparisons with the AIC criterion to be valid, only observations with complete genotype information were used. When all 12 SNPs were included, this procedure searched for models by using 687 observations. The AIC was computed for 64,000 models with 2, 3, or 4 main effects and any combination of 2-way interaction terms. A model was selected from the top 100 models with a small AIC that included SNPs with high rank assigned by the classification procedure random forest.21 The rank or importance of a predictor SNP was determined by the increase in the misclassification error rate when the values of the predictor were randomly permuted. The combinations of SNPs and possible interactions that were statistically significant for rhinitis were then further tested for allergic rhinitis or nonallergic rhinitis and compared with the control group of children without rhinitis and a negative SPT response by using a logistic regression model. Furthermore, these SNPs were tested for association with SPTat age 10 years. We concentrated on the outcomes at age 10 years because of the reliability of the diagnosis.
A bootstrap analysis was performed to explore the proportion of times a genetic association is significant at the 5% level in random subsets. Through a bootstrap analysis,22 we provided a measure of how likely the SNPs and their interactions were to be selected and examined what other factors could have been preferred when some cases or some control subjects were missing. In the bootstrap approach we selected 10,000 random samples (n = 923) with replacement (including missing data in the original set) and counted the number of times a particular SNP or interaction was significant at the 5% level. This proportion indicated whether the significance of a particular SNP relied on the specific set of cases and control subjects or would also be significant in random subsets. Some SNP patterns repeatedly occurred among the top 100 models. A leave-one-out model-selection procedure was performed to check the robustness of the model selected. For this purpose, 1 SNP at a time was excluded, and the same model-selection procedure was performed. To better understand the significance of a GATA3-IL13 interaction, separate models were fitted to rhinitis, allergic and nonallergic rhinitis, and SPT response as outcomes. The selection procedure used the codominant model that compares the rare homozygous or heterozygous variant effects with the common homozygous variant as the reference category, respectively, and the corresponding results of the logistic regression models have been reported. For computational efficiency for cross-validation, we used a dominant model in which the heterozygous and the rare homozygous variants were combined. The selected SNP combination was the same in both the dominant and codominant models.
RESULTS
Phenotype and population characteristics
The characteristics of the study population are shown in Table II. Of the 923 genotyped children who were examined at age 10 years, 162 (18%) had a physician’s diagnosis of rhinitis. Based on SPT responses, children with rhinitis were classified into subgroups of 99 (11%) children with allergic rhinitis and 62 (7%) children with nonallergic rhinitis. These subgroups were compared with a group of 606 (66%) children without rhinitis and with negative SPT responses. More than half of the children with rhinitis were found to have a family history of hay fever; 84 (55%) children with rhinitis had a mother or father with hay fever, and 104 (64%) children with rhinitis had an immediate family member (mother, father, or sibling) with hay fever.
TABLE II.
Population characteristics
| Variable | Initial sample (n = 1373; %) | No. used in variable selection procedure* (n = 687; %) | No. genotyped† (n = 923; %) |
|---|---|---|---|
| Sex | |||
| Male | 696 (51) | 349 (51) | 463 (50) |
| Female | 677 (49) | 338 (49) | 460 (50) |
| Missing | 0 (0) | 0 (0) | 0 (0) |
| Rhinitis at age 10 y | |||
| Yes | 205 (15) | 133 (19) | 162 (18) |
| No | 1157 (84) | 497 (81) | 761 (82) |
| Missing | 11 (1) | 0 (0) | 0 (0) |
| Positive SPT response at age 10 y | |||
| Yes | 279 (20) | 187 (27) | 250 (27) |
| No | 757 (55) | 497 (72) | 668 (72) |
| Missing | 337 (25) | 3 (1) | 5 (1) |
| Maternal hay fever | |||
| Yes | 320 (23) | 164 (24) | 228 (25) |
| No | 998 (73) | 488 (71) | 653 (71) |
| Missing | 55 (4) | 35 (5) | 42 (5) |
| Paternal hay fever | |||
| Yes | 216 (16) | 118 (17) | 153 (17) |
| No | 1100 (80) | 533 (78) | 726 (79) |
| Missing | 57 (4) | 36 (5) | 44 (5) |
| Sibling hay fever | |||
| Yes | 254 (21) | 117 (19) | 174 (21) |
| No | 928 (75) | 471 (76) | 624 (75) |
| Missing | 47 (4) | 31 (5) | 36 (4) |
Number of individuals with complete genotype information to select variables. Complete genotype information is necessary for valid model comparisons.
Number of individuals genotyped and used in the logistic regression model.
Genotyping
All polymorphisms examined were in Hardy-Weinberg equilibrium. Minor allele frequencies exceeded 0.10 in all SNPs, with the exception of rs2229359, which was genotyped despite a low minor allele frequency (0.07) because it was located in an exon (Table III).
TABLE III.
GATA3 and IL13 polymorphisms tested
| SNP | Location | Type | IOW minor allele frequency | dbSNP minor allele frequency* | Genotypes | IOW genotype frequency (n) |
|---|---|---|---|---|---|---|
| GATA3 | Chr 10 | |||||
| rs4143094 | 8129142 | Intergenic | 0.243 | 0.250 | GG/GT/TT | 497/334/46 (877) |
| rs2275806 | 8135346 | 5′ Promoter | 0.456 | 0.373 | AA/AG/GG | 258/482/177 (917) |
| rs2229359 | 8140653 | Exon 3 | 0.066 | 0.065† | GG/GA/AA | 804/114/4(922) |
| rs444762 | 8143266 | Intron 3 | 0.332 | 0.348† | CC/AC/AA | 410/414/99 (923) |
| rs406103 | 8151627 | Intron 5 | 0.234 | 0.172 | GG/AG/AA | 549/315/58 (922) |
| rs1058240 | 8156604 | 3′ UTR | 0.191 | 0.183 | TT/CT/CC | 595/298/27 (920) |
| rs379568 | 8165825 | 3′ UTR | 0.124 | 0.092 | GG/AG/AA | 644/188/10 (842) |
| IL13 | Chr 5 | |||||
| rs1800925 | 132020708 | 5′ Promoter | 0.201 | 0.192 | CC/CT/TT | 557/295/35 (907) |
| rs2066960 | 132022334 | Intron 1 | 0.097 | 0.150 | CC/CA/AA | 729/157/8 (894) |
| rs20541 | 132023742 | Intron 3 | 0.196 | 0.233 | GG/GA/AA | 583/291/32 (906) |
| rs1295685 | 132023863 | Exon 4 | 0.200 | 0.242 | GG/GA/AA | 584/280/41 (905) |
| rs1295686 | 132024344 | 3′ UTR | 0.194 | 0.242 | CC/CT/TT | 483/240/25 (748) |
IOW, Isle of Wight, United Kingdom, 1989–90 birth cohort; dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP; n = number of individuals with genotype data (total number of individuals genotyped = 923).
Data are HapMap CEU minor allele frequencies unless marked with a dagger.
Data are Program for Genomic Applications European Panel minor allele frequencies because HapMap data are not available.
SNP-SNP interactions and set association analyses
Combinations of SNPs and their interactions increased the risk for rhinitis and atopy at age 10 years. The logistic regression model combined GATA3 and IL13 polymorphisms and their interaction and shows statistically significant associations for rhinitis at age 10 years. The adverse effects were due to the heterozygous variant at each locus (Table IV). There were 13 pairs of twins. However, they did not influence the association because there were no major changes when we repeated the analysis without the twins. A model with 3 GATA3 SNPs (rs1058240, rs379568, and rs4143094) and 1 IL13 polymorphism (rs1800925) and their interactions was selected to predict rhinitis and a positive SPT response. rs1058240 was associated with rhinitis and allergic rhinitis (P < .05), and the gene-gene interaction rs1058240:rs1800925 was associated with rhinitis (P < .05). rs379568 (GATA3) and rs1800925 (IL13) were associated with SPT responses. Genotype combinations of these 4 polymorphisms showed an increased risk for rhinitis and a positive SPT response. Based on the information available from these 2 genes, we computed odds ratios and their 95% CIs to provide an estimate of the risk associated with combinations of these genotypes (Table V) and predicted probabilities from the logistic regression model that show the probability of having rhinitis at age 10 years given a specific genotype (Table VI).
TABLE IV.
Association analysis of GATA3 and IL13 SNPs and SNP pairs with rhinitis, allergic and nonallergic rhinitis, and SPT response at age 10 years (n = 923)
| Rhinitis |
Allergic rhinitis† |
Nonallergic rhinitis† |
SPT |
|
|---|---|---|---|---|
| Risk factors* | Affected: n = 162, not affected: n = 761 |
Affected: n = 99, not affected: n = 606 |
Affected: n = 62, not affected: n = 606 |
Affected: n = 250, not affected: n = 668 |
| GATA3 rs1058240 | 0.032 | 0.044 | 0.290 | 0.711 |
| GATA3 rs379568 | 0.128 | 0.109 | 0.154 | 0.012 |
| GATA3 rs4143094 | 0.018 | 0.051 | 0.054 | 0.182 |
| IL13 rs1800925 | 0.168 | 0.228 | 0.083 | 0.033 |
| GATA3 rs1058240:GATA3 rs379568 | 0.004 | 0.005 | 0.428 | 0.557 |
| GATA3 rs1058240:IL13 rs1800925 | 0.043 | 0.112 | 0.244 | 0.211 |
| GATA3 rs379568:GATA3 rs4143094 | 0.019 | 0.016 | 0.228 | 0.091 |
| GATA3 rs379568:IL13 rs1800925 | 0.069 | 0.083 | 0.119 | 0.117 |
| GATA3 rs4143094:IL13 rs1800925 | 0.062 | 0.106 | 0.105 | 0.189 |
The analyses were performed with logistic regression.
The statistical significance is due to the comparison of the heterozygous variant versus the common homozygous variant. Therefore only those P values are listed.
The control group for allergic and nonallergic rhinitis consisted of those children without rhinitis at age 10 years and negative SPT responses.
TABLE V.
Interactions between GATA3 and IL13 genotypes for rhinitis and SPT responses
| GATA3 rs1058240 | GATA3 rs379568 | GATA3 rs4143094 | IL13 rs1800925 | Rhinitis | Rhinitis (P value)* | SPT | SPT (P value)* |
|---|---|---|---|---|---|---|---|
| TT | GA or AA | GT or TT | CA or AA | 0.87 (0.03–5.12) | .868 | 2.14 (0.41–8.77) | .448 |
| CT or CC | GG | GG | CC | 0.49 (0.07–1.78) | .591 | 1.90 (0.76–4.41) | .303 |
| TT | GA or AA | GG | CA or AA | 0.72 (0.16–2.23) | .747 | 1.81 (0.73–4.18) | .324 |
| TT | GG | GG | CC | 1 | NA | 1 | NA |
| TT | GA or AA | GT or TT | CC | 0.70 (0.03–3.92) | .803 | 1.62 (0.32–6.03) | .641 |
| CT or CC | GG | GT or TT | CC | 1.10 (0.55–2.14) | .868 | 1.40 (0.78–2.47) | .409 |
| TT | GG | GT or TT | CA or AA | 1.76 (0.64–4.29) | .542 | 1.47 (0.58–3.45) | .537 |
| TT | GG | GG | CA or AA | 1.26 (0.67–2.33) | .689 | 2.07 (1.23–3.48) | .044 |
| CT or CC | GA or AA | GT or TT | CA or AA | 1.58 (0.48–4.31) | .677 | 1.07 (0.33–2.86) | .927 |
| TT | GA or AA | GG | CC | 1.95 (0.83–4.31) | .280 | 2.58 (1.24–5.25) | .044 |
| CT or CC | GG | GT or TT | CA or AA | 1.39 (0.60–3.00) | .677 | 1.06 (0.48–2.18) | .927 |
| TT | GG | GT or TT | CC | 2.11 (1.05–4.13) | .090 | 2.19 (1.18–4.04) | .044 |
| CT or CC | GG | GG | CA or AA | 7.59 (1.83–33.7) | .012 | 1.26 (0.17–5.62) | .927 |
| CT or CC | GA or AA | GT or TT | CC | 2.58 (1.23–5.28) | .032 | 2.16 (1.08–4.25) | .071 |
| CT or CC | GA or AA | GG | CC | 4.13 (1.27–12.55) | .032 | 3.66 (1.19–10.97) | .044 |
| CT or CC | GA or AA | GG | CA or AA | 8.05 (1.61–45.65) | .014 | 3.16 (0.56–15.72) | .303 |
Odds ratios (ORs) and 95% CIs for the different genotype combinations are shown.
The probabilities were adjusted for multiple testing by using the Benjamini-Hochberg algorithm.
TABLE VI.
Predictive probabilities for rhinitis, allergic rhinitis, and nonallergic rhinitis at age 10 years for given genotype combinations
| No. of individuals with genotype pattern | GATA3 rs1058240 | GATA3 rs379568 | GATA3 rs4143094 | IL13 rs1800925 | Rhinitis (predictive probability [%]) | Allergic rhinitis (predictive probability [%]) | Nonallergic rhinitis (predictive probability [%]) | SPT (predictive probability [%]) |
|---|---|---|---|---|---|---|---|---|
| 9 | TT | GA or AA | GT or TT | CA or AA | 3.2 | 2.2 | 2.2 | 19.8 |
| 29 | CT or CC | GG | GG | CC | 7.7 | 5.4 | 3.9 | 20.4 |
| 30 | TT | GA or AA | GG | CA or AA | 12.7 | 11.5 | 7.1 | 36.3 |
| 209 | TT | GG | GG | CC | 14.0 | 11.1 | 6.3 | 21.4 |
| 11 | TT | GA or AA | GT or TT | CC | 14.5 | 10.4 | 11.7 | 29.4 |
| 100 | CT or CC | GG | GT or TT | CC | 14.7 | 10.8 | 8.8 | 27.4 |
| 32 | TT | GG | GT or TT | CA or AA | 15.3 | 11.8 | 9.3 | 28.5 |
| 119 | TT | GG | GG | CA or AA | 18.5 | 15.0 | 11.7 | 31.8 |
| 25 | CT or CC | GA or AA | GT or TT | CA or AA | 20.7 | 14.6 | 7.5 | 19.3 |
| 42 | TT | GA or AA | GG | CC | 22.2 | 19.4 | 13.2 | 35.9 |
| 55 | CT or CC | GG | GT or TT | CA or AA | 23.0 | 13.5 | 16.4 | 21.0 |
| 67 | TT | GG | GT or TT | CC | 25.1 | 21.0 | 13.7 | 28.6 |
| 9 | CT or CC | GG | GG | CA or AA | 27.3 | 17.2 | 20.1 | 23.7 |
| 51 | CT or CC | GA or AA | GT or TT | CC | 29.3 | 26.0 | 13.4 | 36.3 |
| 15 | CT or CC | GA or AA | GG | CC | 41.1 | 42.0 | 15.0 | 43.4 |
| 7 | CT or CC | GA or AA | GG | CA or AA | 53.2 | 50.1 | 21.8 | 35.6 |
The genotype patterns have been sorted based on the predictive probability for rhinitis at age 10 years (from low to high risk). A predictive probability of .532 indicates a 53.2% chance for an individual with the given genotype pattern to have rhinitis at age 10 years.
This combination of GATA3 and IL13 SNPs was represented in the top-ranked models for rhinitis, allergic rhinitis, and SPT response when applying the model-selection procedure for these outcomes separately. However, a different combination emerges for nonallergic rhinitis, consisting of rs4143094 (GATA3), rs1800925, rs1295686, and rs129585 (all IL13).
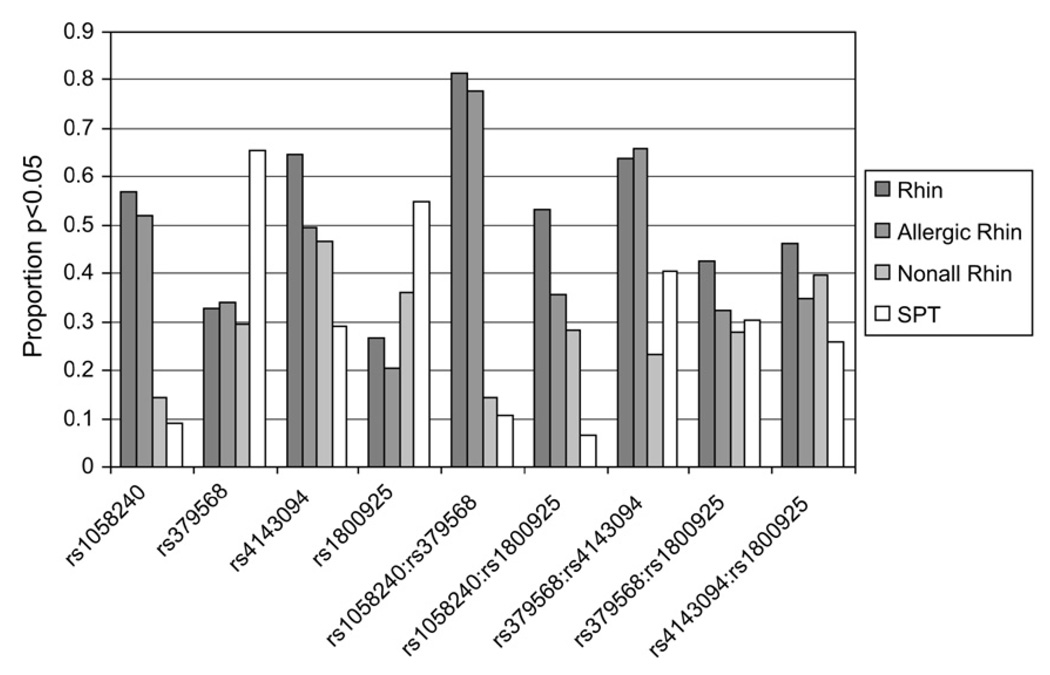
The bootstrap values in Fig 2 indicate the number of times a particular SNP or interaction was significant at the 5% level in 10,000 random samples. This provides a measure of how likely the SNPs and their interactions were to be selected and examines what other factors could have been preferred when some cases or some control subjects were missing. The associations of the SNPs with rhinitis and allergic rhinitis are similar. Fig 2 also highlights the importance of the interactions.
FIG 2.
Proportions of the number of times an SNP or interaction of SNPs has been statistically significant at the 5% level in the logistic regression model in 10,000 bootstrap samples of size 923.
DISCUSSION
The ability to predict the risk for having allergic disease in later childhood is enormously helpful in terms of offering an accurate prognosis to parents and identifying children for investigation of preventative strategies. We have previously reported the development of a predictive index for risk of having persistent asthma up to age 10 years using information available from history and SPT responses.23 The development of allergic disease, including rhinitis, depends on both genetic and environmental factors and their interactions. It has been suggested that heritable factors predominate over shared environmental influences in asthma,24 and the same might be true for rhinitis because both diseases are closely linked.25 Using the genotype information available, we estimated the risk of an individual having rhinitis at age 10 years. As expected, the majority of children fall into the category in which the risk is similar to that of the whole group (around 20%). However, a significant number of children were identified at both ends of the spectrum, having very low or high risk (Table VI).
Because of the critical role of GATA3 in TH2 cell development and its role in regulating expression of IL4, IL5, and IL13, it is important to examine GATA3 along with IL13. Considering the biologic relation between GATA3 and IL13, we sought to study the SNP patterns and interactions between these genes in the context of rhinitis and atopy. GATA3 has been associated with human asthma6 and is located within a quantitative trait locus for allergic asthma in a murine model,7 and IL13 is one of the genes most consistently associated with asthma and IgE-related phenotypes in association studies.8–14 Gene-gene interactions, in particular 2 loci interactions, in the TH2 pathway have been examined for IL13 and IL4/IL4R for asthmatic subjects in Dutch, Chinese, and African American populations.26–28 The GATA3 SNPs for this analysis were chosen to provide information on the entire gene, and we found that a model with 4 loci to be a better predictor than models with 2 loci for rhinitis. To our knowledge, this is the first association analysis for GATA3 and IL13 interactions for rhinitis and atopy.
GATA3 is expressed at high levels in TH2 lineage cells. From a pathogenetic viewpoint, the proteins these genes encode are important in the IgE-mediated immunologic pathway. It can therefore be hypothesized that polymorphisms in GATA3 and IL13 and their interactions are associated with allergic rhinitis. This was indeed the case because a number of SNPs and SNP-SNP interactions had statistically significant associations with allergic rhinitis.
In nonallergic rhinitis symptoms are perennial, and no evidence of IgE-mediated hypersensitivity can be detected. However, in many of these patients, inflammation and cellular infiltration is similar to that seen in allergic rhinitis. The underlying pathogenesis might be T cell–mediated inflammation, in addition to an inherent instability of vascular tone, which responds to various immunologic and nonimmunologic triggers. In allergic rhinitis activation of TH2 cells results in the production of specific IgE to relevant allergens, as evidenced by a positive SPT response. There is the possibility that some subjects with nonallergic rhinitis might be sensitized to an allergen that has not been tested (covert allergy), and in others IgE can be produced locally and SPT responses remain negative.29 Thus in a subgroup of patients with nonallergic rhinitis, TH2 immune responses can be equally important. We did observe an association of GATA3 and IL13 SNPs in those with nonallergic rhinitis, although, as expected, this association was not as strong as that seen for allergic rhinitis.
The SNPs found to have significant associations in this study were located in the flanking regions of the genes with rs1800925 in the 5′ promoter of IL13, rs1058240 and rs379568 in the 3′ UTR of GATA3, and rs4143094 in the 5′ promoter of GATA3. Although no functional consequence has been reported for any of these SNPs, their locations in areas of potential importance for gene regulation make them of interest. Nevertheless, a single SNP that is not a missense mutation might have only a small effect and might not efficiently discriminate between cases and control subjects in a genetic association allergy study. Thus first examining main effects and eliminating some before considering 2-way interactions might miss important epistatic effects. However, patterns of SNPs could contribute to the risk of a complex disease. SNP-SNP interactions provide insight into the relationship between genes and their combined effect. Several algorithms are available to examine SNP combinations for complex diseases.30–33 These methods include dimension reduction, which examines 2-way interactions for a given subset size of the SNPs and chooses the combination that minimizes the classification error of cases and control subjects.32,33 Some analyses use classification scoring functions to identify subsets of SNPs likely associated with disease risk.30 Multivariate logistic regression and bootstrap analyses can be used to select SNP-SNP interactions through stepwise regression.31 There has been great interest in the random forest classification procedure in studies with a large number of SNPs.34 In unbalanced association studies the misclassification rate can be high, and therefore we use the random forest ranking only in combination with other criteria. Use of a 2-step procedure, with model selection based on AIC and a ranking function for the SNPs for validation, reduces the effect of selecting a model that by chance produces the largest difference between cases and control subjects. This approach examines main effects and all possible 2-way interactions. A large number of SNPs would take much computational time.
In summary, using the 2-step method, we have identified combinations of SNPs and their interactions associated with rhinitis and atopy in the Isle of Wight birth cohort. We found that GATA3 polymorphisms and their interactions with IL13 SNPs are associated with rhinitis and atopy. Future studies that include additional genes and environmental factors in a systematic assessment will likely improve the understanding of the interactions of genes in the TH2 pathway in rhinitis and associated phenotypes.
Clinical implications
The ability to predict risk based on set genetic patterns and loci interactions might allow for early intervention and individualized therapy.
Acknowledgments
Supported by the National Institutes of Health (R01 AI061471) and an MSU Foundation Strategic Partnership Grant for the Quantitative Biology and Modeling Initiative.
Disclosure of potential conflict of interest: M. Huebner has received grant support from the National Institutes of Health. The rest of the authors have declared that they have no conflict of interest.
We thank Dennis Shubitowski for technical assistance and Hans Cheng for use of the Pyrosequencing equipment.
Abbreviations used
- AIC
Akaike Information Criterion
- SNP
Single nucleotide polymorphism
- SPT
Skin prick test
- UTR
Untranslated region
REFERENCES
- 1.The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet. 1998;351:1225–1232. [PubMed] [Google Scholar]
- 2.Latvala J, von Hertzen L, Lindholm H, Haahtela T. Trends in prevalence of asthma and allergy in Finnish young men: nationwide study, 1966–2003. BMJ. 2005;330:1186–1187. doi: 10.1136/bmj.38448.603924.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright AL, Holberg CJ, Martinez FD, Halonen M, Morgan W, Taussig LM. Epidemiology of physician-diagnosed allergic rhinitis in childhood. Pediatrics. 1994;94:895–901. [PubMed] [Google Scholar]
- 4.Lundback B. Epidemiology of rhinitis and asthma. Clin Exp Allergy. 1998;28 suppl 2:3–10. [PubMed] [Google Scholar]
- 5.Arshad SH, Kurukulaaratchy RJ, Fenn M, Waterhouse L, Matthews S. Rhinitis in 10-year-old children and early life risk factors for its development. Acta Paediatr. 2002;91:1334–1338. doi: 10.1111/j.1651-2227.2002.tb02830.x. [DOI] [PubMed] [Google Scholar]
- 6.Pykalainen M, Kinos R, Valkonen S, Rydman P, Kilpelainen M, Laitinen LA, et al. Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. J Allergy Clin Immunol. 2005;115:80–87. doi: 10.1016/j.jaci.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, et al. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol. 2000;23:537–545. doi: 10.1165/ajrcmb.23.4.4199. [DOI] [PubMed] [Google Scholar]
- 8.Hunninghake GM, Soto-Quiros ME, Avila L, Su J, Murphy A, Demeo DL, et al. Polymorphisms in IL13, total IgE, eosinophilia, and asthma exacerbations in childhood. J Allergy Clin Immunol. 2007;120:84–90. doi: 10.1016/j.jaci.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 9.DeMeo DL, Lange C, Silverman EK, Senter JM, Drazen JM, Barth MJ, et al. Univariate and multivariate family-based association analysis of the IL-13 ARG130GLN polymorphism in the Childhood Asthma Management Program. Genet Epidemiol. 2002;23:335–348. doi: 10.1002/gepi.10182. [DOI] [PubMed] [Google Scholar]
- 10.Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–513. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 11.Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, Ohshima K, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 12.Howard TD, Whittaker PA, Zaiman AL, Koppelman GH, Xu J, Hanley MT, et al. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–384. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 13.Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, Poulsen LK, et al. Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet. 2003;30:355–359. doi: 10.1046/j.1365-2370.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim HB, Lee YC, Lee SY, Jung J, Jin HS, Kim JH, et al. Gene-gene interaction between IL-13 and IL-13Ralpha1 is associated with total IgE in Korean children with atopic asthma. J Hum Genet. 2006;51:1055–1062. doi: 10.1007/s10038-006-0061-x. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Xing ZM, Lu C, Ma YX, Yu DL, Yan Z, et al. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum Genet. 2003;113:387–390. doi: 10.1007/s00439-003-1001-x. [DOI] [PubMed] [Google Scholar]
- 16.Kurukulaaratchy RJ, Fenn M, Twiselton R, Matthews S, Arshad SH. The prevalence of asthma and wheezing illnesses amongst 10-year-old schoolchildren. Respir Med. 2002;96:163–169. doi: 10.1053/rmed.2001.1236. [DOI] [PubMed] [Google Scholar]
- 17.Arshad SH, Tariq SM, Matthews S, Hakim E. Sensitization to common allergens and its association with allergic disorders at age 4 years: a whole population birth cohort study. Pediatrics. 2001;108:e33. doi: 10.1542/peds.108.2.e33. [DOI] [PubMed] [Google Scholar]
- 18.Vercelli D. Genetics of IL-13 and functional relevance of IL-13 variants. Curr Opin Allergy Clin Immunol. 2002;2:389–393. doi: 10.1097/00130832-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 20.Akaike H. A new look at the statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. [Google Scholar]
- 21.Breiman L. Random Forests. Machine Learn. 2002;45:5–32. [Google Scholar]
- 22.Efron B, Tibshirani R. An introduction to the bootstrap. Boca Raton (FL): Chapman & Hall/CRC; 1994. [Google Scholar]
- 23.Kurukulaaratchy RJ, Matthews S, Holgate ST, Arshad SH. Predicting persistent disease among children who wheeze during early life. Eur Respir J. 2003;22:767–771. doi: 10.1183/09031936.03.00005903. [DOI] [PubMed] [Google Scholar]
- 24.Koeppen-Schomerus G, Stevenson J, Plomin R. Genes and environment in asthma: a study of 4 year old twins. Arch Dis Child. 2001;85:398–400. doi: 10.1136/adc.85.5.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Allergy Organization. [Accessed May 2007];Combined allergic rhinitis and asthma syndrome. Available at: http://www.worldallergy.org/professional/allergy_update/caras/airwayssynopsis.shtml.
- 26.Battle NC, Choudhry S, Tsai HJ, Eng C, Kumar G, Beckman KB, et al. Ethnicity-specific gene-gene interaction between IL-13 and IL-4Ralpha among African Americans with asthma. Am J Respir Crit Care Med. 2007;175:881–887. doi: 10.1164/rccm.200607-992OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan IH, Leung TF, Tang NL, Li CY, Sung YM, Wong GW, et al. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. J Allergy Clin Immunol. 2006;117:127–133. doi: 10.1016/j.jaci.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 28.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;70:230–236. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powe DG, Jones NS. Local mucosal immunoglobulin E production: does allergy exist in non-allergic rhinitis? Clin Exp Allergy. 2006;36:1367–1372. doi: 10.1111/j.1365-2222.2006.02593.x. [DOI] [PubMed] [Google Scholar]
- 30.Goodman JE, Mechanic LE, Luke BT, Ambs S, Chanock S, Harris CC. Exploring SNP-SNP interactions and colon cancer risk using polymorphism interaction analysis. Int J Cancer. 2006;118:1790–1797. doi: 10.1002/ijc.21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onay VU, Briollais L, Knight JA, Shi E, Wang Y, Wells S, et al. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 33.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 34.Bureau A, Dupuis J, Falls K, Lunette K, Hayward B, Keith T, et al. Identifying SNPs predictive of phenotype using random forests. Gen Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]