Abstract
Background
Termination of translation in eukaryotes is controlled by two interacting polypeptide chain release factors, eRF1 and eRF3. While eRF1 recognizes nonsense codons, eRF3 facilitates polypeptide chain release from the ribosome in a GTP-dependent manner. Besides termination, both release factors have essential, but poorly characterized functions outside of translation.
Results
To characterize further the functions of yeast eRF1 and eRF3, a genetic screen for their novel partner proteins was performed. As a result, the genes for γ (TEF4 and TEF3/CAM1) and α (TEF5/EFB1) subunits of the translation elongation factor eEF1B, known to catalyze the exchange of bound GDP for GTP on eEF1A, were revealed. These genes act as dosage suppressors of a synthetic growth defect caused by some mutations in the SUP45 and SUP35 genes encoding eRF1 and eRF3, respectively. Extra copies of TEF5 and TEF3 can also suppress the temperature sensitivity of some sup45 and sup35 mutants and reduce nonsense codon readthrough caused by these omnipotent suppressors. Besides, overproduction of eEF1Bα reduces nonsense codon readthrough in the strain carrying suppressor tRNA. Such effects were not shown for extra copies of TEF2, which encodes eEF1A, thus indicating that they were not due to eEF1A activation.
Conclusion
The data obtained demonstrate involvement of the translation elongation factor eEF1B in modulating the functions of translation termination factors and suggest its possible role in GDP for GTP exchange on eRF3.
Background
Termination of translation of mRNA is governed by stop codons in the ribosomal A-site and polypeptide chain release factors of two classes. Class I release factors RF1 and RF2 in bacteria recognize UAA/UAG and UAA/UGA stop codons, respectively, whereas eukaryotes employ only one such factor, eRF1, which is able to decode all three nonsense codons [1]. The release factors of class I bind to the ribosomal A site, recognize the stop codon, and promote hydrolysis of the P-site peptidyl-tRNA to release completed polypeptide chain from the ribosome. Translation termination is stimulated by class II release factors, RF3 in bacteria, and eRF3 in eukaryotes. Both class II factors are GTPases enhancing the termination efficiency by stimulating activity of class I release factors in a GTP-dependent manner [2-4].
Determination of the crystal structure of human eRF1 has shown that it is composed of three domains and resembles by overall shape and dimensions a tRNA molecule with the N-terminal and middle domains corresponding to the tRNA's anticodon stem and aminoacyl acceptor stem, respectively [5]. eRF3 also has a complex structure and can be divided into at least two regions: a non-conserved N-terminal region and a conserved C-terminal region (domain C), showing a considerable sequence similarity to the translation elongation factor eEF1A, which brings aminoacyl-tRNAs to the ribosomal A site [6,7]. In the yeast Saccharomyces cerevisiae eRF1 and eRF3 are encoded by the essential SUP45 and SUP35 genes.
According to recent data, eRF3 functions in termination by applying its GTPase activity to assist eRF1 with stop codon recognition and ensures efficient hydrolysis of peptidyl tRNA [8,9]. Binding of eRF1 and eRF3·GTP to the pretermination ribosome forms a complex that is not active in peptide release, and further rearrangement, induced by GTP hydrolysis, is required for proper positioning of the GGQ loop of eRF1 in the peptidyl transferase center and triggering peptidyl-tRNA hydrolysis [9,10].
It is known that bacterial ribosomes can accelerate GDP for GTP exchange on RF3 [11]. In contrast, 80S ribosomes do not noticeably influence either binding of guanine nucleotides to the eRF1·eRF3 complex or GDP for GTP exchange on it [12]. In vitro studies have shown that while free eRF3 binds GDP, it binds GTP only in the presence of eRF1 [12-14]. However, kinetic analysis of interaction of eRF3 with guanine nucleotides demonstrated that eRF1 does not act like a classical guanine nucleotide exchange factor (GEF), which increases the dissociation of GDP from a GTPase, but rather as a GTP dissociation inhibitor for eRF3, promoting efficient ribosomal recruitment of its GTP-bound form [12].
Importantly, our knowledge of the mechanism of translation termination is mostly based on in vitro studies. However, genetic approaches allowed to reveal new molecular partners of both eRF1 and eRF3 in yeast [15-18] as well, as their functions unrelated to translation termination [17,19,20]. Here, we present results of the search for additional proteins functionally interacting with yeast eRF1 and eRF3. We found that extra copies of genes which encode the γ (TEF3/CAM1 and TEF4) and α (TEF5/EFB1) subunits of the translation elongation factor eEF1B, known to catalyze the exchange of bound GDP for GTP on eEF1A, suppressed synthetic lethal interaction between some mutant SUP45 and SUP35 alleles. Besides, extra copies of TEF3 and TEF5 relieved temperature sensitivity of some mutants in these genes and reduced nonsense readthrough in one of the sup45 mutants. Overproduction of eEF1Bα also reduced nonsense readthrough in the sup35 mutant and in the strain carrying ochre suppressor tRNA. The described effects most likely were not mediated by an increase of eEF1A activity. Obtained results allowed us to suggest that the observed effects are mediated by the ability of yeast eEF1B to stimulate guanine-nucleotide exchange on eRF3.
Methods
Strains, media, growth conditions and genetic methods
Yeast strains were grown in either YEPD (1% Bacto yeast extract, 2% peptone, 2% dextrose) or defined synthetic complete media (C or C-) supplemented with 2% dextrose as a carbon source. The 5-fluoroorotic acid (5FOA) medium was prepared as described [21]. The final concentration of 5FOA was 0.9 mg/ml. The expression of the tetO2-controlled SUP45 was repressed by incubation of corresponding strains on medium selective for the pCM183-SUP45 plasmid which contained 20 μg/ml doxycycline. LB and 2× YT media were used for bacteria [22]. Appropriate amounts of antibiotics, amino acids, and bases were added when necessary. Yeast cells were grown at 30°C, if not indicated otherwise, and bacteria at 37°C. DNA transformation of lithium acetate-treated yeast cells was performed as described previously [23]. Escherichia coli cells were transformed by the method described in [24]. E. coli strain DH5α [supE44 Δlac U169 (φ 80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used in cloning experiments [22]. The yeast strains used are listed along with their genotypes in Table 1. To construct the strain BY4741-ΔSUP35, the SUP35 gene was deleted in the strain BY4741, with the use of the Eco32I-NotI fragment of pBSS35::H3 as a disruption cassette. PCR was used to prove correct integration of the disruption cassettes.
Table 1.
S. cerevisiae strains
| Strain | Genotype | Source |
| 33G-D373 | MATα ura3-52 leu2-3,112 trp1-289 his7-1 lys9-A21 ade2-144,717 pheA-11 | [25] |
| 33G-D373-rSL23 | MATα ura3-52 leu2-3,112 trp1-289 his7-1 lys9-A21 ade2-144,717 pheA-11 sup45-sl23ts | [20] |
| 33G-D373-r36 | MATα ura3-52 leu2-3,112 trp1-289 his7-1 lys9-A21 ade2-144,717 pheA-11 sup45-36ts | [20] |
| 33G-D373-rSL23-r35C | MATα ura3-52 leu2-3,112 trp1-289 his7-1 lys9-A21 ade2-144,717 pheA-11 sup45-sl23tsSUP35-C | [20] |
| 33G-D373-r36-r35C | MATα ura3-52 leu2-3,112 trp1-289 his7-1 lys9-A21 ade2-144,717 pheA-11 sup45-36tsSUP35-C | [20] |
| 8V-H80 | MATα ade1-14 his7-1 leu2-3,112 ura3-52 trp1-289 lys2-A12 | This work |
| 8V-H80-168 | MATα ade1-14 his7-1 leu2-3,112 ura3-52 trp1-289 lys2-A12 sup35-168 | This work |
| 8V-H80-196 | MATα ade1-14 his7-1 leu2-3,112 ura3-52 trp1-289 lys2-A12 sup35-196 | This work |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| BY4741-ΔSUP35 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sup35::HIS3 [pRG416-SUP35C] | This work |
| 50V-H78 | MATα ade1-14 ura3-52 leu2-3,112 his3-D200 his5-2 lys1-1 met8-1 ilv1-1 SUP4 | This work |
Plasmid that supports viability of the BY4741-ΔSUP35 strain is indicated in square brackets.
Plasmids and nucleic acid manipulation
All DNA manipulations were carried out by standard protocols [22]. The plasmids used are listed along with their essential characteristics in Table 2. The plasmid YEplac195-SUP45 was constructed by inserting the SUP45-containing XbaI-PvuII fragment of the pRG415-SUP45 plasmid into the XbaI and SmaI sites of YEplac195. The mutant alleles of SUP35, namely sup35-168ts and sup35-196ts, were amplified by PCR (primers SUP35prod and SUP35terr; Table 3) using genomic DNA of the strains 8V-H80-168 (sup35-168ts) and 8V-H80-196 (sup35-196ts) as a template. The amplification products were digested with XbaI and SacI and inserted into the same sites of pRS315, thus creating the pRS315-sup35-168 or pRS315-sup35-196 plasmids. The plasmid mutant alleles were sequenced. To construct YEplac181-SUP35C, the EcoRI-XbaI fragment carrying SUP35-C of the pRS315-SUP35C plasmid [20] was inserted between the same sites of YEplac181.
Table 2.
Plasmids
| Plasmid | Characteristics | Source |
| pCM183-SUP45 | Centromeric TRP1 vector containing SUP45 under the control of regulatable tetO2 promoter | [20] |
| YEplac195 | Multicopy URA3 vector | [26] |
| YEplac195-SUP45 | Same as YEplac195, but with SUP45 | This work |
| YEplac195-TEF2 | Same as YEplac195, but with TEF2 | This work |
| YEplac195-TEF4 | Same as YEplac195, but with wild type TEF4 | This work |
| YEplac195-TEF5 | Same as YEplac195, but with wild type TEF5 | This work |
| YEplac195-TEF5Δi | Same as YEplac195, but with TEF5 without intron (TEF5-Δi) | This work |
| YEplac181 | Multicopy LEU2 vector | [26] |
| YEplac181-TEF2 | Same as YEplac181, but with TEF2 | This work |
| YEplac181-TEF5Δi | Same as YEplac181, but with TEF5 without intron (TEF5-Δi) | This work |
| YEplac181-TEF3 | Same as YEplac181, but with TEF3 | This work |
| YEplac181-TEF4Δi | Same as YEplac181, but with TEF4 without intron (TEF4-Δi) | This work |
| YEplac181-SUP35C | Same as YEplac181, but with SUP35-C | This work |
| pRS315 | Centromeric LEU2 vector | [27] |
| pRS315-SUP35 | Same as pRS315, but with SUP35 | [20] |
| pRS315-sup35-168 | Same as pRS315, but with the sup35-168ts allele | This work |
| pRS315-sup35-196 | Same as pRS315, but with the sup35-196ts allele | This work |
| pRS315-SUP45 | Same as pRS315, but with SUP45 | [17] |
| pRS315-sup45-sl23 | Same as pRS315, but with the sup45-sl23ts allele | [17] |
| pRG416-SUP35C | Centromeric URA3 vector containing SUP35-C | [7] |
| pRG415-SUP45 | Centromeric URA3 vector containing SUP45 | [19] |
| pBSS35::H3 | Bacterial vector containing the sup35::HIS3 disruption allele | This work |
Table 3.
Oligonucleotides
| Oligonucleotide | Sequence |
| EFB1E1rev | 5'-CCTTCAATGTATGACTTGT-3' |
| EFB1E2dir | 5'-TACTGCTGTTTCTCAAGCTGA-3' |
| TEF4E1r | 5'-ATAAAATTGGATAGCCAAAGC-3' |
| TEF4E2d | 5'-GTGGCTAATCAAGTTGCCGA-3' |
| CAM1r | 5'-CTGCTCTGCTCAACGGAA-3' |
| CAM1d | 5'-TGCTCTAGACGGGCTGATACGGCCATT-3' |
| TEF2r | 5'-GAGGCCGTCTTTTGTTGA-3' |
| TEF2d | 5'-CGTGGATCCTAGGCGCTTCCCCTGCCG-3' |
| SUP35prod | 5'-ACGAGCTCAAATTATTATTTTTTACTAAG-3' |
| SUP35terr | 5'-AATTCTAGATATATTGAGAGGTGA-3' |
To obtain a yeast genomic library, chromosomal DNA of the 5V-H19 strain [28] was partially digested with Sau3A, fractionated on agarose gel and DNA fragments ranging from 4 to 12 kb were isolated. The ends of chromosomal DNA fragments were partially filled in with Klenow enzyme and ligated to the partially filled in SalI site of the YEplac195 plasmid. A plasmid with the 4.5 kb chromosomal DNA fragment containing TEF4 and RRP14 was isolated from the genomic library. The EcoRI-SmaI TEF4-containing fragment of this plasmid was inserted into the same sites of YEplac195, thus resulting in YEplac195-TEF4. The intronless variant of TEF4 (TEF4-Δi) was obtained as follows. The TEF4-containing PstI-EcoRI DNA fragment of YEplac195-TEF4 was inserted between the same sites of pUC18 [29]. The obtained plasmid was used as a template for PCR with the primers TEF4E1r and TEF4E2d (Table 3). Amplified DNA fragment, representing the plasmid lacking TEF4 intron sequence, was self ligated, resulting in the plasmid bearing TEF4-Δi. The TEF4-Δi-containing PstI-EcoRI DNA fragment of this plasmid was inserted between the same sites of YEplac181 resulting in the plasmid YEplac181-TEF4Δi.
The TEF3 gene was amplified by PCR (primers CAM1r and CAM1d; Table 3) using genomic DNA of the strain 33G-D373 as a template. The amplification product was digested with XbaI and inserted into the XbaI and SmaI sites of YEplac181, thus generating YEplac181-TEF3. The TEF5 gene was amplified by PCR (primers EFB1r and EFB1d; Table 3) using genomic DNA of 33G-D373 as a template. The amplification product was digested with BamHI and inserted into the BamHI and Ecl136II sites of YEplac195, thus generating YEplac195-TEF5. The intronless variant of TEF5 (TEF5-Δi) was obtained as follows. The TEF5-containing BamHI-SacI fragment of YEplac195-TEF5 was inserted between the same sites of the pUC18 plasmid [29]. The obtained plasmid was used as a template for PCR with primers EFB1E1rev and EFB1E2dir (Table 3). Amplified DNA fragment with TEF5 lacking intron sequence, was self ligated to obtain the plasmid bearing TEF5-Δi. The TEF5-Δi-containing BamHI-Ecl136II DNA fragment of this plasmid was inserted between the same sites of YEplac181 and YEplac195, thus resulting in the plasmids YEplac181-TEF5Δi and YEplac195-TEF5Δi, respectively. The presence of the multicopy plasmid YEplac181-TEF5Δi in yeast cells caused approximately 6-fold overproduction of eEF1Bα, as was shown by probing appropriately diluted cell lysates with the mouse polyclonal antibody against eEF1Bα. The TEF2 gene was amplified by PCR (primers TEF2r and TEF2d; Table 3) using genomic DNA of 33G-D373 as a template. The amplification product was digested with BamHI and inserted into the BamHI and Ecl136II sites of YEplac195, thus generating YEplac195-TEF2. The plasmid YEplac181-TEF2 was constructed by inserting the BamHI-SnaBI fragment of YEplac195-TEF2 containing TEF2 into the same sites of YEplac181. To construct the SUP35 disruption cassette (plasmid pBSS35::H3), the Eco105I-MunI internal fragment of the SUP35 gene was replaced with the HIS3 selectable marker.
Determination of the efficiency of nonsense codon readthrough
Following plasmids carrying tandem Renilla and firefly luciferase genes separated by a single in-frame stop codon or a corresponding sense codon control were used to measure the efficiency of nonsense codon readthrough: pDB691 (UGAC), pDB690 (CGAC), pDB723 (UAAC), pDB722 (CAAC), pDB720 (UAGC) and pDB721 (CAGC) [30]. Luciferase assays were performed with a dual luciferase reporter assay system (Promega). Assays were performed as described [31] with minimal modifications using Glomax 20/20 luminometer (Promega). Assays were done in triplicate, and the data are expressed as the means ± the standard errors. The percent readthrough in each strain is expressed as the ratio of Renilla luciferase activity/firefly luciferase activity (nonsense) divided by the ratio of Renilla luciferase activity/firefly luciferase activity (sense) multiplied by 100. For other details, see [32].
Results
TEF3, TEF4 and TEF5 extra copies suppress synthetic lethal interaction between the sup45-sl23ts and SUP35-C mutant alleles
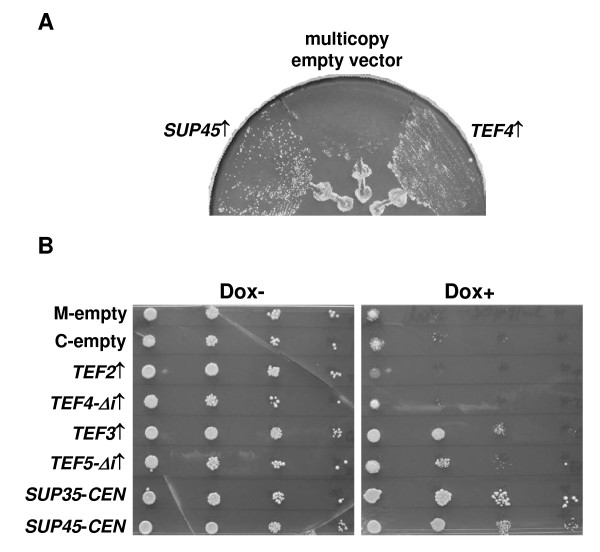
Earlier, we have identified mutations in the SUP45 gene which manifest lethality in combination with the SUP35-C allele, which encodes eRF3 lacking the inessential N-terminal region. The study of these mutations and their lethal interaction with SUP35-C indicated the role of N-terminal region of yeast eRF3 in uncharacterized non-translational function of the eRF1·eRF3 termination complex [20]. To clarify the non-translational mechanisms associated with the eRF1·eRF3 complex we performed a screen for dosage suppressors of this synthetic lethality. To isolate such suppressors, we used the strain 33G-D373-rSL23-r35C which carried the sup45-sl23ts mutation and the SUP35-C allele, as well as the centromeric pCM183-SUP45 plasmid with the TRP1 selectable marker and SUP45 under the control of regulatable tetO2 promoter [20]. This strain, which was unable to grow on medium containing 20 μg/ml doxycycline due to repression of the plasmid wild type SUP45 gene, was transformed with a S. cerevisiae genomic library based on the multicopy plasmid YEplac195, carrying the URA3 selectable marker. The transformants were selected on uracil omission medium, which contained doxycycline. From about one hundred of transformants selected only two were unable to grow on medium containing doxycycline after the loss of the library plasmid on 5FOA medium. Restriction analysis showed that these transformants contained plasmids with identical 4.5 kb inserts of genomic DNA. Both plasmids carried a DNA fragment with the TEF4 and RRP14 genes encoding a subunit of the translation elongation factor eEF1B [33] and the Rrp14 protein involved in ribosome synthesis and positioning of the mitotic spindle [34], respectively, but subsequent deletion analysis indicated that suppression required the presence of TEF4 alone (Figure 1A).
Figure 1.
Extra copies of TEF3, TEF4 and TEF5 suppress synthetic lethality between sup45-sl23tsand SUP35-C. (A) The multicopy plasmids YEplac195-TEF4 (TEF4↑), YEplac195-SUP45 (SUP45↑), with the TEF4 and SUP45 genes, respectively, or the multicopy empty vector YEplac195 were introduced into the strain 33G-D373-rSL23-r35C with the sup45-sl23ts and SUP35-C alleles, bearing the centromeric plasmid pCM183-SUP45. Obtained transformants were streaked on C-Ura medium containing doxycycline and incubated at 30°C for 2 days. (B) Transformants of the strain 33G-D373-rSL23-r35C containing the pCM183-SUP45 plasmid along with the empty multicopy YEplac181 (M-empty) or centromeric pRS315 (C-empty) vectors were used as negative control. Transformants with the centromeric plasmids pRS315-SUP35C (SUP35-CEN) and pRS315-SUP45 (SUP45-CEN) bearing the SUP35 and SUP45 genes, respectively, represented positive control, since a single copy of either one of these genes abolished synthetic lethality [20]. Growth of the control transformants was compared with that of the transformants bearing one of the multicopy plasmids YEplac181-TEF2 (TEF2↑), YEplac181-TEF4Δi (TEF4-Δi↑), YEplac181-TEF3 (TEF3↑) or YEplac181-TEF5Δi (TEF5-Δi↑). Transformants were grown in liquid C-Leu medium at 30°C and diluted to an OD600 of 1.0. Ten-fold serial dilutions were spotted onto C-Leu medium which contained (Dox+) or did not contain (Dox-) doxycycline and grown at 30°C for 3 days.
eEF1B serves as a GEF for eEF1A, accelerating dissociation of its complex with GDP. In contrast to mammals, in which eEF1B is composed of three subunits, α, β and γ, yeast eEF1B contains only two of them, α and γ. Yeast eEF1Bγ is encoded by two inessential genes, TEF3 and TEF4 [33,35], and probably serves as a positive regulator of the catalytic eEF1Bα subunit [36-38], which is encoded by a single essential gene TEF5 [39]. We isolated the genomic TEF3 and TEF5 genes to study the ability of their extra copies to suppress synthetic lethal interaction between sup45-sl23ts and SUP35-C. The TEF4 and TEF5 genes contain intervening sequences encoding the small nucleolar RNAs snR38 and snR18, respectively. The latter RNA was shown to be involved in fine tuning of translation termination in yeast [40]. To determine, whether the overproduction of the TEF4- and TEF5-encoded proteins themselves was responsible for suppression of synthetic lethality, we constructed multicopy plasmids carrying the TEF4 or TEF5 genes without introns (TEF4-Δi and TEF5-Δi, respectively). Surprisingly, a spot test did not reveal any noticeable suppressor effect for TEF4-Δi extra copies, while high dosage of TEF3 and TEF5-Δi suppressed synthetic lethality (Figure 1B). It is noteworthy that the absence of suppression for the TEF4-Δi extra copies was not due to the lack of an intron, since no suppression in a spot test was observed in transformants carrying the plasmid with wild type TEF4 (data not shown). This discrepancy indicates that TEF4 extra copies suppress synthetic lethality less efficiently than extra copies of either TEF3 or TEF5.
The sup45-sl23ts mutation is not the only mutation in the SUP45 gene manifesting synthetic lethality with SUP35-C; the same effect was found for the sup45-36ts mutation [20]. Both mutations altered the N domain of eRF1 and caused substitutions of amino acids located in proximity to each other, Ser30 to Phe replacement in the sup45-sl23ts mutant [20] and Leu34 to Ser in sup45-36ts .[41]. However, TEF5-Δi was unable to act as a dosage suppressor of the lethal interaction between sup45-36ts and SUP35-C indicating the sup45 allele specificity of the suppressor effect.
Temperature sensitivity of sup45 and sup35 mutants can be suppressed by overproduction of eEF1B subunits
The data obtained indicated a role of eEF1B in functioning of the eRF1·eRF3 complex. This raised a question, whether eEF1B overproduction can suppress mutational defects of the individual components of this complex? To address this question, we examined, if temperature sensitivity of the sup45-sl23ts and sup45-36ts mutants and some sup35ts mutants can be alleviated by the identified dosage suppressors. Efficient suppression of temperature sensitivity of the strain 33G-D373-rSL23, carrying the sup45-sl23ts mutation, was revealed only for extra copies of the TEF5-Δi gene. Extra copies of the TEF3 but not of the TEF4 gene caused weak but reproducible alleviation of the sup45-sl23ts temperature sensitivity (Figure 2A). This difference in the suppression efficiency can be expected, since eEFB1γ, encoded by TEF3, is a regulatory subunit which stimulates activity of the TEF5-encoded eEFB1α [36-38]. Importantly, extra copies of TEF5-Δi did not suppress temperature sensitivity of the sup45-36ts mutant 33G-D373-r36, which agrees with the sup45 allele specific suppression of synthetic lethality (data not shown).
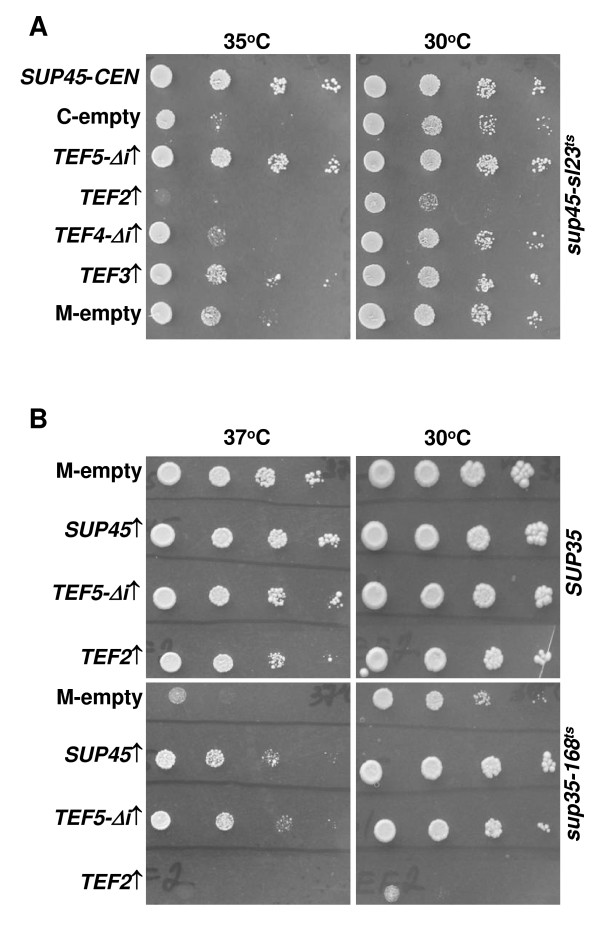
Figure 2.
Effects of extra copies of the TEF3, TEF4 and TEF5 genes on temperature sensitivity of the sup45-sl23ts and sup35-168ts mutants. (A) Growth of transformants of the 33G-D373-sl23 strain with the sup45-sl23ts mutation, bearing the centromeric or multicopy empty vectors pRS315 (C-empty) or YEplac181 (M-empty), respectively (negative controls), or centromeric plasmid pRS315-SUP45 (SUP45-CEN) (positive control), was compared with growth of transformants, bearing one of the multicopy plasmids: YEplac181-TEF2 (TEF2↑), YEplac181-TEF3 (TEF3↑), YEplac181-TEF4Δi (TEF4-Δi↑) or YEplac181-TEF5Δi (TEF5-Δi↑). Transformants were grown as described in legend to Figure 1B, except cell suspensions were spotted on C-Leu without doxycycline and incubated at 30°C or 35°C. (B) The strain BY4741-ΔSUP35 with disrupted SUP35 contained the URA3 SUP35-C centromeric plasmid pRG416-SUP35C, which was shuffled for either the centromeric LEU2 pRS315-SUP35 or pRS315-sup35-168 plasmids bearing the wild type SUP35 or mutant sup35-168ts alleles, respectively. Shuffling for the sup35-168ts plasmid produced transformants unable to grow at 37°C. Then, the strains with either the SUP35 or sup35-168ts plasmids were transformed with one of the multicopy plasmids YEplac195 (M-empty), YEplac195-TEF2 (TEF2↑), YEplac195-TEF5Δi (TEF5-Δi↑) or YEplac195-SUP45 (SUP45↑). The transformants were grown in liquid C-Ura medium and diluted to OD600 of 1.0. Ten-fold serial dilutions were spotted onto C-Ura medium and grown at 30°C or 37°C for 4 days.
Since TEF5 was found to be the most efficient dosage suppressor of temperature sensitivity of the sup45-sl23ts mutant, effects of its extra copies on growth of sup35 mutants were studied further. Examination of 21 sup35ts mutants of the strain 8V-H80 revealed two mutants with phenotypes that depended on the presence of the TEF5-Δi multicopy plasmid: temperature sensitivity of the sup35-168ts mutant was suppressed, while growth of the sup35-196ts was inhibited even at permissive temperature (data not shown). The ability of TEF5-Δi extra copies to suppress temperature sensitivity of sup35-168ts was reproduced in the BY4741-ΔSUP35 strain. Interestingly, extra copies of SUP45 also alleviated temperature sensitivity of the sup35-168ts mutant (Figure 2B). Similar effect of SUP45 overdose was earlier shown for other sup35ts mutants [42].
Sequencing of the sup35ts mutant alleles, manifestation of which depended on TEF5-Δi extra copies, revealed that the mutations altered the C-terminal region of eRF3 located downstream of its GTP-binding domain: sup35-168ts caused replacement of Leu553 to Pro, while nucleotide substitution in the sup35-196ts allele corresponded to change of Thr667 to Pro.
Extra copies of TEF5 and TEF3 reduce efficiency of nonsense codon readthrough
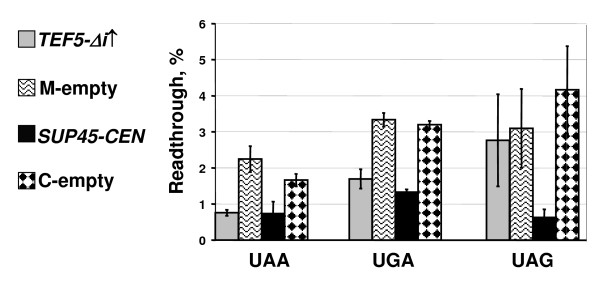
Genetic interactions described above show that eEF1B is functionally related to the yeast release factors, but do not indicate its involvement in translation termination. To test the latter possibility we investigated the influence of a high dosage of the genes for eEF1B subunits on nonsense readthrough in the sup45 and sup35 omnipotent suppressor mutants. Extra copies of TEF5-Δi decreased the levels of UAA и UGA readthrough in the sup45-sl23ts mutant almost to the levels observed in this strain in the presence of wild type SUP45, but did not affect readthrough of UAG (Figure 3). A smaller decrease of UAA readthrough due to eEF1Bα overproduction was found in the strain carrying the sup35-196ts mutation, though this effect was statistically insignificant in the sup35-168ts mutant (Figure 4). The observed effects agree with the fact that extra copies of TEF5 weakened suppression of the his7-1 UAA mutation in the strain with the sup35-25 omnipotent suppressor [43]. However, other results demonstrated that overproduction of eEF1Bα could increase the levels of nonsense readthrough in strains lacking suppressor mutations [44,45].
Figure 3.
High dosage of TEF5 reduces UAA and UGA readthrough levels in the sup45-sl23ts mutant. The 33G-D373-rSL23 strain with the sup45-sl23ts mutation was transformed with one of the following plasmids: multicopy plasmids YEplac181-TEF5Δi (TEF5-Δi↑) or YEplac181 (M-empty), and centromeric plasmids pRS315-SUP45 (SUP45-CEN) or pRS315 (C-empty). The appropriate dual luciferase reporter plasmids were introduced into cells of obtained transformants. The transformants carrying plasmid pairs were grown in liquid C-Leu, Ura medium at 30°C and then readthrough levels were measured as described in Methods. The plasmid YEplac181-TEF5Δi causes a statistically significant decrease of UAA and UGA readthrough levels (P ≤ 0.02 and P ≤ 0.01, respectively) compared to those in transformants carrying YEplac181. Comparison of transformants bearing pRS315-SUP45 and pRS315 revealed statistically significant difference in readthrough of all stop codons: P ≤ 0.1, P ≤ 0.001 and P ≤ 0.05 for UAA, UGA and UAG codons, respectively.
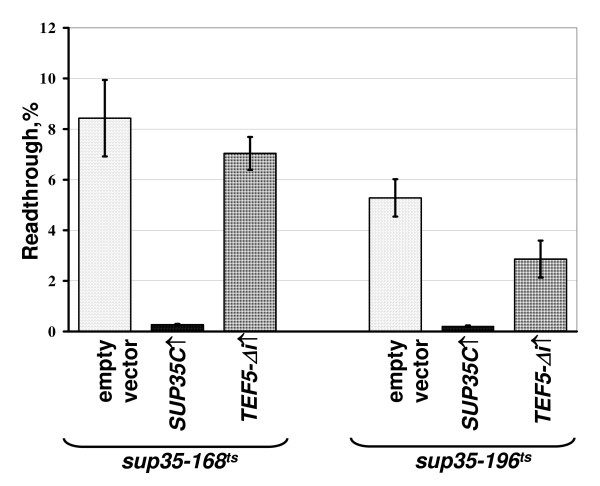
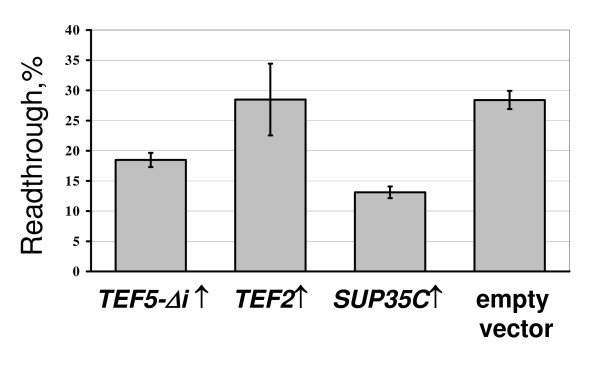
Figure 4.
High dosage of TEF5 reduces efficiency of UAA readthrough in the sup35 mutants. The strains 8V-H80-168 (sup35-168ts) and 8V-H80-196 (sup35-196ts) were transformed with one of the multicopy plasmids YEplac181-TEF5Δi (TEF5-Δi↑), YEplac181-SUP35C (SUP35C↑) or YEplac181 (empty vector) and then with the appropriate dual luciferase reporter plasmids. The transformants with plasmid pairs were grown and readthrough measured as described in legend to Figure 3 and in Methods. The difference in UAA readthrough in the mutant sup35-196ts strain with the YEplac181-TEF5Δi and YEplac181 plasmids is statistically significant (P ≤ 0.1).
In addition to TEF5-Δi, a decrease of UAA readthrough in the sup45-sl23ts mutant was also observed for a high dosage of TEF3, but not of TEF4-Δi (Figure 5). At last, overproduction of eEF1Bα manifested an antisuppressor effect not only in the sup35 and sup45 suppressor mutants, since it reduced the levels of nonsense readthrough caused by the ochre tRNA suppressor SUP4 in the strain 50V-H78. Remarkably, SUP35-C overdose caused in this strain just a slightly stronger antisuppressor effect than extra copies of TEF5-Δi (Figure 6).
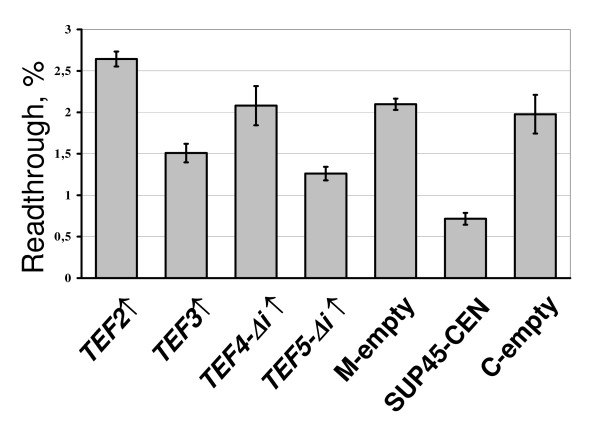
Figure 5.
Extra copies of TEF3, but not of TEF4 reduce efficiency of UAA readthrough in the sup45-sl23ts mutant. The 33G-D373-rSL23 strain was transformed with one of the multicopy plasmids YEplac181-TEF2 (TEF2↑), YEplac181-TEF3 (TEF3↑), YEplac181-TEF4Δi (TEF4-Δi↑), YEplac181-TEF5Δi (TEF5-Δi↑), YEplac181 (M-empty) or centromeric plasmids pRS315-SUP45 (SUP45-CEN) or pRS315 (C-empty). The appropriate dual luciferase reporter plasmids were then introduced into cells of obtained transformants. The transformants with plasmid pairs were grown and readthrough measured as described in legend to Figure 3 and in Methods. The plasmids YEplac181-TEF3, YEplac181-TEF5Δi and pRS315-SUP45 cause a statistically significant (P ≤ 0.02, P ≤ 0.002 and P ≤ 0.01, respectively) decrease of UAA readthrough levels compared to those in transformants carrying corresponding empty vectors. The levels of UAA readthrough are increased in transformants with YEplac181-TEF2 as compared with that in transformants with YEplac181 (P ≤ 0.01).
Figure 6.
Extra copies of TEF5 manifest an antisuppressor effect in the strain with the ochre tRNA SUP4 suppressor. The strain 50V-H78 (SUP4), was transformed with one of the multicopy plasmids YEplac181-TEF5Δi (TEF5-Δi↑), YEplac181-TEF2 (TEF2↑), YEplac181-SUP35C (SUP35C↑) or YEplac181 (empty vector). The appropriate dual luciferase reporter plasmids were then introduced into cells of obtained transformants. The transformants with plasmid pairs were grown and readthrough measured as described in legend to Figure 3 and in Methods. The plasmids YEplac181-TEF5Δi and YEplac181-SUP35C cause a statistically significant (P ≤ 0.01 and P ≤ 0.002, respectively) decrease of UAA readthrough levels compared to those in transformants carrying YEplac181.
Effects of eEF1B overproduction are not mediated by an increase of eEF1A activity
Translation elongation factor eEF1A is a classic G protein involved in the GTP-dependent binding of amino-acylated tRNA, and the eEF1B α subunit, associated with its γ subunit, functions as a GEF for eEF1A [46,47]. This suggests that the effects of overproduction of eEF1B subunits, observed in our work, are mediated by an increased activity of its target protein, eEF1A. However, extra copies of TEF2, one of the two yeast genes encoding eEF1A, neither suppressed synthetic lethality between sup45-sl23ts and SUP35-C (Figure 1), nor alleviated temperature sensitivity of the sup35-168ts or sup45-sl23ts mutants (Figure 2), thus ruling out this possibility. In contrast, extra copies of TEF2 could even inhibit growth of the sup35-168ts and sup45-sl23ts mutants, though in these experiments growth inhibition was not observed when the strains expressed wild type SUP45 (Figure 1B, Dox- panel) or SUP35 (Figure 2B).
Measurement of nonsense readthrough levels confirmed this conclusion: in contrast to TEF5-Δi and TEF3, the overdose of which caused an antisuppressor effect, extra copies of TEF2 either did not reduce UAA readthrough in the SUP4 ochre suppressor-carrying strain (Figure 6), or even stimulated it in the sup45-sl23ts mutant (Figure 5). It is also necessary to stress that in contrast to TEF2, extra copies of SUP35-C caused a decrease of UAA readthrough in the SUP4 strain (Figure 6).
Discussion
In eukaryotes a GTP-bound form of eRF3 is active in termination, because GTP hydrolysis by eRF3 on the ribosome is required for the efficient translation termination [2,4,8,9]. However, in vivo eRF3 should mostly be present in its GDP-bound form, since (i) when termination is completed it is released from the ribosome in a complex with GDP, and (ii) newly synthesized eRF3 has a high affinity to GDP, being able to bind GTP only upon interaction with eRF1 [12-14]. It is also important that though eRF1 is able to enhance the levels of the GTP-bound form of eRF3, it does not act like classic GEFs, which increase the dissociation of GDP from a GTPase [12]. Since no GEF was identified for eRF3, it could be that it is able to dissociate GDP spontaneously similarly to eEF1A which can do this, although with a 700-fold slower rate than with the assistance of eEFB1 [48]. However, despite this, the results of our work suggest that eEFB1 acts as a GEF for eRF3. This suggestion is based on (i) the functional interaction of the α and γ subunits of eEF1B with the release factors and (ii) the reduced nonsense readthrough caused by overproduction of eEF1B subunits. It is also necessary to stress that eEF1Bγ and eEF1Bα are structurally different proteins and the former can stimulate in vitro the nucleotide exchange activity of the latter [36-38]. This additionally relates the observed effects of their overproduction to the role of the eEF1B factor in GDP/GTP exchange on eRF3. It is noteworthy that the effects of an overdose of the TEF3- and TEF4-encoded γ subunit of eEF1B were different. While extra copies of either gene can suppress synthetic lethal interaction between the sup45-sl23ts and SUP35-C mutant alleles, only TEF3 extra copies reduce nonsense readthrough in the sup45-sl23ts mutant and alleviate its growth at restrictive temperature. This indicates a functional difference between TEF3- and TEF4-encoded proteins, which agrees with earlier findings [33]. It may be suggested that the complex of eEF1Bα with the TEF3-encoded protein preferentially stimulates guanine-nucleotide exchange on eRF3, while its complex with the TEF4-encoded eEF1Bγ has a preference for eEF1A.
Since eEF1B acts as GEF for eEF1A, its overproduction should increase the concentration of active (GTP-bound) eEF1A. However, it is unlikely that the effects of eEF1B overproduction observed in this work were mediated by activation of eEF1A, because extra copies of TEF2 did not suppress synthetic lethality between sup45-sl23ts and SUP35-C, as well as temperature sensitivity of the sup35-168ts and sup45-sl23ts mutants and even could inhibit growth of these mutants. It is known that, besides translation, overexpression of eEF1A in yeast also affects actin cytoskeleton which may be the cause of growth inhibition [45]. However, we observed that extra copies of TEF2 caused a noticeable growth inhibition only when mutant sup35 or sup45 alleles were expressed. Similarly to eEF1A, yeast eRF1 and eRF3 have nontranslational functions and their deficiency, as well as mutational inactivation, may inhibit yeast cell growth via perturbations of the cytoskeleton organization [17,19]. Therefore, one can suggest that TEF2 extra copies and sup35 or sup45 mutations act synergistically to affect cytoskeleton. It is also important that extra copies of TEF2 did not influence nonsense readthrough in the strain with the SUP4 tRNA ochre suppressor and even increased it in the sup45-sl23ts mutant. At the same time, in contrast to TEF2, extra copies of SUP35-C, encoding the C domain of eRF3 caused a decrease of UAA readthrough in the SUP4 strain, which also makes eRF3 an appropriate target for the eEF1B action.
It is noteworthy that an overproduction of eEF1Bα suppressed temperature sensitivity of the sup35-168ts mutant, but did not noticeably reduce nonsense codon readthrough indicating that temperature sensitivity of this mutant is not due to a defect of translation termination. This suggests that even slight stimulation of GDP for GTP exchange on eRF3, which did not restore translation termination in the sup35-168ts mutant, could repair defect of the non-translational function of mutant eRF3. Temperature sensitivity of sup45-sl23ts is also unrelated to a high level of nonsense readthrough [20], though in this mutant overproduction of eEF1Bα alleviated the growth defect and decreased UAA and UGA readthrough. Importantly, overproduction of eEF1Bα did not suppress temperature sensitivity of the sup45-36ts mutant. It was shown that at restrictive temperature the sup45-36ts mutation affects cytokinesis due to a defect of the non-translational function of eRF1 mediated by its interaction with the myosin light chain Mlc1. Since the eRF1·Mlc1 complex, does not contain eRF3 [17], it is logical that temperature sensitivity of this mutant did not depend on the levels of eEF1Bα.
Taken together, these data suggest that the guanine-nucleotide exchange activity of eEF1B plays a role in the functioning of release factors in translation termination as well as in their functions outside of termination. Though the mechanisms which underlie suppression of mutational defects of eRF1 and eRF3 by overproduction of the eEF1B subunits are unknown, this suggestion looks natural for the sup35 mutants, since it presumes that eEF1B acts as a GEF for eRF3. However, an overdose of the eEF1Bα can also suppress the mutational defect of eRF1. The suppressible sup45-sl23ts mutation causes amino acid replacement in the eRF1 N-terminal domain which is not implicated in interaction with eRF3. In agreement with this, sup45-sl23ts did not noticeably influence interaction between eRF1 and eRF3 [20]. Therefore, one can suggest that this mutation interferes with the ability of eRF1 to inhibit dissociation of the eRF3·GTP complex. This should decrease the overall levels of the ternary eRF1·eRF3·GTP complex, the abundance of which could be restored by the eEF1B-mediated intensification of exchange of GDP for GTP on eRF3.
Conclusion
The data presented suggest that eEF1B, the nucleotide exchange factor of eEF1A, also catalyzes GDP for GTP exchange on eRF3, a GTPase, the C domain of which is structurally similar to eEF1A [2,3,7]. This reaction is required for eRF3 function both in translation termination and outside of termination. However, it is obvious that in yeast the eEF1B-mediated exchange of nucleotides is more critical for eEF1A than for eRF3. This follows from the observations that while eEF1Bα is normally essential for viability, cells can survive without the protein in the presence of excess eEF1A [49]. One can suppose that the requirement for GEF is less pronounced for eRF3 than for eEF1A, because of a unique property of eRF1 to increase affinity of eRF3 to GTP. Indeed, interaction with eRF1 stabilizes eRF3 in a GTP-bound form after spontaneous exchange of GDP for GTP [12-14]. No such inhibitor of GTP dissociation is known for eEF1A.
Structural similarity suggests that eRF3 has evolved from eEF1A. Our genetic data suggest that the accessory protein, eEF1B, stimulating the activity of elongation factor eEF1A preserved this ability for the termination factor eRF3 as well. Direct biochemical studies are necessary to confirm the role of eEF1B in nucleotide exchange on eRF3.
Authors' contributions
IV performed a screen for dosage suppressors of synthetic lethality between the sup45 and SUP35-C mutations and participated in molecular genetic studies. GF and ES carried out the molecular genetic studies. VS participated in planning of experiments and drafted the manuscript. MT-A designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are grateful to D.M. Bedwell for providing the dual luciferase reporter plasmids and V.V. Kushnirov for the yeast genomic library and critical reading of the manuscript. The work was supported by the grant from Russian Foundation for Basic Research (# 08-04-0062).
Contributor Information
Igor A Valouev, Email: birdie69@mail.ru.
Gleb V Fominov, Email: glebfom@mail.ru.
Elizaveta E Sokolova, Email: juegodevoces@gmail.com.
Vladimir N Smirnov, Email: v.n.smirnov@mtu-net.ru.
Michael D Ter-Avanesyan, Email: mdter@mail.ru.
References
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev LL. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov SG, Kisselev LL, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1 – mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, Inge-Vechtomov SG. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene. 1988;66:45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;197:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CUT, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Mitkevich AV, Kononenko AV, Petrushanko IYu, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1·eRF3·GTP·Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Bailleul PA, Newnam GP, Steenbergen JN, Chernoff YO. Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81–94. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakov VN, Valouev IA, Lewitin EI, Paushkin SV, Kosorukov VS, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol Biol. 2001;2:9. doi: 10.1186/1471-2199-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev IA, Urakov VN, Kochneva-Pervukhova NV, Smirnov VN, Ter-Avanesyan MD. Translation termination factors function outside of translation: yeast eRF1 interacts with myosin light chain Mlc1p, to effect cytokinesis. Mol Microbiol. 2004;53:687–696. doi: 10.1111/j.1365-2958.2004.04157.x. [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- Valouev IA, Kushnirov VV, Ter-Avanesyan MD. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil Cytoskeleton. 2002;52:161–173. doi: 10.1002/cm.10040. [DOI] [PubMed] [Google Scholar]
- Urakov VN, Valouev IA, Kochneva-Pervukhova NV, Packeiser AN, Vishnevsky AYu, Glebov OO, Smirnov VN, Ter-Avanesyan MD. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol Biol. 2006;7:34. doi: 10.1186/1471-2199-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Inge-Vechtomov SG, Derkatch IL, Ptyushkina MV, Tarunina OV, Dagkesamanskaya AR, Ter-Avanesyan MD. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast. 1992;8:489–499. doi: 10.1002/yea.320080702. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marko J, Gao L, Kaenjak-Angeletti A, Bedwell DM. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Reed R, Marciniak RA. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1539–1549. doi: 10.1128/EC.4.9.1539-1549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzy TG, Ripmaster TL, Woolford JL., Jr Multiple genes encode translation elongation factor EF-1γ in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:2703–2707. doi: 10.1093/nar/22.13.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Fatica A, Route MP, Tollervey D. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 2007;35:1354–1366. doi: 10.1093/nar/gkl824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamborius NG, Burke DJ, Creutz CE. Cloning and genetic characterization of a calcium- and phospholipid-binding protein from Saccharomyces cerevisiae that is homologous to translation elongation factor-1γ. Yeast. 1993;9:151–163. doi: 10.1002/yea.320090206. [DOI] [PubMed] [Google Scholar]
- Saha SK, Chakraburtty K. Protein synthesis in yeast. Isolation of variant forms of elongation factor 1 from the yeast Saccharomyces cerevisiae. J Biol Chem. 1986;281:12599–12603. [PubMed] [Google Scholar]
- Janssen GMC, Moller W. Elongation factor 1βγ from Artemia. Purification and properties of its subunits. Eur J Biochem. 1988;171:119–129. doi: 10.1111/j.1432-1033.1988.tb13766.x. [DOI] [PubMed] [Google Scholar]
- Janssen GMC, Moller W. Kinetic studies of the role of elongation factors 1β and 1γ in protein synthesis. J Biol Chem. 1988;263:1773–1778. [PubMed] [Google Scholar]
- Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T. Cloning and characterization of the elongation factor EF-1β homologue of Saccharomyces cerevisiae . EF-1β is essential for growth. FEBS Letters. 1993;316:165–169. doi: 10.1016/0014-5793(93)81208-h. [DOI] [PubMed] [Google Scholar]
- Hatin I, Fabret C, Namy O, Decatur WA, Rousset J-P. Fine tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics. 2007;177:1527–1537. doi: 10.1534/genetics.107.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breining P, Piepersberg W. Yeast omnipotent suppressor SUP1 (SUP45): nucleotide sequence of the wild type and a mutant gene. Nucleic Acids Res. 1986;14:5187–5197. doi: 10.1093/nar/14.13.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Didichenko SA, Kushnirov VV, Dagkesamanskaya AR. SUP35 and SUP45 genes code for ribosome-bound proteins involved in the control of translational fidelity in yeast. In: Brown AJP, Tuite MF, McCarthy JEG, editor. Protein Synthesis and Targeting in Yeast, NATO ASI Series. H 71. Berlin, Heidelberg: Springer-Verlag; 1993. pp. 81–90. [Google Scholar]
- Aksenova A, Munoz I, Volkov K, Arino J, Mironova L. The HAL3-PPZ1 dependent regulation of nonsense suppression efficiency in yeast and its influence on manifestation of the yeast prion-like determinant [ISP+] Genes Cell. 2007;12:435–445. doi: 10.1111/j.1365-2443.2007.01064.x. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A, Valente L, Loik VI, Williams T, Starita LM, Kinzi TG. Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translation fidelity. Mol Cell Biol. 1999;19:5257–5266. doi: 10.1128/mcb.19.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi R, Kandl KA, Carr-Schmid A, Whitacre JL, Adams AEM, Kinzi TG. Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics. 2001;157:1425–1436. doi: 10.1093/genetics/157.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobin LI, Moller W. Purification and properties of an elongation factor functionally analogous to bacterial elongation factor Ts from embryos of Artemia salina. Eur J Biochem. 1978;84:69–77. doi: 10.1111/j.1432-1033.1978.tb12142.x. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Schummer T, Stromgaard A, Knudsen CR, Kinzi TG. Kinetics of the interactions between yeast elongation factors 1A and 1Bα, guanine nucleotides, and aminoacyl-tRNA. J Biol Chem. 2007;282:35629–35637. doi: 10.1074/jbc.M707245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman Y, Valente L, Jeppesen GR, Andersen GR, Patei S. Mg2+ and key lysine modulate exchange activity of eukaryotic translation elongation factor 1Bα. J Biol Chem. 2006;281:19457–19468. doi: 10.1074/jbc.M601076200. [DOI] [PubMed] [Google Scholar]
- Kinzy TG, Woolford JL., Jr Increased expression of Saccharomyces cerevisiae translation elongation factor EF-1α bypasses the lethality of a TEF5 null allele encoding EF-1β. Genetics. 1995;141:481–489. doi: 10.1093/genetics/141.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]