SUMMARY
Extracellular signals and cell-intrinsic transcription factors cooperatively instruct generation of diverse neurons. However, little is known about how neural progenitors integrate both cues and orchestrate chromatin changes for neuronal specification. Here, we report that extrinsic signal retinoic acid (RA) and intrinsic transcription factor Neurogenin2 (Ngn2) collaboratively trigger transcriptionally active chromatin in spinal motor neuron genes during development. Retinoic acid receptor (RAR) binds Ngn2 and is thereby recruited to motor neuron genes targeted by Ngn2. RA then facilitates the recruitment of a histone acetyltransferase CBP to the Ngn2/RAR-complex, markedly inducing histone H3/H4-acetylation. Correspondingly, timely inactivation of CBP and its paralogue p300 results in profound defects in motor neuron specification and motor axonal projection, accompanied by significantly reduced histone H3-acetylation of the motor neuron enhancer. Our study uncovers the mechanism by which extrinsic RA-signal and intrinsic transcription factor Ngn2 cooperate for cell-fate specification through their synergistic activity to trigger transcriptionally active chromatin.
Keywords: Spinal cord, motor neuron, CBP, p300, retinoic acid, RAR, Ngn2
INTRODUCTION
In the embryonic central nervous system (CNS) development, neural progenitors produce a large number of neuronal subtypes with distinct cellular and physiological properties. The specification of neuronal subtype identity is precisely controlled in space and time by cooperative actions between extrinsic signals, which are locally provided, and cell intrinsic transcription factors (Jessell, 2000). Chromatin modifications affect transcription profoundly (Berger, 2007) and thus likely function as critical regulatory points that orchestrate changes of numerous gene expressions during neuronal cell fate specification. However, the molecular mechanisms by which extrinsic and intrinsic cues collaboratively trigger chromatin modifications are poorly understood in CNS development.
To tackle these issues, the specification of spinal motor neurons serves as a good model system, as the role of extracellular signals and transcription factors is relatively well defined in this context. Sonic hedgehog (Shh) secreted from the notochord and floor plate is involved in specifying uncommitted neural progenitors into motor neuron progenitors (pMN cells) (Lee and Pfaff, 2001; Marquardt and Pfaff, 2001). Retinoic acid (RA), a bioactive derivative of vitamin A, is also an extrinsic signal essential for motor neuron differentiation (Appel and Eisen, 2003; Maden, 2007). Paraxial mesoderm surrounding the neural tube expresses retinaldehyde dehydrogenase-2 (Raldh2) enzyme, which converts retinaldehyde to RA, during neural tube formation and the spinal cord development (Novitch et al., 2003). RA secreted by paraxial mesoderm directs progressive steps in motor neuron specification; induction of pMN transcription factors Pax6 and Olig2, promotion of pan-neuronal differentiation and motor neuron specification (Novitch et al., 2003; Sockanathan et al., 2003). Consistently, vitamin A deficiency in quail embryos impairs motor neuron generation and motor axonal projections in the embryonic spinal cord (Maden et al., 1996; Wilson et al., 2004).
RA regulates transcription by binding nuclear hormone receptors retinoic acid receptors (RARs), which form an obligatory heterodimer with their paralogues retinoid X receptors (RXRs) (Maden, 2007). Without RA, RAR suppresses transcription by recruiting corepressor complexes containing histone deacetylases (Glass and Rosenfeld, 2000). Upon RA-binding, RAR undergoes a dramatic structural change, permitting an exchange of corepressors for coactivators such as CBP and p300, which evoke transcriptional activation through their histone acetyltransferase (HAT) activity (Chakravarti et al., 1996; Kamei et al., 1996; Glass and Rosenfeld, 2000). The activation function 2 (AF2) domain of RAR is required for RA-dependent recruitment of coactivators, including CBP/p300, to RAR (Chakravarti et al., 1996; Glass and Rosenfeld, 2000; Kamei et al., 1996). RARs and RXRs are highly expressed in the neural tube (Diez del Corral et al., 2003), suggesting their roles in neural development. Indeed, blocking RAR function prevented the specification of motor neurons in the chick neural tube (Novitch et al., 2003). RA-signaling also triggers neurogenesis in multi-potent mouse embryonic cells, chick neural tube and Xenopus embryos (Diez del Corral et al., 2003; Maden, 2002; Maden, 2007). Despite accumulating evidences that RA-signaling and RARs are involved in motor neuron specification and neurogenesis, the molecular mechanism underlying their function and the downstream target genes of RA-bound RARs during neural development have been elusive.
Proneural basic helix-loop-helix (bHLH) factors play critical roles in triggering neuronal differentiation (Bertrand et al., 2002). For instance, bHLH factors Neurogenin1 (Ngn1, Neurog1) and Neurogenin2 (Ngn2, Neurog2) promote the cell cycle withdrawal and neurogenesis, and simultaneously inhibit astrogenesis (Farah et al., 2000; Lee et al., 2005; Nieto et al., 2001; Sun et al., 2001). Analysis of mutant embryos deficient in Ngn1 and Ngn2 revealed that Ngn1 and Ngn2 are required for neurogenesis and motor neuron fate specification in the ventral spinal cord (Scardigli et al., 2001). Supporting the role of Ngn2 in motor neuron fate decision, we have shown that Ngn2 collaborates with the motor neuron-specifying LIM-complex, containing LIM homeodomain (LIM-HD) transcription factors Isl1 and Lhx3, to specify motor neurons in the embryonic spinal cord and P19 stem cells (Lee et al., 2004; Lee and Pfaff, 2003). Ngn2 binds E-box DNA elements in the enhancer region of a motor neuron-specific gene Hb9 and directly upregulates the expression of Hb9 (Lee et al., 2004; Lee and Pfaff, 2003).
Here we show that RA-signaling and Ngn2 cooperate for motor neuron specification through their synergistic activity to establish transcriptionally active chromatin. CBP plays an essential role in coupling RA-signaling and Ngn2 function for motor neuron development. Specifically, RAR forms a complex with Ngn2 and is thereby recruited to E-boxes of motor neuron genes targeted by Ngn2. The extrinsic signal RA subsequently facilitates the recruitment of CBP to the Ngn2/RAR-complex, which in turn induces chromatin alterations in motor neuron genes, leading to transcriptional activation. Indeed, timely inactivation of CBP in the differentiating motor neurons results in severe defects in motor neuron specification and axon pathfinding, accompanied by reduced active chromatin markers in motor neuron genes. Our findings define a developmental regulatory strategy that directly couples extrinsic signals and intrinsic transcription factors for chromatin changes in neuronal genes and neuronal cell-type determination.
RESULTS
A cross-talk between RA-signaling and Ngn2 via association between Ngn2 and RAR
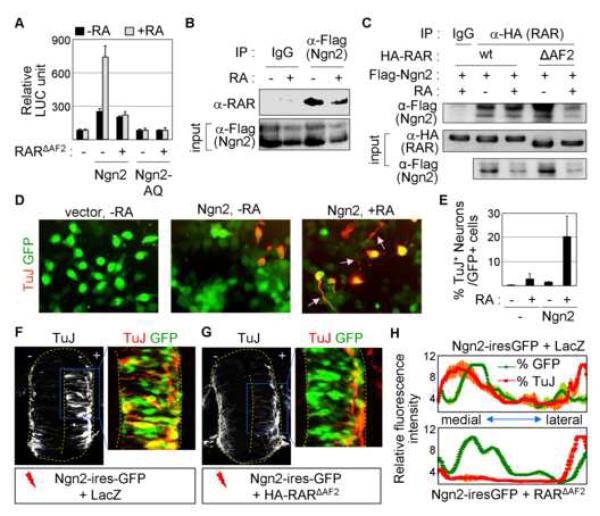
As the extrinsic RA-signaling and the intrinsic transcription factor Ngn2 share the ability to specify motor neurons in the embryonic spinal cord through the transcriptional activator function of Ngn2 and RA-bound RAR, we considered the possibility that a developmental program exists to functionally couple their activities in controlling motor neuron generation. To test this, we examined the effect of RA on Ngn2 transactivation using the Ngn2-responsive luciferase reporter E-box:LUC (Lee et al., 2005). In P19 cells expressing no Ngn2, RA had no effect on this reporter (Fig. 1A), consistent with the fact that the E-box:LUC reporter lacks RAR response elements (RAREs). E-box:LUC was transactivated by Ngn2 but not by Ngn2-AQ, a Ngn2 point mutant unable to bind E-box (Lee and Pfaff, 2003) (Fig. 1A). Strikingly, RA augmented transactivation of this reporter by Ngn2, but not by Ngn2-AQ, suggesting that RA-dependent activation of E-box elements requires the DNA-binding activity of Ngn2.
Figure 1. RA-signal stimulates Ngn2 activity through the formation of the Ngn2/RAR-complex.
(A) RA enhances the transcriptional activity of Ngn2, but not Ngn2-AQ, in E-box:LUC reporter in P19 cells. Ngn2-AQ is a Ngn2 mutant that no longer binds E-box. RARΔAF2, a RAR mutant that is unable to interact with AF2-dependent coactivators, inhibits the stimulating effect of RA on E-box:LUC. (B) Ngn2 binds RAR in a RA-independent manner in HEK293 cells, as determined by immunoprecipitation (IP) of lysates of HEK293 cells expressing Flag-Ngn2 with anti-Flag antibody followed by immunoblotting with anti-RAR antibody. (C) CoIP assays in HEK293 cells transfected with Flag-Ngn2 and either HA-RAR or HA-RARΔAF2. Both RAR and RARΔAF2 interact with Ngn2 in a RA-independent manner. (D) P19 cells transfected with vector or Ngn2-ires-GFP were treated with vehicle or RA, and analyzed for neuronal differentiation using immunostaining with anti-TuJ antibody. Arrows indicate neurite-like processes. (E) % quantification of TuJ+-neurons among GFP+-transfected P19 cells under indicated condition. (F, G) Immunohistochemical analysis of neuronal differentiation (TuJ+-cells) in HH stage 20 chick embryos electroporated with the indicated constructs on the bottom. RARΔAF2 compromises precocious neuronal differentiation triggered by Ngn2 in the medial zone of the chick neural tube. GFP+-cells mark electroporated cells. (H) Quantification of GFP (green) and TuJ (red) fluorescence intensity in the neural tube. The X-axis indicates the most medial to lateral sides of the neural tube. Coexpression of Ngn2-ires-GFP and LacZ leads to the increase in TuJ staining concomitant with GFP expression in the medial zone of the neural tube (upper panel), whereas GFP expression does not correlate with TuJ staining in the medial zone of the neural tube electroporated with Ngn2-ires-GFP and RARΔAF2 (bottom panel). (A, E) The error bars represent the standard deviation.
To define the basis of the RA-dependent enhancement of Ngn2 transactivation on E-box elements, we examined the association between RAR and Ngn2 in cells using coimmunoprecipitation (CoIP) assays. In HEK293 cells transfected with RARα and Flag-tagged Ngn2, RARαwas present in the Ngn2-containing protein complex immunopurified by anti-Flag antibody independently of the presence of RA (Fig. 1B). Ngn2 also interacted with RARβ and RARγ in CoIP assays (S-Fig. 1). Thus, RARs and Ngn2 associate in a RA-independent manner in cells. These results exclude the possibility that RA stimulates Ngn2 transactivation by enhancing the association of RAR and Ngn2. Thus, we asked whether RA potentiates Ngn2 function by recruiting the transcriptional coactivators to the Ngn2/RAR-complex. To test this, we employed RARΔAF2, a RARα mutant lacking the C-terminal AF2 domain that binds the coactivators in the presence of RA. Because RARΔAF2binds RA and its cognate response element RARE but is specifically impaired for binding coactivators in response to RA, it acts as a dominant negative mutant to inhibit RA-dependent RAR transactivation on RARE in cells (Glass et al., 1997). CoIP assays revealed that RARΔAF2 also associates with Ngn2 in a RA-independent manner (Fig. 1C). Interestingly, RARΔAF2 inhibited RA from promoting Ngn2 transactivation in E-box:LUC reporter assays (Fig. 1A), indicating that RA-dependent interaction between RAR and the coactivators via the AF2 domain is required for RA to enhance Ngn2 transactivation.
These reveal that Ngn2 and RA-signaling are engaged in a novel mode of cross-talk, in which Ngn2 tethers RAR to E-box via forming the Ngn2/RAR-complex and subsequently RA facilitates the recruitment of the AF2 domain-dependent coactivators to this complex resulting in enhanced transactivation.
RA-signaling stimulates Ngn2-mediated neurogenesis
To test whether the cross-talk with RA-signaling operates in the proneural activity of Ngn2, we used P19 cells that undergo neurogenesis upon ectopic expression of Ngn2 (Farah et al., 2000; Lee et al., 2005). In P19 cell, Ngn2 expression triggered differentiation of neurons, as labeled by pan-neuronal marker TuJ (Fig. 1D). RA treatment cooperated with Ngn2 in a synergistic manner, resulting in not only greater induction of pan-neuronal gene β-tubulin III expression, as detected by TuJ-staining, but also enhanced neurite outgrowth (Fig. 1D, E).
Next, to test the role of endogenous RA-signaling in Ngn2-induced neurogenesis in the developing nervous system, we performed chick embryo electroporations. Forced expression of Ngn2 led to premature cell cycle exit and upregulation of pan-neuronal gene β-tubulin III in the medial zone of the chick neural tube (Fig. 1F, H, data not shown), indicating that the expression of Ngn2 triggered precocious neurogenesis in the neural tube. Remarkably, coexpression of RARΔAF2, which blocks RA-dependent transactivation by endogenous RAR, severely attenuated the neurogenic activity of Ngn2 in the neural tube (Fig. 1G, H). These show that endogenous RA-signaling plays a crucial role for the neurogenic activity of Ngn2 in the developing spinal cord.
CBP is a key effector for RA-signaling in the cross-talk of RA and Ngn2
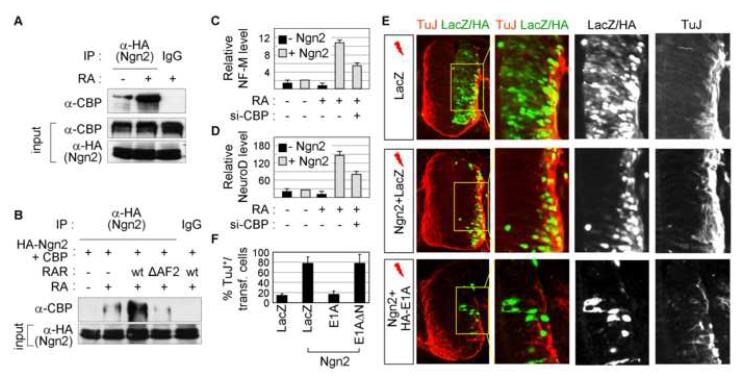
Our results implicate RA-dependent coactivators of RAR in the cross-talk between RA-signaling and Ngn2. Interestingly, CBP and its paralogue p300 function as coactivators of both RARs, by binding RA-bound RAR via the AF2 domain of RAR, and Ngn-family members (Chakravarti et al., 1996; Glass and Rosenfeld, 2000; Kamei et al., 1996; Koyano-Nakagawa et al., 1999; Sun et al., 2001; Vojtek et al., 2003). Thus, we examined whether CBP and p300 serve as key effectors for the RA action in stimulating Ngn2 transactivation. We first tested whether RA affects the association between Ngn2 and CBP/p300 by CoIP assays in HEK293 cells transfected with Ngn2. While Ngn2 bound CBP only weakly, RA greatly enhanced this interaction (Fig. 2A). Expression of RAR further augmented the association of Ngn2 and CBP, whereas RARΔAF2 weakened this interaction (Fig. 2B). Ngn2 also bound p300, but RA did not enhance this interaction (data not shown). These suggest that RA potentiates the recruitment of CBP to the Ngn2/RAR-complex via the AF2 domain of RAR and that CBP is the critical RA-dependent coactivator of RAR involved in the cross-talk of RA-signaling and Ngn2.
Figure 2. CBP is a key effector for RA-signal to promote Ngn2 function.
(A) RA enhances in vivo association between Ngn2 and CBP in CoIP experiments, as tested with HEK293 cells transfected with HA-Ngn2. (B) CoIP assays using IP with anti-HA antibody followed by immunoblotting with anti-CBP antibody in HEK293 cells transfected with HA-Ngn2 and CBP along with either RAR or RARΔAF2. RAR expression further strengthens the association of Ngn2 and CBP, while RARΔAF2 attenuates this interaction. (C, D) Quantitative-RT-PCR for neurofilament M (NF-M) (C) and NeuroD (D) in P19 cells treated as indicated. (E) Immunohistochemical analysis of neuronal differentiation (TuJ+-cells) in HH stage 22 chick embryos electroporated with the indicated constructs on the left. Cells coexpressing Ngn2 and E1A fail to differentiate to TuJ+-neurons, suggesting that Ngn2-mediated neurogenesis is suppressed by E1A. In contrast, E1AΔN does not block neurogenesis in the medial zone of the chick neural tube (data not shown). (F) Quantitative analysis of neuronal differentiation in the chick neural tube. (C, D, F) The error bars represent the standard deviation.
Next, we explored the role of CBP in the cooperative action of Ngn2 and RA for inducing neurogenesis of P19 cells. Consistent with cellular differentiation results (Fig. 1D, E), Ngn2 and RA synergized to stimulate expression of neuronal genes Neurofilament M and NeuroD, as shown by quantitative RT-PCR (Fig. 2C, D). The cooperative induction of these neuronal genes by RA and Ngn2 was attenuated by down-regulating CBP using si-RNAs (Fig. 2C, D, S-Fig. 2), indicating that CBP plays a critical role in neurogenesis triggered by Ngn2 and RA. To further test the role of endogenous CBP in neurogenesis triggered by Ngn2 during development of the spinal cord, we used E1A, which titrates out CBP and thus inhibits CBP function (Arany et al., 1995; Lundblad et al., 1995). While ∼84% of Ngn2-expressing cells differentiate into TuJ+ neurons upon electroporation of Ngn2 and LacZ, coinjection of E1A with Ngn2 markedly inhibited this neurogenic activity of Ngn2 in the spinal cord to ∼14% of Ngn2-expressing cells (Fig. 2E, F). E1AΔN, which no longer binds CBP, had no effect (Fig. 2F, data not shown).
These suggest that CBP is an integral player in the synergistic action between Ngn2 and RA-signaling for driving progenitor cells to a neuronal fate.
RA promotes the motor neuron differentiation through the Ngn2/RAR-complex binding to the motor neuron enhancer
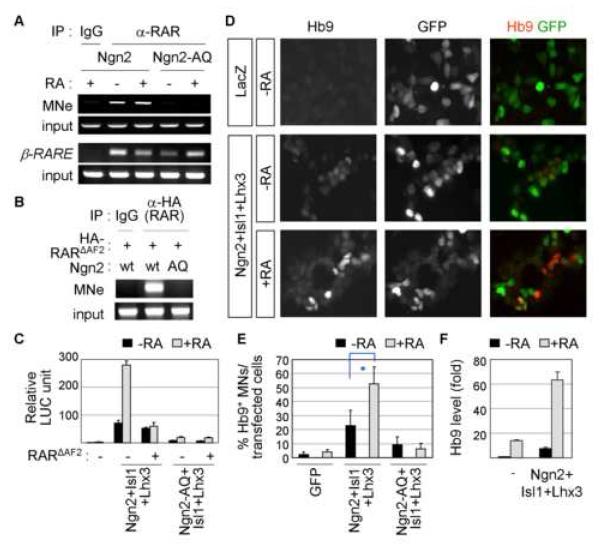
Ngn2 binds the motor neuron enhancer directly and activates the transcription of motor neuron genes (Lee et al., 2005; Lee and Pfaff, 2003). The cross-talk of Ngn2 and RA-signaling raises the possibility that RA promotes a motor neuron fate, at least in part, by upregulating motor neuron genes targeted by Ngn2. To test the RA-mediated transcriptional regulation of Ngn2-target motor neuron genes, we used the motor neuron enhancer (MNe) in a motor neuron gene Hb9. MNe, consisting of E-boxes for Ngn2-binding and response elements for the motor neuron specifying LIM-complex of Isl1 and Lhx3, has been well defined as a genomic target site of Ngn2 (Lee et al., 2008; Lee et al., 2004; Lee and Pfaff, 2003). We examined whether RAR is recruited to MNe in chromatin context, using chromatin immunoprecipitation (ChIP) assays in P19 multi-potent embryonic cells devoid of endogenous Ngn2. RAR bound to MNe in a RA-independent manner in P19 cells transfected with Ngn2, but not with Ngn2-AQ (Fig. 3A). This indicates that RAR is recruited to MNe through forming a complex with Ngn2 that binds to E-box, not through its own DNA-binding activity to RARE. Notably, MNe lacks conventional RARE sequences (data not shown) (Lee et al., 2004). As expected, RAR bound its cognate binding site β-RARE (de The et al., 1990), independently of Ngn2 (Fig. 3A). Like RAR, RARΔAF2 also occupied MNe when Ngn2, but not Ngn2-AQ, is coexpressed (Fig. 3B). Next, we tested whether RA regulates the transcription of MNe:LUC reporter in P19 cells. The expression of Ngn2 with Isl1 and Lhx3 activated the reporter ∼70-fold (Fig. 3C). RA enhanced the activation of MNe:LUC reporter by Ngn2 and Isl1/Lhx3 ∼276-fold, whereas RA was ineffective with Ngn2-AQ (Fig. 3C). The potent stimulation of MNe:LUC by RA was eliminated by blocking RAR-mediated recruitment of coactivator(s) with RARΔAF2 (Fig. 3C). These suggest that RAR is recruited to Ngn2-target motor neuron genes by forming a complex with Ngn2, and that RA enhances the transcription of these motor neuron genes.
Figure 3. RA-signaling promotes the specification of a motor neuron fate by Ngn2.
(A) ChIP assays using IP with anti-RAR antibody in P19 cells transfected with Ngn2 or Ngn2-AQ. DNA-binding activity of Ngn2 is required to recruit RAR to MNe in a RA-independent manner, as shown by occupancy of MNe by RAR in the presence of Ngn2 but not Ngn2-AQ. β-RARE is a cognate DNA response element of RAR. (B) ChIP assays in P19 cells transfected with HA-RARΔAF2 and Ngn2 or Ngn2-AQ. RARΔAF2 is recruited to MNe by Ngn2. (C) RA synergizes with Ngn2 and Isl1/Lhx3 in activating MNe:LUC reporter in P19 cells. (D-F) RA stimulates motor neuron differentiation in P19 cells transfected with Ngn2, Isl1 and Lhx3, as monitored by immunostaining with anti-Hb9 antibody (D, E) and measuring Hb9 mRNA levels in quantitative RT-PCR (F). GFP+-cells mark transfected cells (D). (C, E, F) The error bars represent the standard deviation. (E) Asterisk, p<0.001 in the two-tailed t-test.
To test whether RA promotes motor neuron differentiation triggered by Ngn2, we used multipotent P19 mouse embryonic cells that acquire motor neuron phenotypes upon forced expression of Ngn2, Isl1 and Lhx3 (Lee and Pfaff, 2003). While expression of Ngn2, Isl1 and Lhx3 triggered differentiation of Hb9+ motor neurons in ∼23% of the transfected cells, RA treatment augmented motor neuron differentiation to ∼53% (Fig. 3D, E, S-Fig. 3A). RA failed to cooperate with Ngn2-AQ (Fig. 3E). Likewise, RA synergized with Ngn2, Isl1 and Lhx3 to upregulate expression of motor neuron genes Hb9 and choline acetyltransferase and neuronal gene NeuroD, as monitored by quantitative RT-PCR (Fig. 3F, S-Fig. 3B, data not shown). These show that RA-signaling potentiates the ability of Ngn2 in inducing motor neuron differentiation of multipotent progenitor cells.
Ngn2 requires transactivation by RA-bound RAR to specify motor neurons in the neural tube
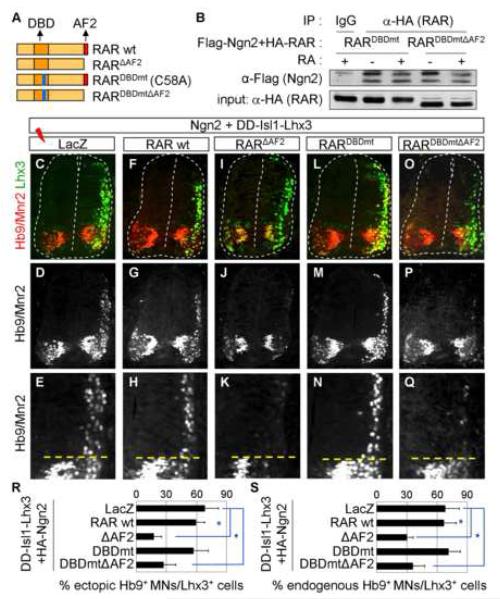
To explore the role of endogenous RA-signaling in motor neuron specification regulated by Ngn2, we investigated motor neuron generation in the chick neural tube. The forced expression of Ngn2 along with Isl1 and Lhx3 triggers motor neuron differentiation in the dorsal spinal cord (Lee and Pfaff, 2003). To test whether transcriptional activation by RA-bound RAR is needed for the ectopic production of motor neurons, we monitored motor neuron generation in the absence and presence of RARΔAF2 that blocks RA-mediated transactivation. The expression of Ngn2 and DD-Isl1-Lhx3, a chimeric molecule that mimics the motor neuron-specifying LIM-complex containing Isl1 and Lhx3 (Lee and Pfaff, 2003), induced ectopic motor neuron differentiation efficiently in ∼70% of the Lhx3+ electroporated cells in the dorsal neural tube, as monitored by α-Hb9 and α-Lhx3 antibodies (Fig. 4C-E, R). Coexpression of RARΔAF2 inhibited the ectopic motor neuron differentiation to ∼17% of the electroporated cells, while coelectroporation of RAR wild-type did not significantly affect motor neuron generation (Fig. 4F-K, R). Expression of neither Ngn2 nor DD-Isl1-Lhx3 appeared to be affected by RARΔAF2 (Fig. 4I, data not shown). These suggest that RA-mediated elicitation of RAR transactivation is necessary for Ngn2 to direct motor neuron specification of neural progenitors. RARΔAF2 may suppress motor neuron specification through inhibiting the activation of RARE by RA-bound RAR, rather than through blocking the activation of E-box elements by the RA-bound RAR/Ngn2 complex. To distinguish between these possibilities, we employed two forms of RARα, mutants, RARDBDmt and RARDBDmtΔAF2 (Fig. 4A). RARDBDmt, a point mutant of 58th RAR residue cysteine to alanine, is specifically impaired in binding RARE (Chen and Lohnes, 2005). The DNA-binding defective RAR mutants have been shown to interfere with RA-dependent activation of RARE, because such mutants lower the intracellular availability of the functional RAR/RXR heterodimer and its coactivators (Shen et al., 1993). RARDBDmtΔAF2 is defective in both RARE-binding and RA-dependent transactivation and, thus, RARDBDmtΔAF2 is ineffective in blocking RA-dependent activation of RARE in P19 cells (Chen and Lohnes, 2005). CoIP assays revealed that both RARDBDmt and RARDBDmtΔAF2 associate with Ngn2 in cells (Fig. 4B), suggesting that RARDBDmt and RARDBDmtΔAF2 can form a complex with Ngn2. Next, we examined whether RARDBDmt or RARDBDmtΔAF2 affects motor neuron differentiation by co-electroporating with Ngn2 and DD-Isl1-Lhx3 to the chick neural tube. RARDBDmt failed to block ectopic motor neuron formation (Fig. 4L-N, R), suggesting that inhibition of RA-dependent activation of RARE does not interfere with motor neuron specification by Ngn2 and DD-Isl1-Lhx3. However, RARDBDmtΔAF2 antagonized the motor neuron generation by Ngn2 and DD-Isl1-Lhx3 (Fig. 4O-R), despite its inability to suppress the activation of RARE. These results demonstrate that the RARE-binding activity is not essential for RARΔAF2 to block motor neuron specification. These suggest that endogenous RA-signaling promotes the ectopic generation of motor neurons through the Ngn2/RAR-complex bound to E-boxes of motor neuron genes, rather than via RAREs.
Figure 4. Transactivation, but not RARE-binding, by RA-bound RAR is required for Ngn2 to specify motor neurons.
(A) Schematic representation of RARα CoIP assays using IP with anti-HA antibody followed by immunoblotting with anti-Flag antibody in HEK293 cells transfected with Flag-Ngn2 and either HA-RARDBDmt or HA-RARDBDmtΔAF2. Both RARDBDmt and RARDBDmtΔAF2 associate with Ngn2 independently of RA. (C-Q) Immunohistochemical analysis of ectopic motor neuron induction or formation of endogenous motor neurons (ventral motor neurons below dotted lines in E, H, K, N, Q) in HH stage 25 chick embryos electroporated with the indicated constructs on the top. (R, S) Quantitative analysis of ectopic motor neuron (MN) generation in the dorsal spinal cord (R) or formation of endogenous motor neurons in the ventral spinal cord (S). RARΔAF2 and RARDBDmtΔAF2, but not RAR wild-type or RARDBDmt, block the specification of motor neuron cell-type. Asterisk, p<0.001 in the two-tailed t-test. The error bars represent the standard deviation.
To further test the physiological involvement of RA-signaling during motor neuron specification by Ngn2, we focused our analysis on the endogenous motor neuron domain in the ventral spinal cord. Previous studies found that inhibition of RA-signaling with RARΔAF2 prevents the expression of Ngn2 and Isl1 in the chick neural tube (Novitch et al., 2003). To test whether RAR activation is required downstream or in parallel with Ngn2 and Isl1, we analyzed motor neuron differentiation of pMN progenitors that express RAR wild-type or mutants along with Ngn2 and DD-Isl1-Lhx3 in the chick neural tube. The expression of RAR wild-type or RARDBDmt displayed little effect on motor neuron generation (Fig. 4F-H, L-N, S). In contrast, RARΔAF2 and RARDBDmtΔAF2 suppressed motor neuron formation in the ventral spinal cord despite high levels of expression of Ngn2 and DD-Isl1-Lhx3 (Fig. 4I-K, O-Q, S, data not shown). These suggest that RAR activation by endogenous RA-signaling is needed for Ngn2 to direct the differentiation of pMN progenitors to motor neurons via RARE-independent mechanism.
Our data establish that RA-signaling and Ngn2 cooperate to induce motor neuron differentiation in the developing spinal cord. Furthermore, these suggest the necessity of the AF2 domain of RAR in recruiting coactivator(s) to the RAR/Ngn2-complex and the dispensability of RARE-binding activity of RAR in the cross-talk of RA-signaling and Ngn2 for the specification of motor neuron fate.
RA-signaling facilitates the recruitment of CBP to the motor neuron enhancer
Given that CBP serves as an effector to integrate RA-signaling and Ngn2 function (Fig. 2), CBP is predicted to play important roles in motor neuron gene regulation. To test this, we examined whether CBP occupies MNe in chromatin using ChIP assays. P19 cells were transfected with Ngn2 and treated with either vehicle or RA. The chromatin fragments recruiting CBP were purified using α-CBP antibody. While CBP binding to MNe was inefficient without RA, RA addition strongly enhanced the recruitment of CBP to MNe (Fig. 5A), consistent with the facilitated association between Ngn2 and CBP in the presence of RA-bound RAR (Fig. 2A, B). CBP-binding to MNe was not detected in P19 cells without forced expression of Ngn2 even with RA (data not shown), indicating that Ngn2 is crucial to recruit CBP to MNe. This agrees with the necessity for the DNA-binding activity of Ngn2 in recruiting RAR to MNe (Fig. 3A). In contrast, p300-binding to MNe was not enhanced by RA (data not shown). Thus, RA facilitates the recruitment of CBP to the motor neuron enhancer occupied by the Ngn2/RAR-complex.
Figure 5. CBP is required for proper motor neuron development.
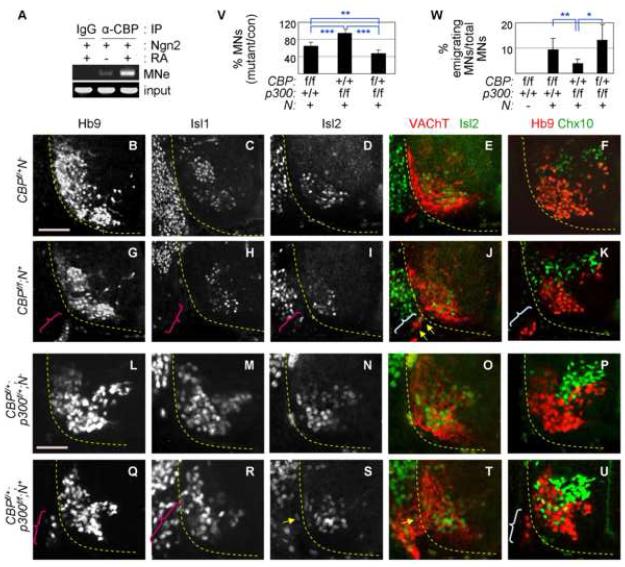
(A) RA facilitates the recruitment of CBP to MNe in P19 cells expressing Ngn2, as shown by ChIP using anti-CBP antibody. (B-U) Immunostaining of motor neuron markers Hb9, Isl1, Isl2, and VAChT and a V2-interneuron marker Ch×10 in E12.0 CBP mutants and E11.5 CBP/p300-compound mutants. The ventral quadrant of the spinal cord is shown. Ectopically emigrating motor neurons outside the spinal cord are marked by parentheses. These emigrating cells coexpress Isl2 and VAChT, as indicated by yellow arrows. (V) Quantification of Hb9+-motor neurons in both inside and outside the spinal cord of CBP/p300 mutants at cervical levels. The number of motor neurons of each genotype over their control littermate is shown in %. (W) Quantification of ectopically emigrating Hb9+-motor neurons outside the spinal cord in CBP/p300 mutants at thoracic levels. The number of emigrating motor neurons over the total number of motor neurons is shown in %. (V, W) One asterisk, p<0.01; two asterisks, p<0.001; three asterisks, p<0.0001 in the two-tailed t-test. The error bars represent the standard deviation. Scale bars, 100 μm (B-K), 50 μm (L-U).
Deletion of CBP in the developing spinal cord results in severely impaired motor neuron specification
Our findings predict that inactivation of CBP in differentiating motor neurons impairs motor neuron development. To test this, we analyzed the embryonic spinal cord in mice deficient in CBP and its paralogue p300. To circumvent the early lethality of CBP- and p300-knockout mice (Tanaka et al., 2000; Yao et al., 1998) and to inactivate CBP and p300 specifically in the developing spinal cord, we bred mice carrying floxed CBP or p300 alleles (Kang-Decker et al., 2004; Kasper et al., 2006) with Nestin-Cre mice (designated as N+ mice), which express Crerecombinase in neuroblasts (Betz et al., 1996). CBP and p300 are widely expressed in embryos but relatively enriched in the neural tube (S-Fig. 4). The CBPf/f;N+ and p300f/f;N+ mice showed normal expression of CBP/p300 by embryonic day E9.5, but their expression was sharply reduced in the developing spinal cord from E10.5 onward (S-Fig. 4, 6, 7, data not shown). Therefore, the removal of CBP/p300 proteins in these mice coincides with the timing in which spinal neurons are being generated in the developing spinal cord.
The relatively wide expression of CBP and p300 led us to first test whether the loss of CBP/p300 affects cell proliferation and/or cell survival and/or progenitor domain patterning within the neural tube. At E11.0-12.0, the Sox2+ progenitor domain was normally established and the number of BrdU+ Sox2+ proliferating progenitors did not significantly change in CBPf/f;N+ and p300f/f;N+ embryos (S-Fig. 5, data not shown). TUNEL assays revealed no change in cell death in the spinal cord of E11.0-12.0 CBPf/f;N+ and p300f/f;N+ mutants (data not shown). The expression of Pax6, Nkx6.1, and Olig2 in the ventral spinal cord of CBPf/f;N+ and p300f/f;N+ mutants was indistinguishable from that in control littermates (S-Fig. 5, data not shown). Thus, the proliferation and survival of progenitors and the establishment of appropriate ventral progenitor cell domains appear to be unaltered in CBP and p300 mutants.
We next tested the generation of motor neurons in CBPf/f;N+ and p300f/f;N+ embryos. At E10.5-13.5, the number of Hb9+ motor neurons in CBPf/f;N+ embryos was substantially reduced compared to control littermates [64.2 ± 8.3% Hb9+ motor neurons of CBPf/+;N- at cervical level of E11.5-12.0 spinal cord] (Fig. 5B, G, V, S-Fig. 6, 7), suggesting a key role of CBP in motor neuron development. The decreased number of Hb9+ motor neurons was not simply due to downregulation of Hb9, a direct target gene of Ngn2 (Lee and Pfaff, 2003), because we also confirmed the reduction in motor neuron generation by immunostaining for additional motor neuron markers Isl1, Isl2 and Lhx3 (Fig. 5C, D, H, I, S-Fig. 8). The number of Hb9+ motor neurons was not significantly altered in p300f/f;N+ embryos (Fig. 5V). However, deletion of one allele of CBP in the p300-inactivated background (i.e. CBPf/+;p300f/f;N+) severely impaired motor neuron specification, as shown by immunostaining for Hb9, Isl1, Isl2 and Lhx3 [46.7 ± 8.4% Hb9+ motor neurons of control littermates (CBPf/+;p300f/+;N-)] (Fig. 5L-V, S-Fig. 9, 10, data not shown). CBPf/+;N+ embryos were indistinguishable from wild-type littermates in the number of motor neurons (data not shown). These suggest that deletion of p300 sensitizes motor neuron phenotypes caused by the loss of one allele of CBP. The reduced number of motor neurons in CBP/p300 mutants is not the outcome of increase in motor neuron death, as TUNEL assays did not reveal any increase in the death of motor neurons (data not shown). Unlike motor neurons, the number of Ch×10+ V2a-interneurons did not decrease in CBPf/f;N+ and CBPf/+;p300f/f;N+ mutants, compared to their littermate controls (Fig. 5F, K, P, U, S-Fig. 8-11), suggesting that the general neuronal differentiation program is relatively intact in these mutants. Interestingly, however, V2a-interneurons were more scattered ventro-laterally in CBP/p300 mutants (Fig. 5F, K, P, U, S-Fig. 6-8).
These establish that CBP and, to lesser degree, p300 are required for the specification of motor neurons.
Motor neurons deficient in CBP show aberrant motor neuron cell body migration
Our analysis of CBP/p300 mutants also revealed some mislocalized motor neuron somata. In E11.5-12.0 CBPf/f;N+ embryos, ∼10% of total Hb9+ motor neurons showed somata in the motor axonal tract outside the spinal cord at thoracic levels (parentheses in Fig. 5G, K, W). These extraspinal Hb9+ cells expressed other motor neuron markers, such as Isl1, Isl2 and vesicular acetylcholine transporter (VAChT), confirming their motor neuron identity (Fig. 5H-J). Approximately 3% and 12% of motor neurons emigrated into the periphery in thoracic levels of E11.5-12.0 p300f/f;N+ and CBPf/+;p300f/f;N+ embryos, respectively (Fig. 5Q, R, U, W, S-Fig. 9, 10). These data suggest that CBP and, to lesser degree, p300 are required for confining cell bodies of motor neurons in the spinal cord.
Neural crest-derived boundary cap cells located at the motor exit point are required for retaining motor neuron somata to the spinal cord (Vermeren et al., 2003). Thus, the extraspinal motor neurons in CBP/p300 mutants may result from the loss of boundary cap cells rather than cell-autonomous defects of motor neurons. Immunostaining with boundary cap cell markers Krox20 and Sox2 revealed that boundary cap cells are formed in the vicinity of motor exit points in CBPf/f;N+ and CBPf/+;p300f/f;N+ embryos (S-Fig. 12), arguing against this possibility. This implies that the extraspinal location of motor neurons in CBP/p300 mutants reflects defects intrinsic to motor neurons, as is the case with mutant embryos lacking motor neuron specific transcription factor Hb9 or Isl2 (Arber et al., 1999; Thaler et al., 1999; Thaler et al., 2004). Thus, some CBP/p300-deficient spinal motor neurons fail to observe the boundary between CNS and the periphery likely due to impaired communication of motor neurons with peripheral signals which constrain motor neuron somata, resulting in erroneous migration of motor neuron cell bodies to the motor axonal tract in the periphery.
Motor neuron axon projection is disorganized in CBP mutants
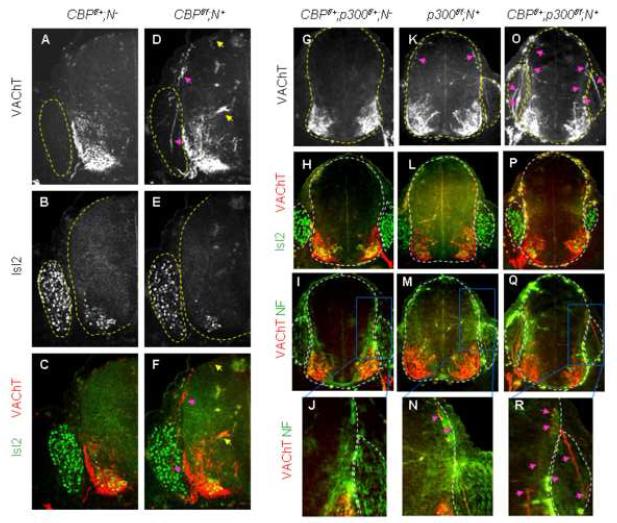
The deficits in initial motor neuron specification in CBP/p300 mutants likely lead to defects in motor axonal trajectory. To test this, we performed immunostaining with VAChT antibody, which labels motor axons specifically. The projection of motor axons was markedly perturbed in E12.0 CBP-inactivated embryos (Fig. 6A-F). Many CBP-inactivated motor axons projected dorsally toward the roof plate within the spinal cord. While some motor axons reached the roof plate (top yellow arrow in Fig. 6D), many motor axons projected to the dorsal root entry zone (DREZ) and, strikingly, often exited the spinal cord through DREZ and invaded to the dorsal root ganglion (DRG) (pink arrows in Fig. 6D, F). In contrast, the motor neuron cell bodies settled only in the ventral spinal cord and did not inappropriately migrate to the dorsal spinal cord (Fig. 6E, F). We also observed that a few CBP-deleted VAChT+ motor axons were extended medially toward the midline and stalled around the central canal (bottom yellow arrow in Fig. 6D). Motor axons of control embryos were extended ventro-laterally and exited only through the ventral motor exit point (Fig. 6A, C). Like the extraspinal motor somata phenotypes, p300-inactivated motor neuron axons displayed similar but milder phenotypes than CBP-deleted motor neurons (Fig. 6K-N). Consistent with the profound disruption in motor neuron specification, motor axon trajectories in CBPf/+;p300f/f;N+ embryos were more severely impaired, as shown by higher incidence of motor axonal projection toward DREZ and multiple VAChT+-motor axonal bundles innervating DRGs (arrows in Fig. 6O, R). This perturbation in initial motor axonal trajectory was accompanied by marked, albeit variable, defasciculation for motor axonal projections in the periphery (data not shown). These demonstrate that the disturbed motor neuron specification in CBP/p300 mutants coincides with their erroneous motor axon projection.
Figure 6. Motor axonal trajectory is profoundly disrupted in CBP/p300 mutants.
Immunostaining analyses of motor neuron markers Isl2 and VAChT in E12.0 CBP mutants (A-F) and E11.5 p300- and CBP/p300-compound mutants (G-R). Spinal cord and dorsal root ganglion (DRG) are outlined by dotted lines. Arrows indicate erroneous projection of VAChT+ motor axons toward dorsal root entry zone (DREZ), roof plate, midline and within DRG.
The motor axon pathfinding defects prompted us to test whether motor connectivity with target muscles in CBP mutant is impaired. Thus, we performed immunostaining with neurofilament antibody to visualize motor nerve innervation and anti-bungarotoxin labeling to detect clusters of acetylcholine receptors (AChRs) on the muscle cells. In our analysis of the neuromuscular junctions between the phrenic motor nerve and the diaphragm muscle, we found that motor axonal projection and branch formation were compromised in CBPf/f;N+ embryos (S-Fig. 13), indicating defects in the muscle innervation by CBP-inactivated motor axons. AChR clusters were also scattered over wider regions of the muscle surface (S-Fig. 13). These indicate that CBP and, to lesser degree, p300 are required for proper motor axon pathfinding and muscle innervation.
Motor neuron-specific deletion of CBP leads to deficits in motor neuron development
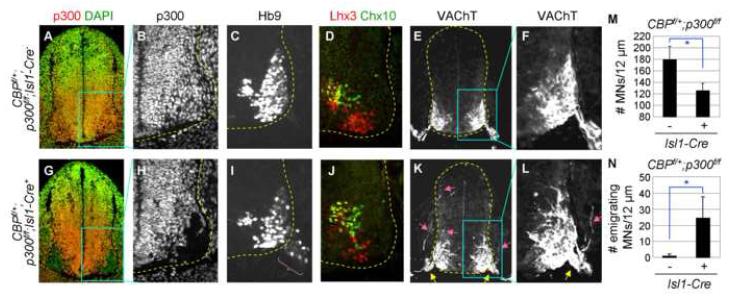
To further test whether CBP is required for Ngn2 and Isl1/Lhx3 to trigger motor neuron fate, we generated mouse mutants in which CBP is specifically deleted in differentiating motor neurons expressing Ngn2, Isl1 and Lhx3 (Lee and Pfaff, 2003). We used an inducible Isl1-MerCreMer line (designated as Isl1-Cre+ mice), in which the Cre recombinase activity is induced by tamoxifen only in Isl1-expressing cells including embryonic motor neurons (Sun et al., 2007). As we have observed profound motor neuron phenotypes in CBPf/+;p300f/f;N+ embryos, we generated CBPf/+;p300f/f;Isl1-Cre+ embryos. We then injected tamoxifen into pregnant dams carrying CBPf/+;p300f/f;Isl1-Cre+ embryos and their control littermates at E10.5 to inactivate CBP and p300 during the period in which motor neurons are being specified by Ngn2 and Isl1/Lhx3. Embryos were harvested 24 hours later at E11.5 for the analyses. In CBPf/+;p300f/f;Isl1-Cre+ embryos, the p300 expression was removed and CBP expression was also compromised specifically in embryonic motor neurons, but not in other ventral spinal interneurons or progenitor cells (Fig. 7G, H, data not shown). Remarkably, this recapitulated the phenotypes observed in CBPf/+;p300f/f;N+ embryos. First, in the ventral spinal cord, the number of motor neurons of CBPf/+;p300f/f;Isl1-Cre+ embryos was reduced to ∼70% of that of the control littermates CBPf/+;p300f/f;Isl1-Cre-, as monitored by immunostaining with α-Hb9 and α-Lhx3 antibodies (Fig. 7C, D, I, J, M). Second, ∼16% of Hb9+ motor neurons in thoracic level of CBPf/+;p300f/f;Isl1-Cre+ embryos emigrated into the periphery (parenthesis in Fig 7I, N). Third, VAChT+-motor axons exiting the spinal cord were severely defasciculated and the motor axon exit point became substantially wider (yellow arrows in Fig. 7K, L). Fourth, motor axons projected dorsally and often exited through DREZ and innervated to DRGs (pink arrows in Fig. 7K, L). Despite profound defects in motor neuron-fate specification, we failed to detect any increase in the death of motor neurons in CBPf/+;p300f/f;Isl1-Cre+ embryos (data not shown), indicating that the reduced number of motor neurons is not due to increased cell death. These indicate that the cell-type specification of motor neurons is impaired upon the timely inactivation of CBP/p300 during cell fate assignment, and thus establish the motor neuron cell-autonomous requirements of CBP/p300. Given the RA-dependent recruitment of CBP to the Ngn2-target motor neuron enhancer, these data support the idea that CBP plays essential roles in motor neuron specification program governed by Ngn2 and RA-signaling.
Figure 7. Timely inactivation of CBP/p300 during motor neuron specification impairs motor neuron development.
(A-L) Immunohistochemical analyses in E11.5 CBPf/+;p300f/f;Isl1-Cre- (A-F) and CBPf/+;p300f/f;Isl1-Cre+ (G-L) embryos. Ectopically emigrating motor neurons outside the spinal cord (parenthesis), widened motor exit points (yellow arrows), and erroneous projection of VAChT+ motor axons toward DREZ, roof plate and within DRG (pink arrows) are marked. (M, N) Quantification of Hb9+-motor neurons in the spinal cord (M) and ectopically emigrating Hb9+-motor neurons outside the spinal cord (N) in 12 μm sections at thoracic levels. Asterisk, p<0.001 in the two-tailed t-test. The error bars represent the standard deviation.
RA triggers the transcriptionally active chromatin in the motor neuron enhancer through the Ngn2/RAR/CBP complex
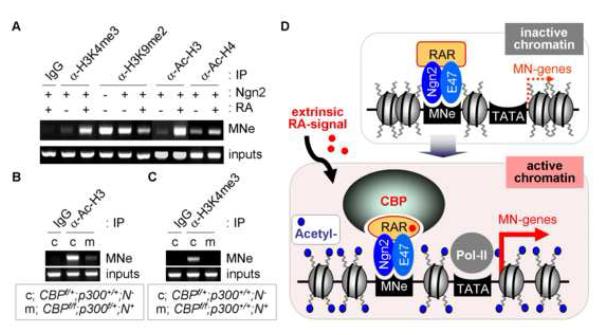
Considering that the Ngn2/RAR-complex facilitates the recruitment of CBP to the motor neuron enhancer in response to RA and that motor neuron generation is impaired in CBP mutants, it is possible that CBP, a prominent HAT enzyme, induces histone acetylation of motor neuron genes in response to RA, thereby playing critical roles in translating RA-signal into the transcriptional activation of target motor neuron genes of the Ngn2/RAR-complex. To test this, we examined the RA-dependent changes in various histone modifications of MNe using ChIP assays in P19 cells. RA-treatment of Ngn2-expressing P19 cells led to a prominent elevation of histones H3 and H4 acetylation in MNe (Fig. 8A), coincident with recruitment of CBP (Fig. 5A). Moreover, RA also markedly enhanced H3-lysine-4-trimethylation, a marker for active chromatin, and reduced H3-lysine-9-dimethylation, a marker for inactive chromatin (Fig. 8A). These suggest that RA-signaling induces transcriptionally active chromatin in motor neuron genes targeted by the Ngn2/RAR-complex.
Figure 8. Transcriptionally active chromatin is established by RA and CBP in MNe.
(A) The histone modifications in MNe upon RA treatment were analyzed by ChIP assays in P19 cells expressing Ngn2. RA facilitates histone H3/H4-acetylation and H3-lysine-4-trimethylation (H3K4me3), while suppressing H3-lysine-9-diemthylation (H3K9me2), in MNe. (B, C) ChIP assays using the spinal cord dissected from mutant embryos of genotypes shown in the boxes. Histone H3-acetylation (B) and H3K4me3 (C) in MNe are impaired in CBP-inactivated E12.5 embryonic spinal cord. (D) The working model. Ngn2 and RAR form a complex in pMN progenitors. The extrinsic signal RA binds RAR and facilitates the association of a chromatin modifier CBP with the Ngn2/RAR-complex. Assembly of the Ngn2/RAR/CBP-complex on Ngn2-target motor neuron enhancers triggers their transcriptionally active open chromatin structure marked by H3/4-acetylation and results in subsequent motor neuron gene expressions, leading to the differentiation to motor neurons.
As CBP is specifically recruited to MNe in response to RA-signaling, it is likely to be a critical enzyme for RA-dependent stimulation of histone H3 and H4 acetylation in MNe. To test this, we performed ChIP assays with E12.5 mouse embryonic spinal cords dissected from CBP mutants. Compared to control littermates, H3-acetylation levels in MNe were markedly reduced in CBP mutants (Fig. 8B). H3-lysine-4-trimethylation in MNe was also diminished (Fig. 8C). H3-acetylation levels in GAPDH promoter were comparable between CBP mutants and their control littermates (data not shown). These demonstrate that CBP plays central roles in coordinating histone modifications to establish transcriptionally active chromatin in motor neuron genes during spinal cord development.
DISCUSSION
In CNS development, various inductive signals and transcription factors controlling cell fate specification have been relatively well defined (Jessell, 2000). However, two key questions remain unclear. Firstly, how are these extrinsic and intrinsic cues coupled for the timely activation of genes for neuronal cell fate specification? Secondly, how is chromatin configuration regulated during neurogenesis? In this study, we addressed these issues in the development of spinal motor neurons. We found that RA-signaling and Ngn2 cooperate to recruit CBP to motor neuron genes, which in turn establishes transcriptionally active chromatin. This study provides a molecular understanding of how chromatin modifying enzymes are selectively recruited to a specific cohort of target genes to promote specification of neuronal subtypes during vertebrate development.
The Ngn2/RAR-complex as a sensor for the extrinsic signal RA
RA-signaling plays critical roles in neurogenesis and sequential phases of motor neuron development (Appel and Eisen, 2003). Blockade of RAR transactivation using RARΔAF2 mutant interfered with motor neuron generation in the chick neural tube, implicating RA-bound RAR as a major transcriptional activator for motor neuron fate specification (Novitch et al., 2003). However, the transcriptional mechanism by which RA-signaling induces motor neuron fate and the downstream target genes of RA-bound RAR in these processes remain unidentified. Our studies reveal a specific mechanism for RA to direct a motor neuron fate, which operates during the transition of Ngn2+ pMN cells into postmitotic motor neurons (Fig. 8D). During spinal cord development, expression of Ngn2 and RAR primes neural progenitors for motor neuron differentiation. The Ngn2/RAR-complex is recruited to motor neuron genes via Ngn2-binding E-boxes, but is transcriptionally inefficient. The arrival of the environmental signal RA then triggers motor neuron differentiation by facilitating recruitment of CBP to the Ngn2/RAR-complex, which in turn establishes transcriptionally active chromatin in motor neuron genes. In this scheme, the Ngn2/RAR-complex in pMN cells acts as a molecular sensor to detect the presence of extrinsic RA-signal, which controls the timing of motor neuron differentiation. This model is consistent with the previous reports that RA deficient quail and mouse embryos as well as Ngn2 mutant mouse embryos show severe defects in motor neuron differentiation (Novitch et al., 2003; Scardigli et al., 2001; Wilson et al., 2004).
Our studies show that RA-bound RAR stimulates Ngn2-dependent motor neuron differentiation by binding and transactivating non-RAREs (i.e., E-boxes), uncovering a key role for RARE-independent action of RA in vertebrate embryonic development. We found that proneural bHLH proteins Ngn1 and Mash1 also associate with RAR in a RA-independent manner (data not shown). Interestingly, a muscle-specific bHLH factor MyoD forms a complex with RAR/RXR heterodimer, recruiting RAR/RXR to specific MyoD-binding E-box elements during RA-induced myoblast differentiation (Froeschle et al., 1998). Thus, functional convergence of cell-type-specific bHLH factors with RA-signaling through formation of a RAR-bHLH protein complex may be involved in a broad range of cell fate decision during development.
The expression level of proneural bHLH factors has been thought to determine the activation of their target genes, as they act as potent transcriptional activators by dimerizing with E-proteins and binding E-boxes (Bertrand et al., 2002). However, recent evidences suggest that the activity of proneural bHLH proteins can be regulated by extrinsic signals. Akt kinases augment the transcriptional activity of Ngn3 by enhancing complex formation between Ngn3 and p300 (Vojtek et al., 2003). The temporal phosphorylation of Ngn2 facilitates the interaction between Ngn2 and a cofactor NLI for motor neuron specification (Ma et al., 2008). Our findings, together with these reports, suggest that extrinsic signals regulate the proneural activity of bHLH factors by controlling the recruitment of coactivators. This may represent important regulatory steps for neuronal subtype specification.
A critical role of CBP for the development of motor neurons
Posttranslational modification of histone tails has been extensively studied as an important regulatory code for gene expression (Berger, 2007). Particularly, histone H3/H4-acetylation and H3-lysine-4-trimethylation have been linked to transcriptionally active or poised chromatin (Ruthenburg et al., 2007). Recent studies suggest that chromatin remodeling serves as an important cell intrinsic mechanism for cell-lineage specification during neural development (Hsieh and Gage, 2005; Lessard et al., 2007). However, little is known about how histone modifications are controlled in a spatially and temporally regulated manner during CNS development and how such chromatin changes affect neuronal lineage specification. Our studies demonstrate an essential histone-modifying mechanism underlying the transition from progenitor cells to motor neurons, which is regulated by extrinsic RA-signaling. RA recruits a HAT enzyme CBP to the Ngn2/RAR-complex that occupies motor neuron genes (Fig. 8D). This may involve a coordinated mobilization of multiple independent interfaces in CBP; at least one for RA-bound RAR and the other for Ngn2. In accordance with its role in facilitating recruitment of CBP to the motor neuron enhancer, RA strongly induces transcriptionally active chromatin on the motor neuron gene Hb9, marked by histone H3/H4-acetylation and H3-lysine-4-trimethylation. Correspondingly, removal of CBP in differentiating motor neurons results in marked deficits in motor neuron specification and axon pathfinding, as well as reduction in histones H3-acetylation and H3-lysine-4-trimethylation in the motor neuron enhancer.
Given that RA-signal triggers both histone H3/H4-acetylation and H3-lysine-4-trimethylation, it is interesting to speculate that RA-signal recruits not only HAT enzyme CBP but also histone methyltransferase complexes mediating H3-lysine-4-trimethylation to motor neuron genes. At least two related H3-lysine-4-methyltransferase complexes are associated with RAR in an RA-dependent manner (Lee et al., 2006). Thus, these complexes may play important roles in RA-induced motor neuron gene expression during the spinal cord development. An intriguing possibility is that RA-signaling functions as an extrinsic cue to tightly couple the activities between HAT-complexes containing CBP and histone methyltransferase complexes in motor neuron genes, thereby establishing a chromatin landscape that favors motor neuron differentiation. Notably, H3-lysine-4-methyltransferase complexes associate with CBP (Ernst et al., 2001; Goto et al., 2002; Petruk et al., 2001). Our finding that the removal of CBP in the spinal cord leads to the reduction in both histone H3-acetylation and H3-lysine-4-trimethylation is consistent with the notion that histone acetylation and H3-lysine-4-trimethylation are coordinately regulated to generate transcriptionally active chromatin.
In conclusion, we present extrinsic RA-signaling as a temporal switch that triggers chromatin changes in motor neuron genes, via a novel cross-talk between RAR and Ngn2, which subsequently induces motor neuron differentiation. Furthermore, we show that, in this cross-talk, a HAT enzyme CBP plays critical roles in RA-dependent chromatin remodeling for motor neuron genes, thereby enabling proper motor neuron development.
EXPERIMENTAL PRODEDURES
Mice generation
The generation of CBPflox, p300flox, Nestin-Cre, and inducible Isl1-MerCreMer mice has been described previously (Betz et al., 1996; Sun et al., 2007; Tanaka et al., 2000; Yao et al., 1998). CBPf/f or p300f/f mice were crossed with Nestin-Cre lines (designated as N+ mice) or Isl1-MerCreMe mice (designated as Isl1-Cre+ mice).
In ovo electroporation and Immunohistochemistry
In ovo electroporation and immunohistochemistry were performed as described (Thaler et al., 1999). Briefly, plasmid DNA was injected into the lumen of the neural tube of HH stage 13 chick embryos which were then electroporated. The embryos were harvested at HH stage 20-26, fixed in 4% paraformaldehyde, embedded in OCT and cryosectioned at 12-18 μm.
Luciferase & P19 cell differentiation assays, Co-immnoprecipitation (CoIP), and Chromatin immunoprecipitation (ChIP) assays
These assays were performed as described previously (Joshi et al., 2009) in P19 mouse carcinoma cells, human embryonic kidney 293 cells, or mouse embryonic spinal cord cells.
For P19 cell differentiation assays, P19 cells were transfected using Lipofectamine 2000 (Invitrogen) and analyzed three days post-transfection by quantitative RT-PCR or immunohistochemistry. 0.5-1 μM all-trans-retinoic acid (Sigma) or vehicle was treated for 48hr prior to cell harvest. Total RNA was extracted with mini-kit (QIAGEN) and reverse transcription (RT) was performed using Superscript III (Invitrogen). The levels of mRNA were determined using quantitative RT-PCR (M×3000P, Stratagene).
Additional experimental procedures and reagent information are provided as supplements.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Paul Overbeek, Adam Antebi, and Huda Zoghbi and the lab members for critical comments on the manuscript; Drs. P. Brindle, J. van Deursen and S. Evans for p300-flox, CBP-flox and Isl1-MerCreMe mice, respectively. This research was supported by grants from NINDS (R01 NS054941), PEW, March of Dimes Foundations and IDDRC P30 HD42064.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Appel B, Eisen JS. Retinoids run rampant: multiple roles during spinal cord and motor neuron development. Neuron. 2003;40:461–464. doi: 10.1016/s0896-6273(03)00688-3. [DOI] [PubMed] [Google Scholar]
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Betz UA, Vosshenrich CA, Rajewsky K, Muller W. Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr Biol. 1996;6:1307–1316. doi: 10.1016/s0960-9822(02)70717-3. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Chen CF, Lohnes D. Dominant-negative retinoic acid receptors elicit epidermal defects through a non-canonical pathway. J Biol Chem. 2005;280:3012–3021. doi: 10.1074/jbc.M411522200. [DOI] [PubMed] [Google Scholar]
- de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Froeschle A, Alric S, Kitzmann M, Carnac G, Aurade F, Rochette-Egly C, Bonnieu A. Retinoic acid receptors and muscle b-HLH proteins: partners in retinoid-induced myogenesis. Oncogene. 1998;16:3369–3378. doi: 10.1038/sj.onc.1201894. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61:839–851. doi: 10.1016/j.neuron.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kang-Decker N, Tong C, Boussouar F, Baker DJ, Xu W, Leontovich AA, Taylor WR, Brindle PK, van Deursen JM. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell. 2004;5:177–189. doi: 10.1016/s1535-6108(04)00022-4. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, Murray PJ, van Deursen JM, Brindle PK. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Wettstein D, Kintner C. Activation of Xenopus genes required for lateral inhibition and neuronal differentiation during primary neurogenesis. Mol Cell Neurosci. 1999;14:327–339. doi: 10.1006/mcne.1999.0783. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci U S A. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Funahashi J, Ruiz EC, Pfaff SL. Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development. 2004;131:3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Supp 1):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- Ma YC, Song MR, Park JP, Henry Ho HY, Hu L, Kurtev MV, Zieg J, Ma Q, Pfaff SL, Greenberg ME. Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron. 2008;58:65–77. doi: 10.1016/j.neuron.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. Retinoid signalling in the development of the central nervous system. Nat Rev Neurosci. 2002;3:843–853. doi: 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Pfaff SL. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Wichterle H, Jessell TM, Sockanathan S. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron. 2003;40:81–95. doi: 10.1016/j.neuron.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Shen S, van der Saag PT, Kruijer W. Dominant negative retinoic acid receptor beta. Mech Dev. 1993;40:177–189. doi: 10.1016/0925-4773(93)90075-9. [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Perlmann T, Jessell TM. Retinoid receptor signaling in postmitotic motor neurons regulates rostrocaudal positional identity and axonal projection pattern. Neuron. 2003;40:97–111. doi: 10.1016/s0896-6273(03)00532-4. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- Vermeren M, Maro GS, Bron R, McGonnell IM, Charnay P, Topilko P, Cohen J. Integrity of developing spinal motor columns is regulated by neural crest derivatives at motor exit points. Neuron. 2003;37:403–415. doi: 10.1016/s0896-6273(02)01188-1. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Taylor J, DeRuiter SL, Yu JY, Figueroa C, Kwok RP, Turner DL. Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. Mol Cell Biol. 2003;23:4417–4427. doi: 10.1128/MCB.23.13.4417-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Gale E, Chambers D, Maden M. Retinoic acid and the control of dorsoventral patterning in the avian spinal cord. Dev Biol. 2004;269:433–446. doi: 10.1016/j.ydbio.2004.01.034. [DOI] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.