SUMMARY
Phenoloxidases (POs) occur in all organisms and are involved in skin and hair coloring in mammals, and initiating melanisation in wound healing. Mutation or overexpression of PO can cause albinism or melanoma, respectively. SDS can convert inactive PO and the oxygen carrier hemocyanin (Hc) into enzymatically active PO. Here we present single particle cryo-EM maps at subnanometer resolution and pseudo atomic models of the 24-oligomeric Hc from scorpion Pandinus imperator in resting and SDS activated states. Our structural analyses lead to a plausible mechanism of Hc enzyme PO activation: upon SDS activation the intrinsically flexible Hc domain I twists away from domains II and III in each subunit, exposing the entrance to the active site; this movement is stabilized by enhanced inter-hexamer and inter-dodecamer interactions, particularly in the central linker subunits. This mechanism could be applicable to other type 3 copper proteins since the active site is highly conserved.
INTRODUCTION
Type 3 copper proteins such as the blue hemocyanins (Hcs) and the enzymes tyrosinase (Ty) and catecholoxidase (CO) contain binuclear copper centers and share similar active site geometry (Decker et al., 2007b; Hazes et al., 1993; Klabunde et al., 1998; Magnus et al., 1994; Matoba et al., 2006; Solomon et al., 1996). Ty catalyses both the o-hydroxylation of monophenols to o-diphenols and the oxidation of o-diphenols to o-quinones, whereas CO only catalyses the second reaction (Decker and Tuczek, 2000; Solomon et al., 1996; Sugumaran, 2002). Both enzymes are commonly subsumed under the name of phenoloxidase (PO) (van Holde et al., 2001). Despite the close relationship between them, Hc functions as an oxygen transporter, whereas PO initiates the synthesis of melanin and is involved in immune response, wound healing, browning and the sclerotisation process in arthropods after molting (Cerenius and Soderhall, 2004; Sugumaran, 2002). PO is also responsible for browning of plants and fruits (Marusek et al., 2006; van Gelder et al., 1997). More importantly, Ty is the first and rate-limiting enzyme in the synthesis of melanin pigments responsible for coloring hair, skin and eyes (Cerenius and Soderhall, 2004; Sugumaran, 2002). Mutation of Ty often decreases melanin production resulting in albinism, whereas an extremely high level of it may induce melanoma, the most deadly skin cancer (Simonova et al., 2000). But to date, the mechanism of PO activity at the molecular level has remained unclear (Garcia-Borron and Solano, 2002; Oetting, 2000; Schweikardt et al., 2007).
Sodium dodecyl sulphate (SDS), commonly known as a denaturant, has been used for decades in an assay to detect PO activity of proteins even though the mechanism is unknown. In this process inactive PO (Kanade et al., 2006; Marusek et al., 2006; Moore and Flurkey, 1990; Robb et al., 1964; van Gelder et al., 1997) is converted to a functionally active PO by incubation with SDS at a concentration above the critical micelle concentration (CMC) (Baird et al., 2007; Jaenicke and Decker, 2008). In recent years weak PO activity was observed in Hcs from both arthropods and molluscs using the above assay (Decker and Rimke, 1998; Decker et al., 2001; Lee et al., 2004; Pless et al., 2003; Salvato et al., 1998; Siddiqui et al., 2006), suggesting a dual role for Hc in vivo. Moreover, it was proposed recently that the monophenoloxidase activity of Hc may be responsible for post mortem black spot formation of crustaceans, which reduces the value of sea food products dramatically (Garcia-Carreno et al., 2008).
Hcs are extracellular proteins found in the hemolymph of arthropods and molluscs (van Holde et al., 2001). Arthropod Hcs occur as integral numbers of hexamers (1×6, 2×6, 4×6, 6×6 and 8×6) depending on the species (Markl and Decker, 1992). Each subunit has a molecular mass of about 72-kD and can reversibly bind one oxygen molecule as peroxide between two copper atoms at its active site, as deduced from crystal structures (Hazes et al., 1993; Magnus et al., 1994). Each Hc subunit folds in three domains (Figure S1): the α-helix rich N-terminal domain I covers the entrance to the active site, followed by domain II containing the active site, and then the C-terminal domain III, consisting of a seven-stranded Greek key β-barrel topology and a huge β-hairpin (Decker and Rimke, 1998; Decker and Tuczek, 2000). Among the three domains, domains II and III dominate the inter-subunit contacts within each hexamer (Figure S3D and Figure S3E), while domain I is involved in inter-hexamer interactions. Each Hc subunit binds oxygen with similar affinity, but heterogeneous subunit types are needed to guarantee self assembly to the native Hc structure (Markl and Decker, 1992). For the chelicerate 2×12-mer Hc (with a molecular mass of about 1.75-MD), seven or eight different subunit types are required since these subunits assume specific positions within the oligomer in order to facilitate self-assembly and cooperative oxygen binding (Markl and Decker, 1992)
In the activation process of Hc and PO it is assumed that the presence of SDS mimics the effects of natural activators or allosteric effectors (Baird et al., 2007; Cerenius and Soderhall, 2004; Jaenicke and Decker, 2008; Kanade et al., 2006; Marusek et al., 2006; Nagai et al., 2001; Sugumaran, 2002) by inducing a conformational switch that displaces a conserved placeholder residue (PHE49 in Limulus Hc, Figure S1) (Baird et al., 2007; Decker and Jaenicke, 2004; Decker et al., 2007b; Decker et al., 2006; Decker and Tuczek, 2000). This allows bulky mono-and di-phenols access to the active site (Decker and Jaenicke, 2004; Decker et al., 2007a; Decker et al., 2006). without denaturing the protein (Baird et al., 2007; Nillius et al., 2008). However, presently there is no direct high resolution structural evidence showing the SDS induced Hc conformational change and the related PO activity conversion.
In this study we present two subnanometer resolution cryo-EM reconstructions of arthropod 24-meric Hc from the scorpion Pandinus imperator in the activated (SDS present) and resting (SDS absent) states. For this Hc system, PO-activity was recently demonstrated in the presence of micellar form of SDS as used in our experimental condition (Nillius et al., 2008). Our cryo-EM maps in combination with pseudo atomic models reveal, for the first time, the structural basis for the enzymatic PO activity of Hc. Our data clearly demonstrate that an SDS concentration above the CMC does not induce a denaturation of Hc. Rather, we observe a collective conformational switch in the intrinsically flexible domain I of each subunit, and this leads to a cooperative quaternary structural rearrangement of the entire oligomer. Our structural observations may be applicable to other type 3 copper proteins since they shire similar active site geometry.
RESULTS
Visualizing the resting and activated conformations of Hc by cryo-EM
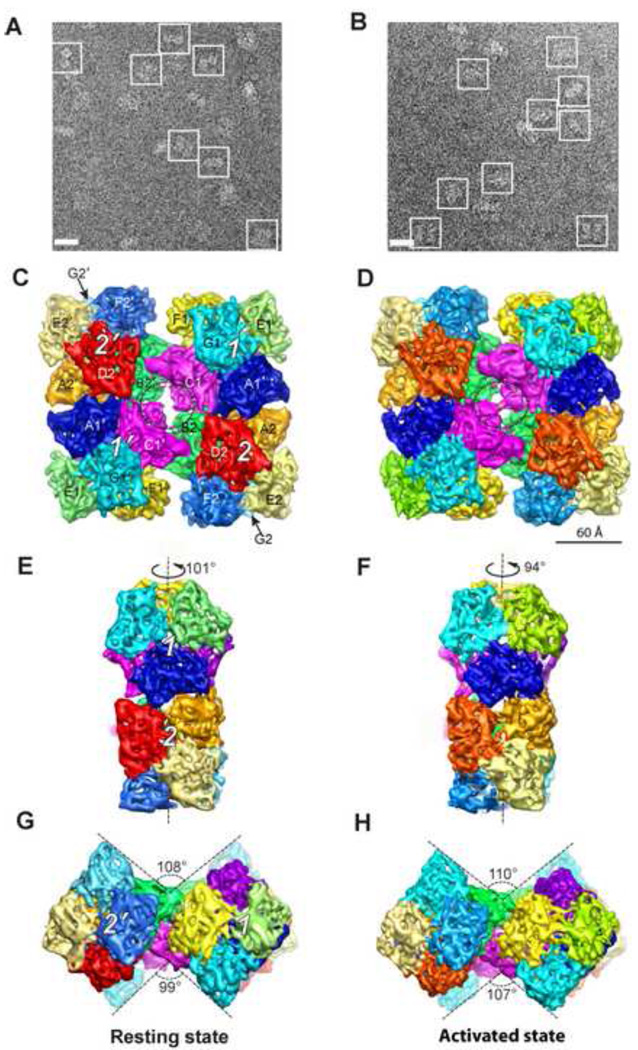
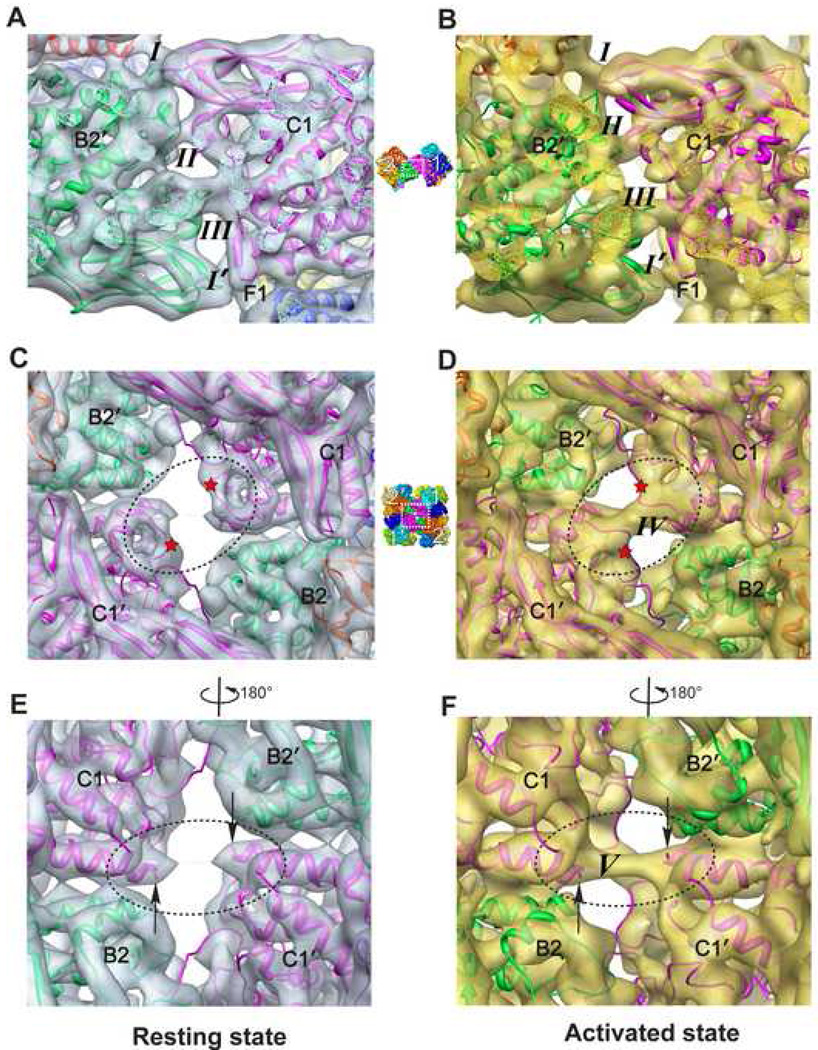
Figure 1A and 1B illustrate typical cryo-EM images of both resting and activated Hcs. Corresponding 3-D density maps were reconstructed independently to 6.8 Å and 8.0 Å resolution, respectively (Figure 1C–1H; Figure S2). Similar to the other 4×6-meric Hc structures (Bijlholt et al., 1982; van Heel and Dube, 1994), the Hc oligomer consists of four hexamers connected by four central linker subunits (subunits C1, C1', B2 and B2' in Figure 1C; Movie 1) (de Haas and van Bruggen, 1994). 2-fold symmetry was imposed in the reconstruction (Experimental Procedures), causing the Hc maps to be comprised of two equivalent asymmetric units. Each asymmetric unit (also called a dodecamer) consists of two hexamers (which are labelled as 1 and 2 and their symmetry mates as 1' and 2' in Figure 1C; Movie 1). The subunits within each hexamer are related to each other by a pseudo D3 symmetry (Figure S3B and S3C; Movie 1) (Hazes et al., 1993; Magnus et al., 1994). The two hexamers in each asymmetric unit are rotated relative to each other by 101°/94° in the resting and activated states, respectively (Figure 1E and 1F; Movie 1). We adopt the conventional subunit designation (Decker et al., 1996) for the 12 subunits in each asymmetric unit labelled as A1, C1, D1, E1, F1, G1, A2, B2, D2, E2, F2, and G2.
Figure 1. Structure of 24-meric Scorpion Hc.
(A)/(B) Typical 300 kV image of ice embedded Hc in the resting/activated state recorded in a JEM3200FSC electron cryomicroscope. Representative particles are highlighted by white boxes. Scale bar represents 245 Å.
(C)/(D) Top view of Hc in the resting/activated state at a threshold corresponding to Hc molecular weight. Different subunits are illustrated in different colors, and labelled. The resolution is 6.8 Å and 8.0 Å for resting and activated states, respectively. The dotted circles highlight the new bridging densities absent/formed in the resting/activated state. The four hexamers are labelled as 1, 2, 1', and 2'. 2-fold symmetry was imposed in the reconstruction. Scale bar in (D) represents 60 Å, which is applicable for (C)–(H).
(E)/(F) Hexamer 3-fold view of Hc in resting/activated state.
(G)/(H) Bottom bow tie view of Hc in resting/activate state.
The overall dimension of the two biochemical states is similar (Movie 2, Figure 1). However, structural differences can be observed between the resting and activated Hcs, from the level of a monomer to the level of the entire 24-meric complex (Movie 2; Movie 3). Collectively, SDS induces a large (~7°) rotation angle difference, i.e. the relative angle between hexamer 1 and 2 within an asymmetric unit decreases from ~101°(resting state, Figure 1E) to ~94°(activated state, Figure 1F). Besides, there is an observable distance decrease between the two dodecamers due to SDS activation (Movie 2, top view morphing between the two models). Moreover, as illustrated in Figure 1G and 1H, the end-on view of the 24-meric Hc has a bow tie shape. Upon SDS activation, the bottom V angle increases by ~8° Possible reasons for these observed differences are discussed below.
Localizations of secondary structure elements of the subunits in both states
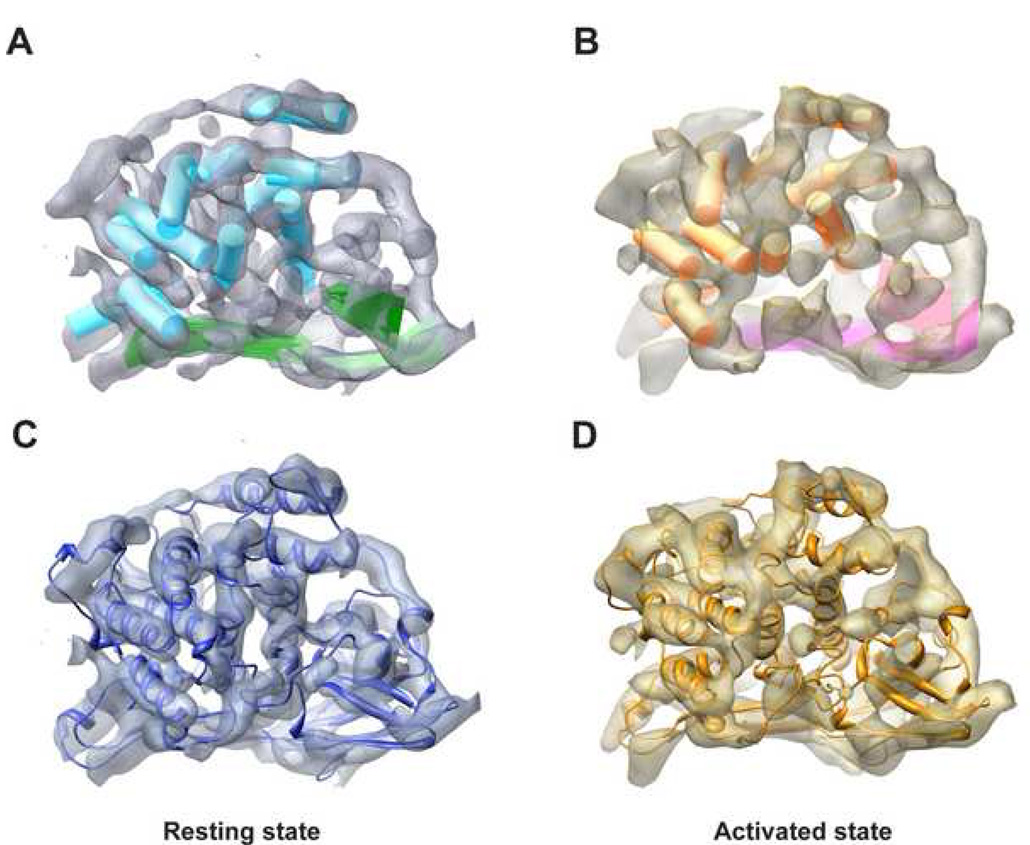
Each of our subnanometer resolution Hc maps resolves not only individual subunits in the 24-meric complex but also secondary structure elements (SSEs). Take the linker subunit C1 as an example, it was segmented from both maps and SSE locations were mapped out (Figure 2A and 2B) by SSEHunter (Baker et al., 2007). In every subunit of the resting state map, all eleven α-helices longer than two turns and three large β-sheets of domain III were identified, while in the activated state, ten or eleven α–helices and three large β–sheets were detected in all subunits. Here in some of the subunit maps of the activated state, one α–helix of three turns could not be identified. The locations of the SSEs identified in our maps are consistent with the crystal structure of a related Limulus Hc (Figure S1) (Hazes et al., 1993; Magnus et al., 1994), which further demonstrates the quality of our maps.
Figure 2. Model of a Hc Monomer.
(A) SSEs of the linker subunit C1 in the resting state identified by SSEHunter. Eleven α-helices longer than 2-turns (cyan cylinders) and three large β-sheets (green surfaces) have been identified in this subunit.
(B) SSEs of the same subunit C1 in the activated state. Ten α-helices (orange cylinders) and three β-sheets (pink surfaces) are identified.
(C)/(D) Pseudo atomic model of C1 subunit fitted into the corresponding map of the resting/activated state where blue/yellow coloring represents the resting/activated state. The same color scheme is utilized in the rest of the figures.
Pseudo atomic model building by comparative modelling and flexible fitting
In order to examine the differences of the two conformational states in more structural detail, we generated a complete 24-mer model for each state (Figure S3; Movie 2) by combination of comparative modelling utilizing MODELLER 9v1 (Sali and Blundell, 1993) and flexible fitting using SITUS (Wriggers and Birmanns, 2001). Details about the model building are described in Experimental Procedures. As demonstrated in Figure 2C and 2D, the models fit well with the segmented subunit density maps. The correlation score between the model and map for C1 subunit is around 0.97/0.95 for the resting/activated state (Experimental Procedures). Moreover, the locations and orientations of alpha helices detected in the map agree accurately with our model (Figure S4).
Conformational variations among different subunits in single biochemical state
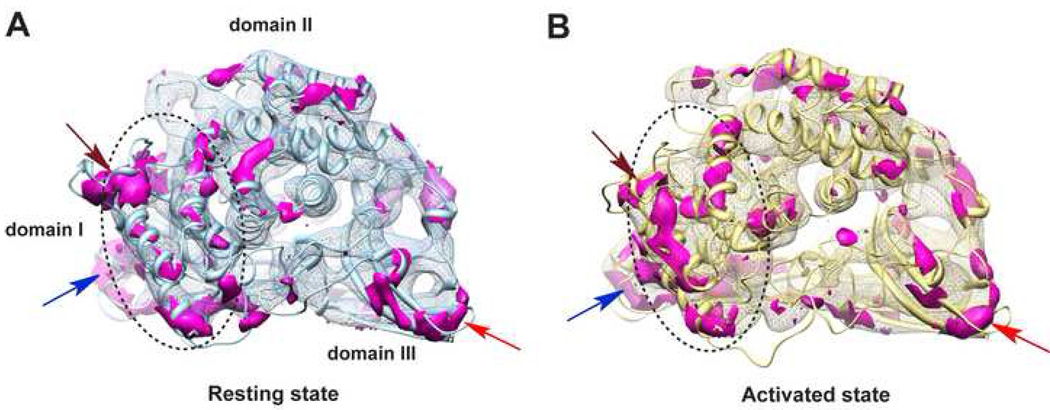
In order to visualize local conformational variations among different subunits in a single state, we calculated the 3D variance maps of the 12 segmented subunit density maps within an asymmetric unit for the resting (Figure 3A) and activated (Figure 3B) states, respectively. Details about the 3D variance map calculation are described in Experimental Procedures. In addition, the corresponding 12 models were aligned and illustrated in Figure S5. Interestingly, our data reveal that the α-helix rich N-terminal domain I (defined in Figure S1) as a whole shows more pronounced local variation than that observed in the other two domains for both biochemical states (dashed circular line in Figure 3 and Figure S5), which demonstrates the intrinsic flexibility of domain I in Hc. Reasons for this are presented in the Discussion. Moreover, this flexible domain I plays an essential role in the activation of Hc, which will be demonstrated later.
Figure 3. Conformational Variations among the 12 Subunits within an Asymmetric Unit.
(A) 3-D variance map (purple) among the 12 subunit map segments within an asymmetric unit of the resting state illustrating their conformational variations. The variance map is shown in the frame of the corresponding average-map, with a model in light blue ribbon shown to orient the figure. Notably, domain I (highlighted by dotted circle) as a whole shows more pronounced variation than the other two domains. Overall, the outer exposed area tends to have more variations as compared to the interior, especially at the inter-subunit contacting regions indicated by arrowheads (brown, blue, and red).
(B) Variance map among the 12 subunits within an asymmetric unit of the activated state with the same visualization style and labels as in (A).
Other relatively large variances are present at the perimeter of the subunits, which are the regions involved in inter-subunit contacts in both states (highlighted by arrows in Figure 3). Notably, the above identified flexible domain and regions have relatively high B-factors in the template crystal structure 1NOL (Hazes et al., 1993) for our comparative model building.
Furthermore, there is a loop region not resolved in the X-ray structure 1NOL (residues V568-S573, location shown by red arrow in Figure S1), most probably due to its structural flexibility. Even though this loop remains invisible in most of the subunits in our maps, it is resolved clearly in the linker subunit B2 of both states (highlighted by ellipsoid in Movie 1), where the loop is stabilized owing to its involvement in an inter-dodecamer bridge formation with subunit C1' (discussed in later section). This indicates that the plastic loop regions might play a significant role in the allosteric network communication in macromolecular assembles, and consequently causes the multiple subunits to function cooperatively. This also demonstrates the utility of cryo-EM studies in capturing critical structural details of macromolecular machines, which are missing in the crystal structure.
Conformational changes between the two biochemical states
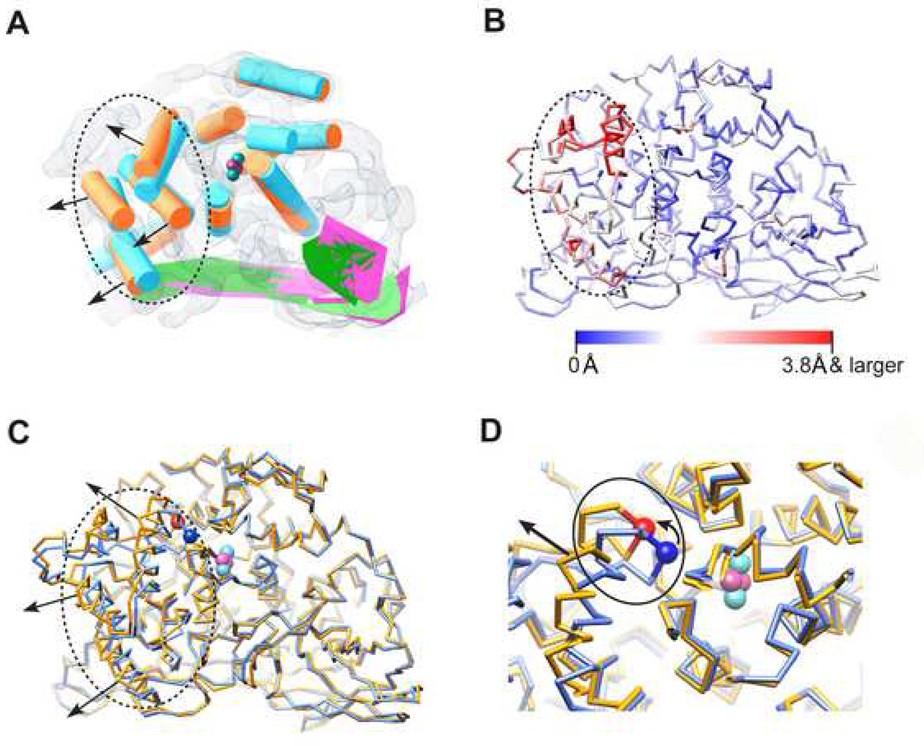
To assess the conformational changes induced by SDS we examined the relative movements of the SSEs detected directly from the cryo-EM maps. By superposition of the SSEs of the C1 subunit of both states (Figure 4A), we observe that activation causes a more pronounced movement of the intrinsically flexible domain I (highlighted by dotted circle in Figure 4A) than those observed in domains II and III. More importantly, the direction of the movement of domain I is to twist away from domains II and III (arrow head direction in Figure 4A), which appears as a slight expansion motion of the entire subunit. This motion potentially leaves the active site (cyan and magenta spheres in Figure 4A) more accessible for the bulky PO substrate.
Figure 4. Conformational Variations between the Two Biochemical States.
(A) Conformational variation analysis using deduced SSEs of the two states (as in Figure 2A and 2B, same colour code adopted), which are aligned together and illustrated in the frame of the resting state map. The black dotted circle highlights the domain I region. Black arrows show the twisting direction of domain I as induced by SDS activation. The cyan and magenta spheres demonstrate the putative locations of the Cu and oxygen atoms forming the active site.
(B) Cα movement between the averaged models of the two states, mapped onto the activated state averaged Cα model. Red demonstrates larger variance (Cα movement 3.8 Å and larger) while blue demonstrates smaller variance.
(C) The aligned averaged Cα models of resting state (light blue) and activated state (gold). The same illustration style as in (A) (dotted circle, black arrow and active site) is adopted. Location of the Cα of the placeholder residue PHE49 is illustrated using blue (resting state) and red (activated state) spheres.
(D) Zoom in view of the active site with solid line circle illustrating the location of the loop region containing PHE49, which also connects to one of the domain I α-helices (indicated by black straight arrow). The small curved arrow indicates the moving direction of PHE49 as a result of SDS activation.
Furthermore, to examine the average Hc domain I movement caused by SDS activation, we calculated the averaged pseudo atomic model (Cα only) of all the 12 subunits in an asymmetric unit of each biochemical state. The Cα movement between the two averaged models (Figure 4B) elucidates that domain I Cα atoms undergo relatively larger movements (up to 4.7 Å) as compared with the other two domains (Movie 3). Particularly, on average Hc domain I is twisting away from the active site (Figure 4C; Movie 3), which is consistent with that seen in subunit C1 by SSEs analysis (Figure 4A). It has been shown in numerous cases that difference Fourier maps in crystallography can show finer details than the resolution of individual maps (Henderson and Moffat, 1971). The difference we observe here is likely a genuine effect of the chemical activation of Hc by SDS. The consequence of the difference is to allow more space for the substrate to slide into the active site of Hc.
Conformational switch at the active site
By zooming into the active site of the averaged pseudo atomic model, one may observe a loop region (highlighted by solid line circle in Figure 4D) consisting of the conserved placeholder residue PHE49 (blocking the active site, Cα as blue/red sphere in Figure 4D, location also illustrated in Figure S1) (Decker and Jaenicke, 2004; Decker et al., 2007a; Decker et al., 2007b; Decker and Tuczek, 2000). This loop also closely connects to one of the domain I α-helices (indicated by black straight arrow in Figure 4D). Upon SDS activation, as domain I twists away from the active site (Movie 3; Figure 4C), this loop region could be dragged away meanwhile pulling the PHE49 residue outwards, which would expose the active site for the bulky PO substrate (Figure 4C and 4D; Movie 3). On average, the Cα of PHE49 moves away from its original location (by ~3.5 Å) upon SDS activation (blue sphere to red sphere in Figure 4D), which is potentially the most critical conformational change in the active site.
Inter-dodecamer linking and interaction strength change upon activation
The characteristics of oligomeric complexes especially mega-enzymes are, to a great extent, determined by the nature of the interactions and linking of functional subunits. It has long been a point of interest to study how the building blocks of Hc (hexamers) interact with each other in order to establish higher-order quaternary structure (van Holde et al., 2001), and in particular, how they communicate to behave cooperatively upon enzymatic activation or oxygenation. In addition, it is appealing to trace the collective interaction strength adjustment in the building block interface induced by the domain I twisting motion of individual subunits. A detailed analysis of the 48-mer Limulus polyphemus Hc inter-hexamer interaction bridges for a single biochemical state has been made (Martin et al., 2007). Here, our cryo-EM maps of Hc in two biochemical states reveal new interaction bridges in those interfaces.
We first examine the inter-dodecamer linking taking the interface between hexamer 1 and 2′ (locations indicated in Figure 1C) as an example. The lower interface between hexamer 1′ and 2 (Figure 1C) has exactly the same linking bridges due to the 2-fold symmetry in this Hc structure (Experimental Procedures). In the resting state we can clearly observe four bridges, i.e. bridges I, II, III, and I' (Figure 5A; Table S1), among which I and I' are of the same type. Of note, our map allows us to identify bridges II and III, which were not seen or not well defined in a previous study (Martin et al., 2007). These structural differences could be attributable to differences in resolution and also in species. Interestingly, most of the bridges are formed by turns and loop regions rather than other SSEs.
Figure 5. Connecting Bridges in the Inter-dodecamer Interface.
(A) Overall view of the four bridges (three types) in the interface between hexamers 1 and 2' in the resting state. The visualization angle and location with respect to the entire Hc complex are illustrated using a small reference panel in the middle column.
(B) View of the four interaction bridges at the same location as in (A), but for the activated state. Maps for both states are visualized at a threshold corresponding to the molecular mass of Hc.
(D) Bridge IV (front bridge) formed across the central cavity connecting the opposite linker subunits C1 and C1' in the activated state, whereas no such bridge exists in the resting state as illustrated in (C). The red stars in (C) and (D) indicate the conformational change loop regions involved in the formation of bridge IV due to activation.
(F) Bridge V (back bridge) in the activated state across the central cavity connecting the N-terminal ends of the opposite two linker subunits C1 and C1'. The black arrows in (E) and (F) point to the N-terminal ends of subunits C1 and C1'. This bridge is extremely weak and almost invisible in the resting state (E).
Upon SDS activation the four bridges (I, II, III and I' in the resting state, Figure 5A) connecting the two dodecamers remain unchanged (Figure 5B). Strikingly, the activated state map clearly depicts two additional bridge densities between linker subunits C1 and C1' across the central cavity (dotted circle in Figure 1D, front bridge IV in Figure 5D and back bridge V in Figure 5F), establishing new interaction pathways between hexamers 1 and 1'. In this central cavity, bridge V (Figure 5F) connects the exact same N-terminus of the opposite linker subunits C1 and C1'. This bridge is tenuous in the resting state (Figure 5E, almost invisible in the visualization threshold corresponding to Hc’s molecular mass, similar as in a previous Hc study (Martin et al., 2007)), while it becomes pronounced in the activated state. Furthermore, bridge IV is formed between exactly the same loop region in the opposing linker subunits C1 and C1' (location indicated by red stars in Figure 5C and 5D). This bridge formation is induced by a loop conformational change triggered by SDS activation. Such conformational plasticity is commonly found in proteins (Chen and Sigler, 1999) and may play an essential role in Hc allosteric communication and activation. In brief, SDS activation induced Hc conformational changes can establish new paths for allosteric network communication between its two asymmetric units.
Inter-hexamer linking and interaction strength change upon activation
By inspecting the interaction interface between hexamers 1 and 2 in an asymmetric unit in the resting state (location illustrated in Figure 1C), 11 bridges are observed; whereas in the activated state, 3 more bridges (14 bridges total) are observed, at a threshold that reflects Hc’s molecular mass (Table S2). Overall, the additionally observable bridges in both inter-dodecamer and inter-hexamer interfaces suggest strengthened interactions in these interfaces upon SDS activation (discussed below).
DISCUSSION
It has been reported that SDS above the CMC can convert Hc into an enzyme to perform the PO activity, which associates with a putative conformational shift (Nillius et al., 2008; Baird et al., 2007; Jaenicke and Decker, 2008). However, the mechanism of PO activity is still not fully understood. In this study, we obtained 6.8 Å and 8.0 Å cryo-EM density maps of the 24-meric scorpion Pandinus imperator Hc in resting and SDS activated states (Figure 1), respectively. We carried out a secondary structure element analysis (Baker et al., 2007) and pseudo atomic model building based on comparative modelling (Sali and Blundell, 1993) and flexible fitting (Wriggers and Birmanns, 2001) (Figure 2, Experimental Procedures). In both states, the agreement of the SSEs detected in the cryo-EM map with the pseudo atomic model confirms the quality of the reconstructed maps (Figure S4). Of note, the maps in both biochemical states clearly demonstrate that, even above the CMC, SDS does not cause structural degradation of the Hc assembly, rather it induces conformational changes (Figure 1; Movie 2), as often stated but never proven structurally (Jaenicke and Decker, 2008). The conformational changes associated with SDS activation extend from tertiary to quaternary structural levels (Movie 2; Movie 3), which is consistent with the changes observed in the oxy- and deoxygenated state of Hc by SAXS methods (Hartmann and Decker, 2002).
At the individual subunit level, the variance maps (Figure 3) and the pseudo atomic models (Figure S5) both demonstrate the intrinsic flexibility of domain I compared to the other two domains. This characteristic flexibility of domain I can be attributed to the fact that it is free of contact with any other subunits within a hexamer; whereas domains II and III dominate the inter-subunit contacts (Figure S3D and S3E). More excitingly, our data reveals that SDS activation pulls the intrinsically flexible domain I (consisting of the placeholder residue PHE49 covering the active site) away from domains II and III in a twisting motion, leaving the enzyme active site more exposed to the bulky PO substrate (Figure 4; Movie 3). This provides the first structural evidence of enzyme activation in the type 3 copper protein family. In particular, the domain blocking the entrance of the substrate into the active site in the resting state must be moved away or be removed in order to expose the active site.
We also found that SDS activation facilitates the formation/strengthening of bridges IV and V across the central cavity (Figure 5C–5F), and causes the distance between the two dodecamers to shorten slightly (Movie 2). These two bridges are unique in the entire complex owing to the specific special arrangement of linker subunits C1 and C1'. In particular, in these two subunits the induced movement of domain I by SDS activation is restrained by the interaction of the opposing N-terminal ends through the strengthened bridge V (Figure 5F). The above observation is consistent with the previous hypothesis (Decker and Jaenicke, 2004; Decker et al., 2007a; Decker and Rimke, 1998; Decker et al., 2001; Decker et al., 2006; Decker and Tuczek, 2000) and the observation, based on biochemical experiments, that the linker subunits show strong PO activity (Decker and Rimke, 1998; Decker et al., 2001).
In addition, our data reveal an enhancement of interactions in the inter-hexamer and inter-dodecamer interfaces. Domain I is only involved in the inter-subunit interactions in the above two interfaces (Figure S3D and S3E). As a consequence, the twisting motion of domain I, as demonstrated above, could result in closer and more intimate interactions in these interfaces. This is supported by our observation that more interaction bridge densities in the inter-hexamer interface (Table S2) as well as in the inter-dodecamer interface (Figure 5; Table S1) are detectable in the activated state but not in the resting state (Figure 5C–5F). This suggests that the collective tertiary domain I motion of multiple subunits may play an important role in the establishment of new interaction bridges, which may strengthen a direct allosteric communication between the hexamers and induce a quaternary conformational change facilitating Hc PO activity.
Quaternary structural change upon SDS activation is evidenced by the reorientation of the subunits relative to each other (Movie 2), emerging as an overall 7° decrease in the rotation angle between the two hexamers within a dodecamer (Figure 1E and 1F), in addition to a slight decrease in the distance (~4 Å) between the two dodecamers (Movie 2). Large conformational changes have also been suggested for 24-meric tarantula Hcs (Decker et al., 1996; Hartmann and Decker, 2002) and for a crustacean dodecameric Hcs in the oxy-state based on SAXS (Hartmann et al., 2001). In a broader view, for large macromolecular assemblies such as multimeric enzymes, such a cooperative quaternary structural change is a recurring mechanism used by oligomeric proteins to regulate their functions (Maier et al., 2006).
In summary, we present for the first time a structural mechanism at the molecular level showing that a denaturing agent such as SDS can modulate the structure of native proteins without destroying them, especially when occurring above the CMC as micelles (Baird et al., 2007; Decker and Jaenicke, 2004; Jaenicke and Decker, 2008; Kanade et al., 2006; Nillius et al., 2008). Thus, SDS seems to act as an allosteric effector (Baird et al., 2007; Nillius et al., 2008). This mechanism is strongly supported by the observation that binding of a serine protease by Tachypleus Hc in a 1:1 stoichiometry also promotes PO activity without any proteolytic activity (Nagai and Kawabata, 2000). Since the active sites of the type 3 copper proteins are highly conserved, our deduced mechanism of activation should be general for this family of proteins.
EXPERIMENTAL PROCEDURES
Purification of the scorpion Pandinus imperator Hc
The Hc of the scorpion Pandinus imperator (animals from Tropenhaus Marxen, Hamburg, Germany) was collected by dorsal punctuation of the membrane between the fifth and sixth mesosomal tergites. The emanating hemolymph was collected into an eppendorf cap with a stabilisation buffer to prevent hemolymph coagulation and protein precipitation. The buffer contained 100 mM TRIS/HCl at pH 7.8, 10 mM CaCl2 and 10 mM MgCl2, was degassed and sterilely filtered.
The hemolymph / buffer solution was centrifuged at 32,000×g in a cooling centrifuge Universal 16R (Hettich Zentrifugen, Tuttlingen, Germany) for 30 minutes to get rid of cellular debris and tissue contaminations. Hereafter, the blue supernatant was subjected to an analytical size exclusion chromatography using a Biologic-FPLC system (Bio-Rad, Munich, Germany) with a Sephacryl S 300 column (HiPrep 26/60, Pharmacia Biotech, Freiburg, Germany) to purify the Hc and to separate it from other hemolymph proteins. The elution was carried out using the same stabilisation buffer at a constant flow rate of 0.4ml·min-1. The fractions containing pure Hc were joined and concentrated with centrifugal filter units (Biomax 30K, Millipore, Eschborn, Germany) in the same centrifuge at 4,000×g and 4°C. Protein concentration was estimated spectroscopically at 278nm using the standard value for proteins: ε278 = 1.0 ml·mg−1·cm−1. The resulting stock solution had a concentration of 23 mg·ml−1 and was kept at 4°C until further use.
SDS activation of Hc
For activation of the Hc we used 2mM SDS. A solution of Hc from the stock (~ 23 mg·ml−1) was diluted with an SDS-buffer-solution (0.1 M TRIS/HCl, pH 7.8 and 2.5 mM SDS) at a ratio of at least 1:30 (v/v). This also avoids the precipitation of Ca-SDS. The Hc will be activated after a very short time and will remain so for several hours (Baird et al., 2007; Jaenicke and Decker, 2008; Nillius et al., 2008).
Electron cryo-microscopy
Hc samples were prepared for cryo-EM studies by dilution of the purified Hc to 0.5 mg·ml−1. Resting state samples were frozen immediately after dilution. Activated state Hc samples were achieved by the procedure described above with SDS final concentration at ~2mM followed by ~5 minutes activation, which were then frozen within ~15 minutes. 3ul of sample was deposited onto a glow-discharged 400-mesh Quantifoil holey grid with 1.2×1.3 µm hole size (Quantifoil Micro Tools GmbH, Jena Germany). The grid was flash frozen in liquid ethane using a Vitrobot (FEI, Hillsboro, Oregon, USA). Data was recorded on a JEM 3200FSC microscope equipped with a field-emission gun operated at 300 kV and an in-column energy filter (using a 20eV slit). The specimen temperature was at 101K during data collection. Images were recorded at 50,000× nominal magnification on Kodak SO-163 films. The specimen dose was 18 electrons·Å−2. The images were digitized on a Nikon Super CoolScan 9000 ED scanner with 6.35µ m per pixel scanning interval. Micrographs with a defocus range of 1~3.5µ m were used in the final reconstructions.
Image processing, 3-D reconstruction, segmentation and variance map calculation
The particles were boxed out using EMAN (Ludtke et al., 1999; Ludtke et al., 2004) and SwarmPS (Woolford et al., 2007). Contrast transfer function parameter estimation was carried out using the EMAN program ctfit. All 3-D reconstructions were completed using EMAN 1.8 with C2 symmetry imposed. Such symmetry imposition was based on previous evidence produced by negative stain electron microscopy (Boisset et al., 1990; van Bruggen et al., 1980; van Heel and Dube, 1994) and other X-ray crystallography and cryo-EM studies on arthropod Hcs (de Haas and van Bruggen, 1994; Fiser et al., 2000; Martin et al., 2007). In addition, a recently developed 2-D fast rotational matching (FRM2D) algorithm (Cong et al., 2005; Cong et al., 2003) for image alignment, available in EMAN 1.8+, was adopted in the final refinement steps. The resolutions of the resting and activated state maps were evaluated to be 6.8 Å and 8.0 Å (Figure S2), respectively, using the 0.5 FSC criterion (Harauz and van Heel, 1986). There were 17,500 and 13,400 particles in the final reconstructions in the resting and activated states, respectively.
Segmentations of the individual 12 subunits within an asymmetric unit (a dodecamer) for each state were done utilizing Amira (http://www.amiravis.com; TGS) and CHIMERA (http://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004). The segmentation is straightforward and without ambiguity at this resolution. All visualizations were done with CHIMERA (Pettersen et al., 2004).
The correlation score, reflecting the similarity between the model and map, was measured using a normalized correlation coefficient after the pseudo atomic model was filtered to the same resolution of the map. For each state, we normalized and aligned the 12 segmented subunit maps within an asymmetric unit using EMAN program proc3d and CHIMERA “fit into map” module, respectively. Then a 3D variance map as well as an average map were calculated among these 12 subunit maps using EMAN program mapvariance.
Comparative model building
To build a pseudo atomic model for a single subunit of Hc, comparative modelling was performed using MODELLER 9v1 (Sali and Blundell, 1993). Since no amino acid sequence was available for the Hc from scorpion Pandinus imperator, we chose the sequence of the Hc As6 subunit from scorpion Androctonus australis (Buzy et al., 1995), which is very close to our system based on sequence analysis of As6 with other typ3 copper protein subunits (Burmester, 2001), as well as immunological crossreactivity (Markl et al., 1984). Moreover, the 2.18 Å crystal structure of oxygenated Limulus polyphemus subunit II Hc (PDB ID: 1NOL (Hazes et al., 1993), Figure S1) was used as template, which has ~60% sequence identity to As6. We used the standard single template modelling protocol implemented in the ‘model’ module of MODELLER 9v1. 50 models were generated, and the best one was selected based on the multivariate model assessment score (Eramian et al., 2006).
In addition, for the missing loop regions in the template structure 1NOL (including residues 21–29, 134–138, 527–530, and 569–572 as indicated by arrows in Figure S1), 200 loop models were generated by the loopmodel class in MODELLER 9v1 (Sali and Blundell, 1993). The best loop model was chosen by a combination of the DOPE statistical potential score (Shen and Sali, 2006) and the cross-correlation score between the loop and the density map (Topf et al., 2006).
Complete 24-mer pseudo atomic model building by flexible fitting
To build a complete model for our 24-meric Hc, using the CHIMERA (Pettersen et al., 2004) “fit into map” module, we first rigid-body fitted the comparative model of a single subunit into the segmented subunit (i.e. the linker subunit C1) for both states. There are noticeable conformational changes between the model and the map especially for the activated state.
To capture the conformational changes revealed by the cryo-EM maps, SITUS (Wriggers and Birmanns, 2001) flexible fitting procedure was used to flexibly fit the comparative model into the map. Taking the resting state C1 subunit as an example, flexible fitting by energy minimization was carried out using X-PLOR (Brunger, 1992) with the default parameters of the CHARMM united-atom force field (Brooks et al., 1983), version 19. In the energy minimization we included six pairs of positional markers determined by SITUS (Wriggers and Birmanns, 2001), with the starting markers from the model and the ending markers from the map. We used the same procedure as described in (Cong et al., 2008): by adding a constraining energy function using landmarks as if they were NOE constraints in X-PLOR so as to force the model to move towards the ending conformation as in the cryo-EM map. The constrained structure was subjected to 10000 steps of Powell energy minimization. Hookean potentials with force constants of 310 kcal· mol−1·Å−2 were employed (Wriggers and Birmanns, 2001). In this way, the comparative model was flexibly fitted into the map of the C1 subunit. We repeated the same flexible fitting procedure to build all the 12 pseudo atomic models in one asymmetric unit in the resting state.
Afterwards, by applying the 2-fold symmetry of this Hc structure we then obtained a complete 24-mer Hc pseudo atomic model for the resting state (Figure S3A). Still, in this complete model, several side chain clashes occurred in the interfaces of adjacent subunits. To eliminate those clashes, a local refinement procedure by energy minimization utilizing X-PLOR was carried out. The clashing regions were subjected to 2000 steps of Powell energy minimization while all other residues were maintained in their original coordinates. As to the activated state, the same flexible fitting procedure described above was applied to obtain a complete 24-mer pseudo atomic model.
Supplementary Material
Supplemental Data include five additional figures, two tables and three movies are available online at ***. The cryo-EM density maps and corresponding models are available from the EMDB and PDB. Their accession numbers are EMD-5100 and PDB ID 3IXV (for the Hc resting state) and EMD-5101 and PDB ID 3IXW (for the activated state), respectively.
Structural organization of the 24-meric Hc, exemplified by the resting state Hc cryo-EM map with different subunits in different colors. The representative views of the 24-mer, dodecamer, hexamer and monomer are illustrated. The B2 subunit map together with the corresponding pseudo atomic model are shown in the last portion of the movie, with the well resolved loop region in the cryo-EM map (not resolved in the crystal template structure 1NOL, discussed in the text) highlighted by ellipsoid.
Conformational change of Hc induced by SDS activation, illustrated by the Hc density maps as well as the pseudo atomic models. Pseudo atomic model morph between the two states clearly elucidates that the 24-oligomeric Hc complex undergoes a tertiary to collective quaternary conformational change upon SDS activation. There is a slight decrease in distance (~4 Å) between the two dodecamers due to SDS activation as illustrated during time period 0:08~0:17.
Averaged Hc monomer Cα model morph between the resting and the activated states. The most mobile domain I is illustrated in deep pink, while the other two domains in yellow and light green. The cyan and magenta spheres demonstrate the putative locations of the Cu and oxygen atoms forming the active site, and the blue sphere illustrates the Cα of the placeholder residue PHE49. This movie clearly demonstrates that Hc domain I has a more pronounced movement than domains II and III; more importantly, its moving direction is to twist away from the other two domains leaving the active site more exposed.
ACKNOWLEDGEMENTS
We thank Dr. Christian Meesters for manuscript reading and comments; Mr. Jonathan L. Respress for involvement in data pre-processing. This research has been partially supported by NIH grants through the National Center for Research Resources (P41RR02250), NIGMS (R01GM08139) and the Roadmap Initiative for Medical Research (2PN2EY016525); and the German Research Foundation and Research Center for Immunology Mainz (HD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baird S, Kelly SM, Price NC, Jaenicke E, Meesters C, Nillius D, Decker H, Nairn J. Hemocyanin conformational changes associated with SDS-induced phenol oxidase activation. Biochim Biophys Acta. 2007;1774:1380–1394. doi: 10.1016/j.bbapap.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlholt MM, van Heel MG, van Bruggen EF. Comparison of 4 X 6-meric hemocyanins from three different arthropods using computer alignment and correspondence analysis. J Mol Biol. 1982;161:139–153. doi: 10.1016/0022-2836(82)90283-2. [DOI] [PubMed] [Google Scholar]
- Boisset N, Taveau JC, Lamy J, Wagenknecht T, Radermacher M, Frank J. Three-dimensional reconstruction of native Androctonus australis hemocyanin. J Mol Biol. 1990;216:743–760. doi: 10.1016/0022-2836(90)90396-4. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4:187–217. [Google Scholar]
- Brunger A. X-PLOR (version 3.1): A System for X-ray crystallography and NMR. The Howard Hughes Medical Insttitute and Department of Molecular Biophysics and Biochemistry, Yale University; 1992. [Google Scholar]
- Burmester T. Molecular evolution of the arthropod hemocyanin superfamily. Mol Biol Evol. 2001;18:184–195. doi: 10.1093/oxfordjournals.molbev.a003792. [DOI] [PubMed] [Google Scholar]
- Buzy A, Gagnon J, Lamy J, Thibault P, Forest E, Hudry-Clergeon G. Complete amino acid sequence of the Aa6 subunit of the scorpion Androctonus australis hemocyanin determined by Edman degradation and mass spectrometry. Eur J Biochem. 1995;233:93–101. doi: 10.1111/j.1432-1033.1995.093_1.x. [DOI] [PubMed] [Google Scholar]
- Cerenius L, Soderhall K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–126.22. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Sigler PB. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- Cong Y, Jiang W, Birmanns S, Zhou ZH, Chiu W, Wriggers W. Fast rotational matching of single-particle images. J Struct Biol. 2005;152:104–112. doi: 10.1016/j.jsb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Cong Y, Kovacs JA, Wriggers W. 2D fast rotational matching for image processing of biophysical data. J Struct Biol. 2003;144:51–60. doi: 10.1016/j.jsb.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Cong Y, Topf M, Sali A, Matsudaira P, Dougherty M, Chiu W, Schmid MF. Crystallographic conformers of actin in a biologically active bundle of filaments. J Mol Biol. 2008;375:331–336. doi: 10.1016/j.jmb.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas F, van Bruggen EF. The interhexameric contacts in the four-hexameric hemocyanin from the tarantula Eurypelma californicum. A tentative mechanism for cooperative behavior. J Mol Biol. 1994;237:464–478. doi: 10.1006/jmbi.1994.1248. [DOI] [PubMed] [Google Scholar]
- Decker H, Hartmann H, Sterner R, Schwarz E, Pilz I. Small-angle X-ray scattering reveals differences between the quaternary structures of oxygenated and deoxygenated tarantula hemocyanin. FEBS Lett. 1996;393:226–230. doi: 10.1016/0014-5793(96)00887-3. [DOI] [PubMed] [Google Scholar]
- Decker H, Jaenicke E. Recent findings on phenoloxidase activity and antimicrobial activity of hemocyanins. Dev Comp Immunol. 2004;28:673–687. doi: 10.1016/j.dci.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Decker H, Jaenicke E, Hellmann N, Lieb B, Meissner U, Markl J. Minireview: recent insights in the structure, function and evolution of hemocyanins. Integrative and Comparative Biology. 2007a;47:631–644. doi: 10.1093/icb/icm063. [DOI] [PubMed] [Google Scholar]
- Decker H, Rimke T. Tarantula hemocyanin shows phenoloxidase activity. J Biol Chem. 1998;273:25889–25892. doi: 10.1074/jbc.273.40.25889. [DOI] [PubMed] [Google Scholar]
- Decker H, Ryan M, Jaenicke E, Terwilliger N. SDS-induced phenoloxidase activity of hemocyanins from Limulus polyphemus, Eurypelma californicum, and Cancer magister. J Biol Chem. 2001;276:17796–17799. doi: 10.1074/jbc.M010436200. [DOI] [PubMed] [Google Scholar]
- Decker H, Schweikardt T, Nillius D, Salzbrunn U, Jaenicke E, Tuczek F. Similar enzyme activation and catalysis in hemocyanins and tyrosinases. Gene. 2007b;398:183–191.23. doi: 10.1016/j.gene.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Decker H, Schweikardt T, Tuczek F. The first crystal structure of tyrosinase: all questions answered? Angew Chem Int Ed Engl. 2006;45:4546–4550. doi: 10.1002/anie.200601255. [DOI] [PubMed] [Google Scholar]
- Decker H, Tuczek F. Tyrosinase/catecholoxidase activity of hemocyanins:structural basis and molecular mechanism. Trends Biochem Sci. 2000;25:392–397. doi: 10.1016/s0968-0004(00)01602-9. [DOI] [PubMed] [Google Scholar]
- Eramian D, Shen MY, Devos D, Melo F, Sali A, Marti-Renom MA. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Borron JC, Solano F. Molecular anatomy of tyrosinase and its related proteins: beyond the histidine-bound metal catalytic center. Pigment Cell Res. 2002;15:162–173. doi: 10.1034/j.1600-0749.2002.02012.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Carreno FL, Cota K, Navarrete Del Toro MA. Phenoloxidase activity of hemocyanin in whiteleg shrimp Penaeus vannamei: conversion, characterization of catalytic properties, and role in postmortem melanosis. J Agric Food Chem. 2008;56:6454–6459. doi: 10.1021/jf800839x. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Hartmann H, Decker H. All hierarchical levels are involved in conformat ional transitions of the 4 x 6-meric tarantula hemocyanin upon oxygenation. Biochim Biophys Acta. 2002;1601:132–137. doi: 10.1016/s1570-9639(02)00459-4. [DOI] [PubMed] [Google Scholar]
- Hartmann H, Lohkamp B, Hellmann N, Decker H. The allosteric effector l-lactate induces a conformational change of 2x6-meric lobster hemocyanin in the oxy state as revealed by small angle x-ray scattering. J Biol Chem. 2001;276:19954–19958. doi: 10.1074/jbc.M010435200. [DOI] [PubMed] [Google Scholar]
- Hazes B, Magnus KA, Bonaventura C, Bonaventura J, Dauter Z, Kalk KH, Holx WG. Crystal structure of deoxygenated Limulus polyphemus subunit II hemocyanin at 2.18 A resolution: clues for a mechanism for allosteric regulation. Protein Sci. 1993;2:597–619. doi: 10.1002/pro.5560020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Moffat JK. The difference Fourier technique in protein crystallography: errors and their treatment. Acta Cryst. 1971;B27:1414–1420. [Google Scholar]
- Jaenicke E, Decker H. Kinetic properties of catecholoxidase activity of tarantula hemocyanin. Febs J. 2008;275:1518–1528. doi: 10.1111/j.1742-4658.2008.06311.x. [DOI] [PubMed] [Google Scholar]
- Kanade SR, Paul B, Rao AG, Gowda LR. The conformational state of polyphenol oxidase from field bean (Dolichos lablab) upon SDS and acid-pH activation. Biochem J. 2006;395:551–562. doi: 10.1042/BJ20051509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Eicken C, Sacchettini JC, Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee BL, Soderhall K. Processing of crayfish hemocyanin subunits into phenoloxidase. Biochem Biophys Res Commun. 2004;322:490–496. doi: 10.1016/j.bbrc.2004.07.145. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtkex SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure. 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Magnus KA, Hazes B, Ton-That H, Bonaventura C, Bonaventura J, Hol WG. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- Markl J, Decker H. Molecular structure of the arthropod hemocyanins. Advances in Comparative and Environmental Physiology. 1992;13:325–376. [Google Scholar]
- Markl J, Gebauer W, Runzler R, Avissar I. Immunological correspondence between arthropod hemocyanin subunits. I. Scorpion (Leiurus, Androctonus) and spider (Eurypelma, Cupiennius) hemocyanin. Hoppe Seylers Z Physiol Chem. 1984;365:619–631. doi: 10.1515/bchm2.1984.365.1.619. [DOI] [PubMed] [Google Scholar]
- Martin AG, Depoix F, Stohr M, Meissner U, Hagner-Holler S, Hammouti K, Burmester T, Heyd J, Wriggers W, Markl J. Limulus polyphemus hemocyanin: 10 A cryo-EM structure, sequence analysis, molecular modelling and rigid-body fitting reveal the interfaces between the eight hexamers. J Mol Biol. 2007;366:1332–1350.25. doi: 10.1016/j.jmb.2006.11.075. [DOI] [PubMed] [Google Scholar]
- Marusek CM, Trobaugh NM, Flurkey WH, Inlow JK. Comparative analysis of polyphenol oxidase from plant and fungal species. J Inorg Biochem. 2006;100:108–123. doi: 10.1016/j.jinorgbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- Moore BM, Flurkey WH. Sodium dodecyl sulfate activation of a plant polyphenoloxidase. Effect of sodium dodecyl sulfate on enzymatic and physical characteristics of purified broad bean polyphenoloxidase. J Biol Chem. 1990;265:4982–4988. [PubMed] [Google Scholar]
- Nagai T, Osaki T, Kawabata S. Functional conversion of hemocyanin to phenoloxidase by horseshoe crab antimicrobial peptides. J Biol Chem. 2001;276:27166–27170. doi: 10.1074/jbc.M102596200. [DOI] [PubMed] [Google Scholar]
- Nillius D, Jaenicke E, Decker H. Switch between tyrosinase and catecholoxidase activity of scorpion hemocyanin by allosteric effectors. FEBS Lett. 2008;582:749–754. doi: 10.1016/j.febslet.2008.01.056. [DOI] [PubMed] [Google Scholar]
- Oetting WS. The tyrosinase gene and oculocutaneous albinism type 1 (OCA1): A model for understanding the molecular biology of melanin formation. Pigment Cell Res. 2000;13:320–325. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pless DD, Aguilar MB, Falcon A, Lozano-Alvarez E, Heimer de la Cotera EP. Latent phenoloxidase activity and N-terminal amino acid sequence of hemocyanin from Bathynomus giganteus, a primitive crustacean. Arch Biochem Biophys. 2003;409:402–410. doi: 10.1016/s0003-9861(02)00615-x. [DOI] [PubMed] [Google Scholar]
- Robb DA, Mapson LW, Swain T. Activation of the Latent Tyrosinase of Broad Bean. Nature. 1964;201:503–504. [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Salvato B, Santamaria M, Beltramini M, Alzuet G, Casella L. The enzymatic properties of Octopus vulgaris hemocyanin: o-diphenol oxidase activity. Biochemistry. 1998;37:14065–14077. doi: 10.1021/bi980879j. [DOI] [PubMed] [Google Scholar]
- Schweikardt T, Olivares C, Solano F, Jaenicke E, Garcia-Borron JC, Decker H. A three-dimensional model of mammalian tyrosinase active site accounting for loss of function mutations. Pigment Cell Res. 2007;20:394–401. doi: 10.1111/j.1600-0749.2007.00405.x. [DOI] [PubMed] [Google Scholar]
- Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui NI, Akosung RF, Gielens C. Location of intrinsic and inducible phenoloxidase activity in molluscan hemocyanin. Biochem Biophys Res Commun. 2006;348:1138–1144. doi: 10.1016/j.bbrc.2006.07.172. [DOI] [PubMed] [Google Scholar]
- Simonova M, Wall A, Weissleder R, Bogdanov A., Jr Tyrosinase mutants are capable of prodrug activation in transfected nonmelanotic cells. Cancer Res. 2000;60:6656–6662. [PubMed] [Google Scholar]
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper Oxidases and Oxygenases. Chem Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Sugumaranx M. Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigment Cell Res. 2002;15:2–9. doi: 10.1034/j.1600-0749.2002.00056.x. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- van Bruggen EFJ, Bijlholt MMC, Schutter WG, Wicherjes T, Bonaventura J, Bonaventura C, et al. The role of structurally diverse subunits in the assembly of three cheliceratan hemocyanins. FEBS Letters. 1980;116:207–210. [Google Scholar]
- van Gelder CW, Flurkey WH, Wichers HJ. Sequence and structural features of plant and fungal tyrosinases. Phytochemistry. 1997;45:1309–1323. doi: 10.1016/s0031-9422(97)00186-6. [DOI] [PubMed] [Google Scholar]
- van Heel M, Dube P. Quaternary structure of mult ihexameric arthropod hemocyanins. Micron. 1994;25:387–418. [Google Scholar]
- van Holde KE, Miller KI, Decker H. Hemocyanins and invertebrate evolution. J Biol Chem. 2001;276:15563–15566. doi: 10.1074/jbc.R100010200. [DOI] [PubMed] [Google Scholar]
- Woolford D, Ericksson G, Rothnagel R, Muller D, Landsberg MJ, Pantelic RS, McDowall A, Pailthorpe B, Young PR, Hankamer B, Banks J. SwarmPS: rapid, semi-automated single particle selection software. J Struct Biol. 2007;157:174–188. doi: 10.1016/j.jsb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Birmanns S. Using situs for flexible and rigid-body fitting of multiresolution single-molecule datag. J Struct Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include five additional figures, two tables and three movies are available online at ***. The cryo-EM density maps and corresponding models are available from the EMDB and PDB. Their accession numbers are EMD-5100 and PDB ID 3IXV (for the Hc resting state) and EMD-5101 and PDB ID 3IXW (for the activated state), respectively.
Structural organization of the 24-meric Hc, exemplified by the resting state Hc cryo-EM map with different subunits in different colors. The representative views of the 24-mer, dodecamer, hexamer and monomer are illustrated. The B2 subunit map together with the corresponding pseudo atomic model are shown in the last portion of the movie, with the well resolved loop region in the cryo-EM map (not resolved in the crystal template structure 1NOL, discussed in the text) highlighted by ellipsoid.
Conformational change of Hc induced by SDS activation, illustrated by the Hc density maps as well as the pseudo atomic models. Pseudo atomic model morph between the two states clearly elucidates that the 24-oligomeric Hc complex undergoes a tertiary to collective quaternary conformational change upon SDS activation. There is a slight decrease in distance (~4 Å) between the two dodecamers due to SDS activation as illustrated during time period 0:08~0:17.
Averaged Hc monomer Cα model morph between the resting and the activated states. The most mobile domain I is illustrated in deep pink, while the other two domains in yellow and light green. The cyan and magenta spheres demonstrate the putative locations of the Cu and oxygen atoms forming the active site, and the blue sphere illustrates the Cα of the placeholder residue PHE49. This movie clearly demonstrates that Hc domain I has a more pronounced movement than domains II and III; more importantly, its moving direction is to twist away from the other two domains leaving the active site more exposed.