Abstract
Insects defend themselves against infectious microorganisms by synthesizing potent antimicrobial peptides. Drosophila has appeared in recent years as a favorable model to study this innate host defense. A genetic analysis of the regulation of the antifungal peptide drosomycin has demonstrated a key role for the transmembrane receptor Toll, which prompted the search for mammalian homologs. Two of these, Toll-like receptor (TLR)2 and TLR4, recently were shown to play a critical role in innate immunity against bacteria. Here we describe six additional Toll-related genes (Toll-3 to Toll-8) in Drosophila in addition to 18-wheeler. Two of these genes, Toll-3 and Toll-4, are expressed at a low level. Toll-6, -7, and -8, on the other hand, are expressed at high levels during embryogenesis and molting, suggesting that, like Toll and 18w, they perform developmental functions. Finally, Toll-5 is expressed only in larvae and adults. By using chimeric constructs, we have tested the capacity of the signaling Toll/IL-1R homology domains of these receptors to activate antimicrobial peptide promoters and found that only Toll and Toll-5 can activate the drosomycin promoter in transfected cells, thus demonstrating specificity at the level of the Toll/IL-1R homology domain. In contrast, none of these constructs activated antibacterial peptide promoters, suggesting that Toll-related receptors are not involved in the regulation of antibacterial peptide expression. This result was independently confirmed by the demonstration that a dominant-negative version of the kinase Pelle can block induction of drosomycin by the cytokine Spaetzle, but does not affect induction of the antibacterial peptide attacin by lipopolysaccharide.
The insect host defense involves synthesis of antimicrobial peptides by the fat body and secretion into the hemolymph (1). Seven distinct antimicrobial peptides (plus isoforms) have been described in Drosophila. Interestingly, they appear to have distinct target specificities, and induction of the expression of the various peptides was found to depend on the type of challenging infectious agent. Fungal infection, for instance, results in a strong induction of the antifungal peptide drosomycin, whereas the antibacterial peptides drosocin and diptericin are only weakly induced (2). Conversely, a challenge with Gram-negative bacteria strongly induces the antibacterial peptide genes, but has a less marked effect on drosomycin expression (2). These data indicate that Drosophila can discriminate between various groups of microorganisms and mount a somewhat adapted response. A genetic analysis further revealed that the expression of the antifungal and antibacterial peptides was controlled by distinct intracellular signaling cascades, albeit with some crosstalk between these cascades. Indeed, the spaetzle/Toll/tube/pelle/cactus gene cassette controls the expression of the antifungal peptide drosomycin (3). Spaetzle codes for a secreted protein of the cysteine-knot family of growth factors, which is activated by proteolytic cleavage (4, 5). Processed Spaetzle is thought to bind to and activate the transmembrane receptor Toll, although direct interaction between the two proteins has not been reported to date. Toll activation then is transduced through the adapter molecule Tube and the Ser/Thr kinase Pelle, and this leads to the phosphorylation and subsequent degradation of the inhibitor Cactus (6). Cactus degradation frees Dif (a member of the Rel family of transcription factors), which translocates to the nucleus where it is thought to bind to and activate the drosomycin promoter (7–9). The same genetic analysis revealed that expression of the antibacterial peptides drosocin and diptericin is independent of Toll.
The discovery of the key role played by Toll in the Drosophila host defense led in 1997 to the description of the first mammalian Toll homolog, now referred to as Toll-like receptor 4 (TLR4) (10), and was followed over the past 2 years by the description of several additional homologs (11–13). The most conserved region between these molecules and Drosophila Toll consists of a 150-aa intracytoplasmic domain that is also shared by members of the IL-1R family and the signal transducer MyD88 (11), and which is named the Toll/IL-1R homology (TIR) domain. In addition, mammalian TLRs share with Toll extracellular leucine-rich repeats flanked by characteristic cysteine-rich regions, whereas ectodomains from IL-1R-related molecules are characterized by three Ig domains. Recent genetic data have implicated two of these receptors, TLR2 and TLR4, in the response to bacterial infections. Importantly, the tlr4 gene was shown to be defective in the C3H/HeJ and C57BL:10ScCR strains of mice (lps locus), which have impaired response to lipopolysaccharide (LPS) derived from Gram-negative bacterial cell wall (14, 15). More recently, TLR2 was shown to play a parallel role in response to peptidoglycan derived from the Gram-positive bacterial cell wall (16). Altogether, these results implicate mammalian Toll homologs in antibacterial immunity and suggest that in Drosophila Toll homologs or Toll itself may function as signaling receptors in response to bacterial infection. We have searched for TIR domain-containing molecules in Drosophila, and we report here the identification of six Toll-related genes (Toll-3 to -8) in addition to Toll and its previously described homolog 18-wheeler (18w) (17).
Materials and Methods
Computational Sequence Analysis.
Toll-related genes were identified as genomic or expressed sequence tag clones sequenced by the Berkeley Drosophila Genome Project (http://www.fruitfly.org/) by using the blast program (18). The TIR domain from Toll, IL-1R, or MyD88 served as query sequences. A genomic clone containing partial sequences from Toll-3 had been reported under the name of MstProx because the gene is adjacent to the Mst84D locus (19). Similarities between the short genomic tag STSDm2245, corresponding to Toll-4, and TIR domains had been noticed by the same authors. Rapid amplification of cDNA ends was performed with the Marathon kit (CLONTECH) to obtain incomplete cDNA sequences. Toll-5 and Toll-8 were both initially identified as genomic clones sequenced by Berkeley Drosophila Genome Project and have been recently described under the names of tehao (GenBank accession no. AF140019) (20) and Tollo (GenBank accession no. AF204158), respectively. Toll-6 and Toll-7 were identified as expressed sequence tag clones (LD08841 and LD03945, respectively) sequenced by the Berkeley Drosophila Genome Project, and the 5′ end of the ORF from both genes was identified in genomic clones sequenced by Celera (Rockville, MD) and cloned by reverse transcription–PCR (RT-PCR). Sequencing of RT-PCR products revealed that like Toll, Toll-3 and Toll-5 contain one intron in the coding sequences, whereas 18w, Toll-6, Toll-7, and Toll-8 do not. Signal peptides were identified in all Toll-related proteins for which complete cDNAs were obtained and sequenced by using the SignalP World Wide Web server (http://www.cbs.dtu.dk/services/SignalP/). Sequences were aligned by using the clustal method in the lasergene 99 package (DNAstar, Madison, WI). LRR motifs as well as C- and N-flanking motifs were identified by using the pfam database (http://www.sanger.ac.uk/Software/Pfam/).
Northern Blot and RT-PCR Analysis.
RNA was isolated with Trizol reagent (GIBCO/BRL) according to the manufacturer's instructions. Poly(A)+ RNA was prepared by using oligo(dT)-coupled beads (Oligotex, Qiagen, Chatsworth, CA), and analyzed by Northern blot with standard procedures (21). All probes specifically reacted with the different Toll-related molecules, as shown by the different sizes of the transcripts. The primers used for RT-PCR analysis correspond to nucleotides 51–71 and 392–370 of Toll-3 (GenBank accession no. AF247769) and nucleotides 1527–1545 and 2006–1988 of Toll-4 (GenBank accession no. AF247768). RT-PCR was performed as described (22), and the PCR bands were sequenced to ensure that they correspond to the expected cDNAs.
Construction of Reporter Plasmids and Expression Vectors.
Reporter constructs are based on the pGL3 vector (Promega) and were constructed by using standard methods (21). The diptericin (23), drosomycin (24), drosocin (25), defensin (26), and cecropin (27) promoters have been described. The attacin promoter (J.-L.I., unpublished results) was inserted in pGL3 as a 2.4-kb fragment (GenBank accession no. AE003813; nucleotides 149,339–146,924).
All expression vectors are based on pPAC (actin 5C promoter and polyadenylation signal) (28). The TollΔLRR allele was constructed by PCR exactly as described (29), except that the Toll signal peptide was replaced by the preprotrypsin leader fused to the Flag epitope derived from the expression vector pFLAG-CMV-1 (Eastman Kodak). The unique BamHI site created at the deletion site was used to fuse the extracytoplasmic domain of TollΔLRR to the transmembrane and intracytoplasmic domains of 18w (GenBank accession no. S76155; nucleotides 3296–5417); Toll-3 (GenBank accession no. AF247769; nucleotides 1778–2359); Toll-4 (GenBank accession no. AF247768; nucleotides 1818–2432); Toll-5 (GenBank accession no. AF247767; nucleotides 1768–2388); Toll-6 (GenBank accession no. AF247766; nucleotides 3118–4545); Toll-7 (GenBank accession no. AF247765; nucleotides 3094–4340); or Toll-8 (GenBank accession no. AF247764; nucleotides 2941–4041). The dominant-negative Pelle construct was created by deletion of the catalytic domain and was a gift of R. Medzhitov (Yale University, New Haven, CT). The modified spaetzle cDNA in which sequences coding for the signal peptide are fused to the sequences encoding the carboxyl terminus 106 aa of the protein was a gift of K. Anderson (Sloan–Kettering Institute, New York) (30). All PCR fragments were sequenced. Details about the constructions can be obtained on request.
Transfection Assays and Western Blot Analysis.
S2 cells were purchased from Invitrogen and grown to 80% confluence at 25°C in Schneider's medium (Sigma) supplemented with 10% FCS/105 units/liter penicillin/100 mg/liter streptomycin. Cells were transfected in 6-cm diameter dishes by the calcium phosphate precipitation technique with 0.1 μg of reporter plasmid, 0.1 μg of β-galactosidase expression vector pACH110 (31), and 1 μg of expression vector. After 48 h, cells were lysed in reporter lysis buffer and luciferase activity was measured in a luminometer (BCL Book, Promega) immediately after addition of the substrate (luciferin, Promega). β-galactosidase activity in the cell lysates was measured by using O-nitrophenyl-β-d-galactoside as substrate, and the values were used to normalize variability in the efficiency of transfection. LPS (E. coli serotype 055:B5; Sigma) was added onto cells at 10 μg/ml for 16 h. For Western blot analysis of protein expression, cells were washed in PBS and resuspended in Laemmli buffer, and proteins were separated by 7.5% denaturing SDS/PAGE. After migration, proteins were electroblotted onto a nitrocellulose filter. The membrane was probed with the monoclonal FLAG-M2 antibody (Sigma) in Tris-buffered saline-Tween 0.05% (21). The second antibody was a donkey anti-mouse horseradish peroxidase conjugate (Dako), and the chimeric proteins were detected with enhanced chemiluminescence, as recommended by the manufacturer (Amersham Pharmacia).
Results
A Family of Toll Receptors in Drosophila.
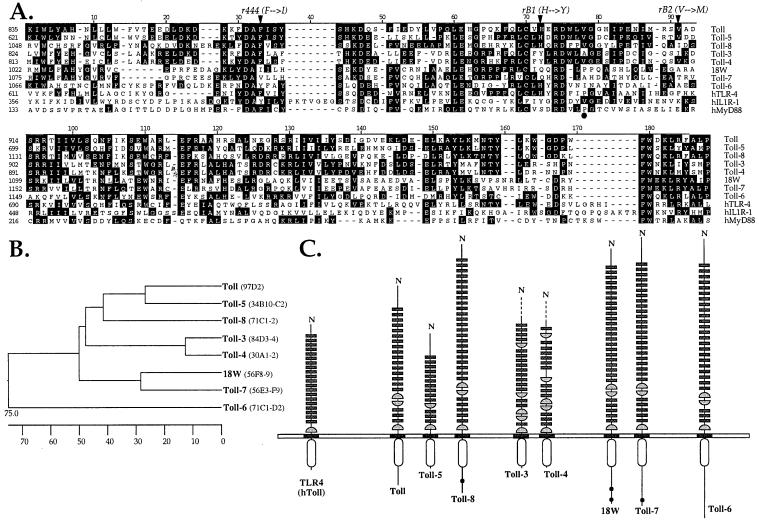
We have searched for Toll-related receptors in the genome of Drosophila melanogaster. Because the TIR domain is the most conserved region between Toll and its mammalian homologs, and it is shared by other molecules critical for the control of innate immunity such as MyD88 and the IL-1 and IL-18 receptors, we have looked for gene sequences encoding TIR domains in the Drosophila genome by using the blast program (18). This search revealed the existence of six genes encoding receptors containing a TIR domain, in addition to Toll and 18w. We will refer to the proteins encoded by these genes as Toll-3 to -8. Alignment of the TIR domains of the Drosophila molecules with the mammalian type I IL-1R, the human Toll homolog TLR4 and MyD88 shows a high degree of conservation throughout a stretch of ≈150 aa, with many identical residues in all sequences (Fig. 1A). The eight Drosophila Toll-related proteins share higher similarity between them than with any mammalian TLR, suggesting that these two groups of proteins have evolved independently (11, 20). The Drosophila Tolls fall into several subsets, as illustrated in Fig. 1B. With regard to the TIR domains, Toll-3 and Toll-4 share 79% identity, Toll and Toll-5 have 60% identity, as is also the case for 18W and Toll-7. Toll-6 is less related to the other seven Drosophila Tolls.
Figure 1.
The Drosophila Toll family. (A) Alignment of the TIR domains from Drosophila Toll-related molecules and human TLR4, IL-1 receptor type I (IL-1RI), and MyD88. Sequences were aligned by using the clustal method. Conserved residues are shaded in black. Residues affected in the loss-of-function r444, rB1, and rB2 alleles of Toll are indicated by arrowheads (46). The black dot points to the position of the Pro residue critical for TLR4 signaling. (B) Phylogenetic relationship between Drosophila Toll molecules based on the alignment shown in A and generated by the neighbor-joining method. The scale beneath the tree measures the distance between the sequences. The chromosomal localization of the Toll-related genes is indicated (Right). (C) Schematic representation of domain organization of Drosophila Toll-related molecules and human TLR4. The receptors are grouped according to conservation of their TIR domains. The leucine-rich repeats are indicated by small rectangles, whereas cysteine-rich carboxyl-flanking motifs and cysteine-rich amino-flanking motifs are represented by half-circles. Black dots indicate polyglutamine stretches. Incomplete sequences of Toll-3 and Toll-4 are indicated by dashed lines.
Examination of the full amino acid sequence of the mammalian and insect TIR-containing proteins reveals that they all contain a putative transmembrane domain and an extracellular domain composed of leucine-rich repeats (LRRs), flanked by characteristic cysteine-rich motifs (32). However, the arrangement of these Cys motifs differ between mammalian and Drosophila Tolls (Fig. 1C). Whereas the mammalian TLRs described to date only contain a membrane-proximal cysteine-rich flanking motif at the C-terminal end of the LRRs, Drosophila Tolls contain C- and N-flanking cysteine-rich motifs. The extracellular regions of Drosophila Toll-related molecules are less conserved than the TIR domains, and the arrangement of the C- and N-flanking motifs is different, suggesting that some of these receptors may activate common targets in response to different signals.
Finally, the TIRs of Toll, 18W, and Toll-6, -7, -8 are followed by a C-terminal extension. These extensions do not present any obvious motif that could hint to a function, with the exception of polyglutamine stretches in 18W, Toll-7, and Toll-8. In the other Tolls (Toll-3 to Toll-5), a stop codon is present a few residues after the TIR domain, as in mammalian TLRs and members of the IL-1R family.
Differential Expression of Toll-Related Molecules in Drosophila.
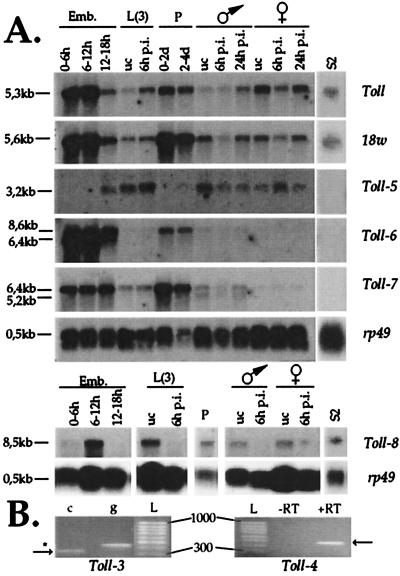
We next examined the expression of all Toll family genes during Drosophila development and in immune-challenged larvae and adults. Toll-6 to Toll-8 were found to be expressed at high levels in embryos, but also in pupae and at a lower level in larvae and adults (Fig. 2A). Toll and 18w are also expressed at high levels in embryos and pupae. In apparent contradiction to the previous report of Williams et al. (33), but in agreement with the initial papers of Eldon et al. (17) and Chiang and Beachy (34), we observed a single transcript of 5.6 kb for 18w at all developmental stages. For Toll-6, two transcripts (8.6 and 6.4 kb) were detected of which only the larger was present in pupae, larvae, and adults. For Toll-7, a major transcript (6.4 kb) was observed in embryos, larvae, and pupae and an additional transcript was present in adult males (5.2 kb). Toll-5 was expressed in larvae and adults, but not in the early embryo or pupae, in contrast to the other Toll genes. Finally, no signals could be observed in Northern blots for Toll-3 and Toll-4. Expression of these two genes could, however, be detected by RT-PCR, indicating that these genes are expressed at low levels or in a limited number of cells (Fig. 2B).
Figure 2.
Northern blot and RT-PCR analysis of Toll-related gene expression. (A) Poly(A)+ RNA extracted from embryos (Emb.), third-instar larvae [L(3)], pupae (P, 0–2 or 2–4 days), adult male or female Drosophila, and the cell line S2 were submitted to Northern blot analysis and hybridized to probes derived from Toll, 18w, and Toll-3 to Toll-8 gene sequences. No signal could be observed for the Toll-3 and Toll-4 probes. A probe recognizing RNA coding for ribosomal protein 49 (rp49) was used to ensure that comparable amounts of RNA were loaded in all lanes. uc, unchallenged; p.i., postinfection with a mixture of Gram-negative and Gram-positive bacteria; and d, days. (B) RT-PCR analysis of Toll-3 and Toll-4 expression. Primers specific for the Toll-3 gene were designed to flank a 104-bp intron sequence in the genomic DNA, resulting in the amplification of a 342-bp cDNA (c)-derived fragment (arrow) or a 446-bp genomic DNA (g)-derived fragment (*). Primers for the Toll-4 gene were designed to amplify a 480-bp intronless fragment. In this case, RT was omitted in a control reaction to ensure that the amplified band is derived from RNA. cDNA was prepared from mRNA derived from pupae (Toll-3) or third-instar larvae (Toll-4). L, 100-bp ladder.
In immune-challenged larvae and adults, we observed that Toll expression was noticeably up-regulated (2- to 5-fold), as shown (3). In contrast to Toll, the expression of all of the other Toll-related genes, including 18w, was only modestly up-regulated in larvae (1.5- to 2-fold) and no up-regulation was detectable in adults. LPS has been reported to induce the expression of the genes encoding the antimicrobial peptides in the macrophage-like Schneider and l(2)mbn cell lines (see ref. 22 for references), indicating that these cells express functional LPS receptors. We therefore examined expression of Toll-related genes in these cell lines. As illustrated in Fig. 2, Northern blot analysis reveals that S2 cells express Toll, 18w, and Toll-8. Similar results were observed with l(2)mbn cells (data not shown).
Induction of Antimicrobial Peptides by Toll-Related Receptors.
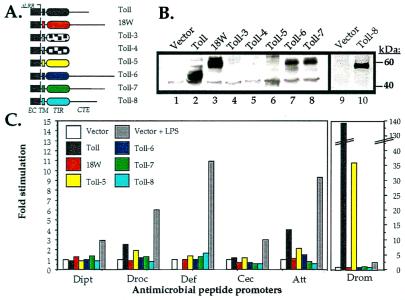
Genetic studies have shown that in a Toll gain-of-function mutant, the gene encoding drosomycin is expressed constitutively whereas expression of the antibacterial peptide genes is not affected (3). Deletion of the leucine-rich repeats of the Toll ectodomain results in a constitutively active receptor (TollΔLRR) (29). We have transfected S2 cells with an expression vector encoding TollΔLRR and observed a strong (more than 100-fold) activation of the drosomycin promoter (Fig. 3C), resulting in a 5- to 10-fold higher level of expression of luciferase than after transfection with a wild-type Toll construct (data not shown). Significantly in these experiments, transfection of TollΔLRR did not activate expression of diptericin, defensin, and cecropin, and only weakly activated the drosocin and attacin promoters (Fig. 3C). To investigate whether the various Toll family members can activate antimicrobial peptide gene expression, we have constructed recombinant chimerae in which the truncated ectodomain from TollΔLRR was fused to the transmembrane and intracytoplasmic domains of 18W and Toll-3 to -8 (Fig. 3A). A Flag epitope was inserted immediately after the signal peptide to monitor expression of the recombinant proteins. As shown in Fig. 3B, the dominant Toll allele and the 18w, Toll-6, Toll-7, and Toll-8 chimeric alleles were efficiently expressed whereas only weak expression of Toll-5, and no expression of the Toll-3 or Toll-4 chimerae was detected. Interestingly, the chimerae that were poorly or not expressed correspond to those Toll proteins that lack a C-terminal extension after the TIR domain (Fig. 1C).
Figure 3.
Induction of antimicrobial peptide promoters by chimeric Toll receptors. (A) Schematic representation of the chimeric molecules. The constructs are based on a constitutively active version of Toll in which all LRR motifs have been deleted. The truncated ectodomain of Toll (black rectangle) is fused to the transmembrane and intracytoplasmic domains of the Toll-related molecules. EC, ectodomain; TM, transmembrane domain; CTE, C-terminal extension. (B) Expression of the chimeric proteins in transfected cells. Protein extracts prepared from S2 cells transfected by expression vectors either empty (Vector) or expressing TollΔLRR (Toll) or the various chimerae (18W, Toll-3 to -8) were submitted to Western blot analysis with an anti-FLAG mAb. Expected sizes for the various molecules are as follows: Toll, 50 kDa; 18W, 63 kDa; Toll-3, 40 kDa; Toll-4, 41 kDa; Toll-5, 41 kDa; Toll-6, 69 kDa; Toll-7, 63 kDa; and Toll-8, 57 kDa. (C) Induction of antimicrobial peptide promoters in transfected cells. S2 cells were cotransfected with 1 μg of expression vector either empty or expressing TollΔLRR (Toll) or the various chimerae (18W, Toll-5 to -8) and 0.1 μg of reporter plasmid encoding luciferase under the control of the diptericin (Dipt), drosocin (Droc), defensin (Def), cecropin (Cec), attacin (Att), or drosomycin (Drom) promoters. Transfections were repeated twice in S2 cells and l(2)mbn cells with identical results. A representative experiment is shown.
The data presented in Fig. 3C show that only the Toll-5 chimeric protein efficiently induced the drosomycin promoter. In contrast, neither the 18W nor the Toll-6 to Toll-8 chimerae were able to induce drosomycin, despite the fact that they contain a conserved TIR domain and are expressed at high levels. Significantly, none of the chimeric proteins induced expression of the antibacterial peptide genes, although they all can be induced by treatment with LPS. Similar results were obtained when the chimeric Tolls were transfected into l(2)mbn cells (data not shown).
The Toll Pathway Mediates Response to the Cytokine Spaetzle but Not to LPS in Drosophila S2 Cells.
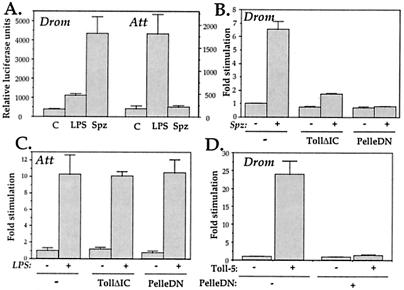
Genetic evidence in Drosophila larvae and adults indicates that Toll is activated by a processed form of the secreted protein Spaetzle. This is intriguing in light of the genetic data in mice showing that the Toll homolog TLR4 is activated by LPS. We have addressed here the well-defined S2 cell line of Drosophila to compare possible inductions of antimicrobial peptide genes by Spaetzle and LPS. As illustrated in Fig.4A, the processed form of Spaetzle markedly induced expression of drosomycin, but only minimally that of a selected antibacterial peptide, attacin. Conversely, LPS had a strong effect on attacin expression, but poorly induced the drosomycin reporter gene. Interestingly, when dominant-negative constructs of the Toll pathway (Toll deleted of its intracytoplasmic domain: TollΔIC; and Pelle deleted of its kinase domain: PelleDN) were cotransfected, only the Spaetzle-induced expression of drosomycin was affected (Fig. 4B), whereas attacin induction by LPS was not modified (Fig. 4C), indicating that the Toll pathway selectively controls the antifungal response in Drosophila cell lines and is not involved in LPS response. Coexpression of PelleDN also completely abolished induction of the drosomycin promoter by Toll-5, indicating that Toll-5 and Toll use similar signaling components (Fig. 4D).
Figure 4.
Activation of antimicrobial peptide promoters by Spaetzle and LPS in S2 cells. (A) S2 cells were cotransfected with 1 μg of either an empty expression vector or a vector expressing the carboxyl-terminal 106 aa of Spaetzle (Spz) (4, 30) and 0.1 μg of reporter constructs expressing luciferase under the control of the drosomycin (Drom) or attacin (Att) promoters. Cells transfected with the empty expression vector were either left untreated or exposed to LPS (10 μg/ml) for 16 h before harvesting and determination of luciferase activity in cell extracts. (B) S2 cells were cotransfected with 0.1 μg of drosomycin-luciferase reporter construct and 0.5 μg of expression vector encoding the C106 processed form of Spaetzle (Spz) together with 0.5 μg of expression vector empty or encoding a truncated Toll version deleted of its intracytoplasmic domain (TollΔIC) or a mutant Pelle version deleted of its kinase domain (PelleDN). (C) S2 cells were cotransfected with 0.1 μg of attacin-luciferase reporter construct and 1 μg of expression vector empty or encoding TollΔIC or PelleDN. Twenty-four hours after transfection, cells were either left untreated or exposed to LPS (10 μg/ml) for 16 h before harvesting and determination of luciferase activity in cell extracts. (D) S2 cells were cotransfected with 0.1 μg of drosomycin-luciferase reporter plasmid and 0.5 μg of expression vector encoding the TollΔLRR-Toll5 chimeric protein together with 0.5 μg of expression vector either empty or encoding PelleDN. All transfections were done in triplicate, and results represent means ± SD.
Discussion
The Drosophila Toll Gene Family.
We describe here a family of transmembrane molecules related to Toll. This family comprises eight members. In the human, six TLRs have been reported to date and the estimate is that the mammalian genome encodes about 20 different Toll-like receptors (10–13). Several plant disease-resistance genes presenting sequence similarities to Toll also have been cloned. Interestingly, some of these genes are found in complex loci containing arrays of related genes, a situation that is thought to favor meiotic instability and generation of new alleles with novel binding specificity for pathogen-derived ligands (ref. 35 and references therein). In Drosophila, the eight Toll genes are disseminated on chromosomes 2 and 3 (with the exception of 18w and Toll-7, which both map to 56E–F, and Toll-6 and Toll-8, which map to 71C–D), which is evocative of the situation observed for mammalian TLRs that are dispersed on several chromosomes (11).
Analysis of the Drosophila genome did not show the presence of homologs of IL-1R, but revealed a gene encoding a putative MyD88-related gene (gene CG2078 in Flybase Gadfly; http://flybase.bio.indiana.edu:82/). In contrast, the Caenorhabditis elegans genome appears to encode one Toll-related gene but no IL-1R- or MyD88-related molecules (36). Because MyD88 plays a critical role in IL-1, IL-18, and LPS signal transduction, it will be interesting to see whether this gene is involved in the Drosophila immune response and interacts with any of the Toll-related receptors described here. A Pro residue has been shown to play a critical role in TLR4 and TLR2 signaling in mammalian cells. Mutant molecules in which this Pro residue has been changed to a His are unable to trigger NF-κB activation (15, 37). Interestingly, whereas most mammalian Toll and IL-1R-related receptors harbor a Pro at this position, only 18W in Drosophila presents this conserved Pro. Another difference between Drosophila Toll-related proteins and their mammalian counterparts is the presence of an intracytoplasmic C-terminal extension after the TIR domain. Interestingly, we observed a clear correlation between the presence of these sequences and the level of expression of the chimeric Toll constructs (Fig. 3B), suggesting that these sequences may be necessary for the stability of the proteins.
Function of Toll-Related Receptors in Drosophila.
Toll and 18w were first described for their developmental functions. Toll plays a critical role in the dorsoventral axis formation of the Drosophila embryo by activating the Rel protein Dorsal (38). Later in development, Toll is expressed in a complex spatial and temporal pattern in many tissues (39), and most of Toll−/− zygotes die as first or second instar larvae. Like zygotic Toll, 18w is expressed in many tissues, often in cells undergoing extensive migration (17, 34), and about 90% of 18w mutant larvae die as second or third instar larvae, without any obvious major defect. The high level of expression of Toll-6 to -8 in embryo and pupae suggests that the proteins they encode also may be involved in development, and like Toll and 18w, function as adhesion molecules (17, 40). Alternatively, these transmembrane proteins may be activated by a secreted factor, such as, for example, a Spaetzle-related molecule.
In mammals, several molecules containing TIR domains have been shown to activate immune-inducible genes through the transcription factor NF-κB. In Drosophila, Toll activates the Rel proteins Dorsal during embryonic development and Dif during immune response in adults (7–9, 38). In addition, inducible expression of the attacin gene has been shown to be selectively abrogated in 18w-deficient larvae, suggesting that this Toll-related receptor also controls activation of Drosophila Rel proteins (33). By using chimeric constructs, we have found that only the TIR domains from Toll and Toll-5 can activate the drosomycin promoter in S2 cells. This demonstrates a specificity at the level of the TIR domain. Interestingly, drosomycin recently has been shown to be inducible in a Toll-independent manner in the tracheae of Drosophila (24), suggesting that Toll-5 may be involved in the control of this local immune response. In contrast to drosomycin, none of the antibacterial peptide gene promoters could be activated by the chimerae in this transfection assay. In our opinion, this negative result cannot be attributed to improper synthesis or folding of the chimeric receptors for the following reasons: (i) all constructs yielded detectable protein expression, with the exception of the Toll-3 and Toll-4 chimerae; (ii) the Toll-5 chimera was able to strongly activate the drosomycin promoter, although its level of expression was much weaker than that of the other proteins; and (iii) a chimera in which the truncated ectodomain from 18W was fused to the transmembrane and intracytoplasmic domain of Toll was able to activate the drosomycin promoter (data not shown). Because attacin induction is severely impaired in 18w loss-of-function mutants (33), we expected that the attacin gene might be up-regulated by the chimeric TollΔLRR-18W construct. The failure to observe any up-regulation is in keeping with the data obtained with transfections of full-length 18w cDNA or mutant versions encoding 18wΔLRR or 18w10bgain-of-function alleles. These results suggest that 18w may not be sufficient for induction of attacin in S2 cells. Alternatively, some of the transducers relaying activation from the plasma membrane to the nucleus may be absent in the cell lines that we used. Because the attacin promoter can be efficiently activated in S2 and l(2)mbn cells by LPS treatment, this raises the question of the receptors and signal transduction pathways activated by LPS in these cells.
LPS Signal Transduction in Drosophila.
Although it recently has been shown that the Rel protein Relish plays a critical role in the immune inducibility of Drosophila antibacterial peptides (41), the nature of the receptors triggering antibacterial response at the plasma membrane remains unknown. Because several molecules containing TIR domains are known to activate Rel proteins, and because the mammalian Toll-like receptors, TLR2 and TLR4, are involved in antibacterial response in mice, it is tempting to speculate that the antibacterial response in Drosophila also involves Toll-related molecules. Expression of three of these, i.e., Toll, 18w, and Toll-8, can be detected by Northern blot in the macrophage-like cell lines S2 and l(2)mbn, which respond to LPS. We have shown here that overexpression of a dominant-negative version of Toll blocks induction of the drosomycin promoter by Spaetzle, but does not affect attacin promoter induction by LPS, confirming that Toll functions as a cytokine receptor and not as a pattern recognition receptor in the Drosophila host defense. In mammals, TLR4 activates the Ser/Thr kinase IRAK (IL-1 receptor associated kinase) and transfection experiments in human cells have shown that expression of dominant-negative forms of this kinase interfere with LPS signaling (42–44). We have tested the involvement of Pelle, which appears to be the sole IRAK homolog encoded in the Drosophila genome, in LPS signal transduction. Again, we observed that overexpression of a dominant-negative version of Pelle did not affect induction of the attacin promoter by LPS in transfected S2 cells, although it abolished induction of the drosomycin promoter by Spaetzle. This result extends previous genetic work indicating that pelle-deficient flies, although showing a dramatically increased susceptibility to fungal infection, have normal resistance to bacterial infection (3). Altogether, these data indicate that the Toll signaling pathway is not involved in LPS signal transduction in Drosophila S2 cells, and suggest that, unlike in mammals, Toll-related receptors may not be part of the LPS receptor complex. Genetic screens recently have allowed the identification of several mutants with a deficient antibacterial response (ref. 45; and D. Ferrandon, personal communication). Cloning of the corresponding genes will undoubtedly clarify the present picture and eventually lead to the identification of the receptors involved in host defense against bacterial infection.
Acknowledgments
We thank R.-O. Clauss and T. Thahouly for technical assistance, D. Ferrandon and J. Royet for comments on the manuscript, M. Capovilla and S. Bolshakov for chromosomal localization of Toll-6 and Toll-7, J.-M. Reichhart, R. Medzhitov, K. Anderson, and P. Simpson for providing plasmids, and our colleagues from UPR9022 for stimulating discussions and advice. This work was supported by grants from Centre National de la Recherche Scientifique (Program Biologie Cellulaire) and National Institutes of Health Grant 1PO1 AI44220–02. E.J. is supported by a grant from the Ligue contre le Cancer.
Abbreviations
- TLR
Toll-like receptor
- TIR
Toll/IL-1R homology
- LPS
lipopolysaccharide
- RT-PCR
reverse transcription–PCR
- LRR
leucine-rich repeat
Note Added in Proof.
The recent annotation of the Drosophila genome shows the presence of an additional Toll-related gene that we propose to name Toll-9 (gene CG5528 in Flybase Gadfly). This gene, which is located in 77B4-5 on the left arm of the third chromosome, is unusual in that it has two introns within the sequences coding for the TIR domain.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The cDNA sequences reported in this paper have been deposited in the GenBank database (accession nos. AF247769, Toll-3; AF247768, Toll-4; AF247767, Toll-5; AF247766, Toll-6; AF247765, Toll-7; and AF247764, Toll-8).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180130797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180130797
References
- 1.Hoffmann J, Reichhart J. Trends Cell Biol. 1997;7:309–316. doi: 10.1016/S0962-8924(97)01087-8. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Reichhart J, Hoffmann J. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemaitre B, Nicolas E, Michaut L, Reichhart J, Hoffmann J. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 4.DeLotto Y, DeLotto R. Mech Dev. 1998;72:141–148. doi: 10.1016/s0925-4773(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 5.Levashina E A, Langley E, Green C, Gubb D, Ashburner M, Hoffmann J A, Reichhart J M. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 6.Imler J, Hoffmann J. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 7.Meng X, Khanuja B S, Ip Y T. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manfruelli P, Reichhart J M, Steward R, Hoffmann J A, Lemaitre B. EMBO J. 1999;18:3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutschmann S, Jung A, Hetru C, Reichhart J, Hoffmann J, Ferrandon D. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 10.Medzhitov R, Preston-Hurlburt P, Janeway C. Nature (London) 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 11.Rock F, Hardiman G, Timans J, Kastelein R, Bazan J. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhary P M, Ferguson C, Nguyen V, Nguyen O, Massa H F, Eby M, Jasmin A, Trask B J, Hood L, Nelson P S. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 13.Takeuchi O, Kawai T, Sanjo H, Copeland N G, Gilbert D J, Jenkins N A, Takeda K, Akira S. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 15.Poltorak A, He X, Smirnova I, Liu M, Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 17.Eldon E, Kooyer S, D'Evelyn D, Duman M, Lawinger P, Botas J, Bellen H. Development (Cambridge, UK) 1994;120:885–899. doi: 10.1242/dev.120.4.885. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Mitcham J, Parnet P, Bonnert T, Garka K, Gerhart M, Slack J, Gayle M, Dower S, Sims J. J Biol Chem. 1996;271:5777–5783. doi: 10.1074/jbc.271.10.5777. [DOI] [PubMed] [Google Scholar]
- 20.Luo C, Zheng L. Immunogenetics. 2000;51:92–98. doi: 10.1007/s002510050017. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Dimarcq J, Imler J, Lanot R, Ezekowitz R, Hoffmann J, Janeway C, Lagueux M. Insect Biochem Mol Biol. 1997;27:877–886. doi: 10.1016/s0965-1748(97)00072-6. [DOI] [PubMed] [Google Scholar]
- 23.Reichhart J, Meister M, Dimarcq J, Zachary D, Hoffmann D, Ruiz C, Richards G, Hoffmann J. EMBO J. 1992;11:1469–1477. doi: 10.1002/j.1460-2075.1992.tb05191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrandon D, Jung A, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann J. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlet M, Lagueux M, Reichhart J, Hoffmann D, Braun A, Meister M. Eur J Biochem. 1996;241:699–706. doi: 10.1111/j.1432-1033.1996.00699.x. [DOI] [PubMed] [Google Scholar]
- 26.Dimarcq J, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart J, Hoffmann J. Eur J Biochem. 1994;221:201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 27.Engstrom Y, Kadayalil L, Sun S, Samakovlis C, Hultmark D, Faye I. J Mol Biol. 1993;232:327–333. doi: 10.1006/jmbi.1993.1392. [DOI] [PubMed] [Google Scholar]
- 28.Krasnow M A, Saffman E E, Kornfeld K, Hogness D S. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 29.Winans K A, Hashimoto C. Mol Biol Cell. 1995;6:587–596. doi: 10.1091/mbc.6.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morisato D, Anderson K. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- 31.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 32.Buchanan S G, Gay N J. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- 33.Williams M, Rodriguez A, Kimbrell D, Eldon E. EMBO J. 1997;16:6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang C, Beachy P A. Mech Dev. 1994;47:225–239. doi: 10.1016/0925-4773(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 35.Meyers B C, Dickerman A W, Michelmore R W, Sivaramakrishnan S, Sobral B W, Young N D. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 36.Tan M, Ausubel F. Curr Opin Immunol. 2000;3:29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 37.Underhill D M, Ozinsky A, Hajjar A M, Stevens A, Wilson C B, Bassetti M, Aderem A. Nature (London) 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 38.Belvin M P, Anderson K V. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 39.Gerttula S, Jin Y S, Anderson K V. Genetics. 1988;119:123–133. doi: 10.1093/genetics/119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keith J, Gay N. EMBO J. 1990;9:4299–4306. doi: 10.1002/j.1460-2075.1990.tb07878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedengren M, Asling B, Dushay M, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:1–20. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 42.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 43.Yang R B, Mark M R, Gurney A L, Godowski P J. J Immunol. 1999;163:639–643. [PubMed] [Google Scholar]
- 44.Zhang F X, Kirschning C J, Mancinelli R, Xu X P, Jin Y, Faure E, Mantovani A, Rothe M, Muzio M, Arditi M. J Biol Chem. 1999;274:7611–7614. doi: 10.1074/jbc.274.12.7611. [DOI] [PubMed] [Google Scholar]
- 45.Wu L P, Anderson K V. Nature (London) 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 46.Schneider D, Hudson K, Lin T, Anderson K. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]