Abstract
A synopsis of the 2007 annual progress report for the Center for Structures of Membrane Proteins, a specialized center of the Protein Structure Initiative.
Keywords: Membrane proteins; Structural biology; X-ray crystallography; NMR, 2-D crystallography
Mission of the CSMP research center (csmp.ucsf.edu)
The goal of the Center for Structures of Membrane Proteins (CSMP) is to express, purify and determine the structures of representative members of membrane protein classes, aiming for coverage of membrane proteins of novel folds, and novel functions. Nearly 30% of all eukaryotic proteins are membrane proteins, and these include protein targets for over 40% of all drugs in use today. There is little understanding of the mechanisms and atomic interactions of any one of these. This is primarily because they are membrane proteins, where preparation in a structurally homogeneous and functionally active-state, and subsequent structure determination has been extremely challenging. Where classes of membrane proteins are represented in prokaryotes, it is likely that structures for a homolog will be determined first for prokaryotic or archaeal members, so target proteins will be first sought from these organisms. Our plan proposes structure determination of membrane protein targets with two or more membrane crossings, from prokaryotic, archaeal, extremeophile, and human genomes. Many human proteins have no good homologs in prokaryotes or archaea, thus human membrane targets are also included with the goal of coverage of the folds of the membrane proteome. The human genes are the only ones from a eukaryote, selected because their structures can instruct in structure-based drug design.
Overall organization of the U54 research center
CSMP (csmp.ucsf.edu) supports an integrated program aimed at facilitating the cloning, expression purification, crystallization, and structure determination of membrane proteins with the aim of coverage of the membrane proteome. The core capabilities provide for fermentation, cell culture, gene cloning, expression, protein purification, characterization, X-ray diffraction at the Advanced Light Source (ALS) in Berkeley, structure determination by 2-D electron microscopy, and structure determination by Nuclear Magnetic Resonance techniques (NMR).
The executive council consists of the seven principal investigators. Delegated areas of responsibility include responsibility over the projects. Dr. Sali directs the effort in dynamic target selection, and also the final step in using structures determined in the Center to model the structures of homologs. Dr. Choe leads the effort to optimize expression of human membrane proteins in E. coli using the MISTIC system that he discovered. Dr. Stroud will manage the standardized effort to express bacterial and archeal membrane proteins in E. coli. Drs. Choe, Minor and Stroud will oversee the purification and characterization of proteins produced in their projects. In almost all cases, prokaryotic and human membrane protein targets are investigated through the CSMP and all the other eukaryotic membrane protein targets are investigated through MPEC. However, the funding of the MPEC center did not provide funds for crystallography or structure determination equipment or personnel. Therefore, promising eukaryotic membrane protein targets that move through the Stroud portion of the MPEC protein expression and purification pipeline to a pure, homogeneous, and stable preparation state are then incorporated into the CSMP crystallography and structure determination pipeline.
The Principal Investigators derive from four institutions, all located in California. The institutions are UCSF (Stroud, Sali, Minor, and Holton), Salk Institute (Choe, Riek, and Kwiatkowski), UCDavis (Stahlberg), and UCLA (Kaback). The PIs and members of the team meet each week in televised conference (1.00 pm–3.00 pm each Tuesday) to present progress in rotation. After 1.5 years we held a self-evaluation to identify weak projects and to plan for any redirection of effort, and resources. Each aim will now be gauged by 3-monthly milestones that will be used to adjust direction and necessary funding. This will be reviewed annually to assure direction.
The principal investigators
The investigators are in close touch with each other via an intranet, phone, and direct contacts. The institutions are UCSF (Stroud, Sali, Minor, and Holton), Salk Institute (Choe, Riek, and Kwiatkowski), UCDavis (Stahlberg), and UCLA (Kaback). Each principal investigator has played a key role in the discovery of major membrane protein biology, and/or in the development of procedures to understand membrane proteins at the molecular level.
Robert Stroud and his group have studied many membrane protein systems seeking characterizations and crystal structures. So far they have determined 7 high-resolution structures of membrane proteins at atomic resolution, one to 1.35 Å resolution, currently the highest resolution of any membrane protein structure. They also report the mechanism of targeting membrane proteins to the endoplasmic reticulum membrane by the signal recognition particle and its receptor complex interaction.
Dr. Andrej Sali is a leader in development of methods for protein structure analysis and prediction. He has played a key role in the New York Structural Genomix Research Consortium aimed at soluble proteins.
Senyon Choe’s group has elucidated mechanisms of potassium channels from structure analysis of domains of these channels. Dr. Choe has pioneered the MISTIC method for production of membrane proteins from all species, in E. coli.
Daniel Minor has also played a key role in understanding the regulation of potassium and calcium channels. He determined key structures that help elucidate the mechanisms of these channels. He has also developed schemes to test for membrane stability.
Ronald Kaback is world renowned for his extensive studies of the Lactose permease. Over many years he has been a pioneer in membrane biology. Kaback recently was coauthored with Iwata of the high-resolution structure determination of the Lac permease. Thus his extensive mutational analysis of Lac permease is legendary and can now be interpreted.
Henning Stahlberg is a world leader in electron and atomic force microscopy of membrane proteins. He beautifully elaborated the structures of the GlpF and other aquaporin channels in their membrane environment.
Dr. Roland Riek is an inventor of TROSY and an expert in the use of NMR for three-dimensional structure determination focused on membrane proteins. He studied under the tutelage of Dr. Kurt Wuthrich.
Dr. James Holton (a key person at the ALS) is one of the world leaders in development of synchrotron and storage ring sources for high throughput X-ray diffraction. He has made fundamental contributions to the operations of beamline 8.3.1 that is currently the model for others as they come on line at the ALS. He is a highly innovative developer. For example he developed a robotic mounting system for crystals at beamline 8.3.1 for under $5,000.
Scientific progress
Progress in the development of new methods, technology, approaches, and ideas for protein production and structure determination
This PSI Specialized Center focuses on membrane proteins. We target prokaryotic proteins expressed in prokaryotic organisms as the most amenable system for producing proper expression and targeting of these proteins to the cellular membranes. We believe that these will provide the most efficacious pathway to determining the structures of new membrane protein folds and proteins. We believe that archaebacterial and human membrane proteins provide an intermediate pathway to provide new folds and proteins since they may be evolutionarily intermediate between prokaryotic and eukaryotic membrane proteins. We also focus on human membrane proteins, while they represent the most technically difficult class of targets; they also have the highest potential value for structure based drug design. Finally, we use all of our skills and technology at our disposal to also target selected membrane protein complexes for expression and subsequent purification, crystallization, and structure determination.
Molecular biology of selected targets
With over 700 E. coli targets to modify for the Ligation Independent Cloning (LIC) method and test for proper insertion of the target gene into the various expression vectors, we were right on the border of being able to efficiently using medium to high throughput molecular biology as opposed to purely manual methods. Melissa Del Rosario was instrumental in developing methods and techniques for using vacuum manifold based techniques on 96 well plates. These techniques permitted the screening of 288 constructs/day and allowed the efficient incorporation of our target genes into a variety of expression vectors.
Expression
The initial bottleneck for determining structure is membrane protein expression. Expression in bacteria is being carried out utilizing a novel scheme developed and applied by Senyon Choe, termed MISTIC, and by standardized procedures in E. coli that have already yielded high-resolution membrane protein structures in the Stroud and Kaback laboratories. We are also using codon optimization to improve expression of non-native proteins in our expression host cells, and Cell-Free Synthesis to target proteins that resist in vivo expression methods.
Human membrane proteins that are not expressed adequately or correctly folded by the above procedures will also be expressed through the NIH Roadmap Center directed by Stroud (GM-73210, Membrane Protein Expression Center, MPEC) funded in October 2004. The Membrane Protein Expression Center focuses specifically on developing novel methods for expression of eukaryotic (only) membrane proteins, and synergism with that center is expected to leverage results with this Structural Center.
Purification and characterization
Purification, solubilization and characterization are carried out as core functions at UCSF. The Protein Analysis Core at the Salk run by Kwiatkowski is developing High Throughput SELDI mass spectroscopy for molecular mass identification, NMR Analysis for detergent exchange, and ultracentrifugation to determine quaternary structure. As we approach the 4th year’s operation of CSMP, many targets are emanating from the expression efforts and entering the Purification and Characterization cores. In our hands, automation of purification is still premature as an option for membrane proteins primarily because of the need for hand-tuned optimization of extraction, purification stability, and solubility, in which each step involves screening many different detergents. Thus, as our expression pipeline is becoming efficient, protein purification to a pure homogeneous and stable form is the current bottleneck. We have addressed this bottleneck by setting up 5 HPLC stations, 2 Tetra Detector HPLC Analysis stations and 5 single detector isocratic or gravity chromatography stations several with auto-injectors and auto fraction collectors.
Crystallization and structure determination
High throughput crystallization methods and trials are being developed at UCSF. James Holton (UCSF and the Advanced Light Source (ALS) in Berkeley) is addressing bottlenecks in X-ray data collection and data analysis for 3D structure determination. Henning Stahlberg, UCDavis, is developing methods for streamlining EM structure determination, including automation of data collection and sample preparation. Electron crystallography is also being used to validate that the structures of 3-D crystalline samples are congruent with those in the 2D bilayer. Some membrane proteins may evade both structure determination schemes. NMR will be applied in these cases by Roland Riek for small size targets in addition to its own set of targets from E. coli.
MISTIC homologs (Senyon Choe, Salk)
Mistic-based over-expression of bacterial kinase receptors has provided a significant advantage of over-producing this class of target proteins over other methods. But for mammalian integral membrane protein targets, it still needs significant optimization. Apart from using Mistic for protein expression, we have also made efforts to understand the function of the Mistic protein in the Bacillus organism. We have focused on the role by up-regulating the expression of Mistic in Bacillus. It led to a surprising consequence. It promotes the formation of biofilm and endows antibiotic resistance in Bacillus. To some extent, the same physiological results can be reproduced when E. coli is transfected and up-regulated by Mistic expression. Several Mistic homologs have been studied for their structures. One such homolog we termed M2 behaves well, and a 3.5 Å dataset has been obtained from M2 crystals. The structure solution has not yet been obtained. Another such homolog we termed M1 behaves well in solution, for which high-quality NMR spectra have been obtained. Near complete assignment of M1 spectral peaks has been accomplished. These homologues have 62–93% residue identity, and are all 84 residues in length, corresponding to the three helices minus the first N-terminal helix of B. subtilis Mistic. In every case, these Mistic homologs have an overlapping gene read-out with a downstream gene that resembles a potassium channel protein. The functional relationship between the two genes is unknown.
Mistic technology has a limited use for eukaryotic targets. In order to explore the possible use of Mistic for eukaryotic targets, we have initiated using Mistic in lieu of a signal sequence present at the N-terminus of typical eukaryotic membrane protein genes. Initial expression constructs have been designed and will be compared to a comparable designer vector containing only the first alpha helical domain in place of the entire Mistic sequence. Once the designed vectors are available for tests, we will use the human integral membrane protein library that consists of ~3,270 target genes that have been constructed as a Gateway-adapted entry vectors.
In vitro expression (Robert Stroud)
Reconstituted cell-free (CF) protein expression systems hold the promise of overcoming several of the traditional barriers associated with in vivo systems. To evaluate the potential of cell-free expression, we cloned 120 membrane (<30 kDa) proteins from E. coli and compared their expression profiles in both an E. coli in vivo system and an E. coli derived cell-free system. Our results indicated that cell-free was a robust system able to express 63% of the targets compared to 44% in vivo (reported in Yr. 2). However, when evaluating an expression system, many factors, such as cost, complexity, throughput, reaction efficiency and duration, personnel time, and protein quality, must be taken into account. While cell-free is a robust expression system capable of expressing more proteins than an in vivo system and suitable for production of membrane proteins at the milligram level for a relatively small cost when producing the S30 fraction in house; the in vivo system is more streamlined, able to be scaled up much easier with our equipment, and more economical in all cases where expression is successful. We are investigating the addition of lipids and/or detergents into the cell-free reaction mixtures to investigate the potential to increase expression and perhaps address the reaction duration issues. If we select a high value target that is not amenable to in vivo expression systems, we will try the cell free approach, but even if the expression level in vivo is quite low (<0.1 mg/l), our Sartorius fermentor units (2 × 15 l and 1 × 75 l) can usually provide enough protein to perform useful experiments.
In year 3, we tested all of the phase 1 constructs that were positive for in vivo expression in larger growth volumes and prepared them for insertion into our regular in vivo expression, purification, and crystallization pathway.
Gene redesign and codon optimization (Robert Stroud UCSF)
Expression of functional protein targets in heterologous hosts is a key element of our efforts within the center. Utilization of codon optimized synthetic genes has been demonstrated in hundreds of published cases to increase, often significantly, the levels of expressed material for soluble proteins. Our efforts in CSMP focus on attempts to translate this observation to expression of integral membrane proteins. Within the past year we have generated codon-optimized genes for 21 different proteins that have lead to seven targets now in crystallization trials. Of these, three have crystallized (Tacid1, SecY and PfAQP) and have been optimized. Codon optimization for each of these proteins was essential for expression success and moving forward to crystallization trials. For instance, the native PfAQP protein did not express, likely owing to the AT rich character of the Plasmodium genome. Following optimization we are now obtaining ~42 g of purified protein/liter of culture. As evident from our initial expression results, we generally observe an improvement in protein expression levels. PfAQP has now had its structure determined to 2.05 Å (PDB: 3C02).
The Stroud laboratory now has a collaboration with DNA2.0 Inc. to pursue a study of the efficacy of codon-optimized genes. Dr. Franklin Hays has provided DNA 2.0 with our hENT1 genes and with the collaboration of Dr. Claus Gustafsson at DNA2.0, DNA2.0 will produce 20–25 different codon-optimized genes for CSMP to expression test and prepare for eventual crystallization trials (Table 1).
Table 1.
Progress table for the codon usage experiments
| Gene | Species | Design | Opt. For | Status | Native attempted | Improvement |
|---|---|---|---|---|---|---|
| Tacid1 | Thermus acidophilus | DNA2.0 | S. cer | Crystals | Yes | ~10× |
| PfAQP | P. falciparum | Kosan | E. coli | PDB 3CO2 | Yes | ~10× |
| SecY | Pyrococcus furiosus | Kosan | P. fur. | Crystals | Yes | ~4×, essential |
| HPV-2B | HPV | Kosan | E. coli | Crystal trials | No | – |
| HPV-3A | HPV | Kosan | E. coli | Crystal trials | No | – |
| rGamma2 | Rattus norvegicus | DNA2.0 | E. coli | Crystal trials | Yes | ~3× |
| rGamma5 | Rattus norvegicus | DNA2.0 | E. coli | Crystal trials | Yes | ~3× |
| Tacid2 | T. acidophilus | DNA2.0 | S. cer | Purification | Yes | ~10× |
| Afmdr | Archaeoglobus fulgidus | DNA2.0 | E. coli | Purification | Yes | ~2× |
| hABCB1 | Human | Codon Devices | S. cer | Solubilization | Yes | ~3× |
| hα7 | Human | Kosan | E. coli | Solubilization | No | – |
| h3a | Human | Kosan | E. coli | Solubilization | No | – |
| hDAT | Human | Kosan | E. coli | Solubilization | No | – |
| hNET | Human | Kosan | E. coli | Solubilization | No | – |
| hSERT | Human | Kosan | E. coli | Solubilization | No | – |
| dSERT | Human | Kosan | E. coli | Solubilization | No | – |
| hABCA1 | Human | Codon Devices | S. cer | Expression | Yes | No, still blank on western |
| hRhAG | Human | GENEART | S. cer | Expression | In progress | No, slight decrease |
| hRhBG | Human | GENEART | S. cer | Expression | In progress | No |
| hRhCG | Human | GENEART | S. cer | Expression | In progress | Yes, detectable on western |
| C5aR | Human | Kosan | S. cer | Expression | Yes | Yes, detectable on western |
LIC vectors (Daniel Minor, Robert Stroud)
In order to facilitate the large amount of cloning and construct screening required for the efforts of the labs involved in this PSI center, the Stroud and Minor labs have developed a suite of expression vectors that implement Ligation Independent Cloning (LIC) strategies. This approach obviates the use of restriction enzymes for joining target protein DNA to the vector and the need for enzymatic ligation steps in clone preparation. The method is efficient and yields ~90–100% positive clones. We have constructed LIC E. coli vectors bearing two N-terminal affinity tags in series, His6 and maltose binding protein (MBP), followed by a tobacco etch virus (TEV) protease site (termed HMT). The His6- and MBP-tags allow for two orthogonal affinity steps to be used in purification. The TEV site permits specific cleavage of the tags following purification. LIC cassettes with different combinations of tags and utilizing the 3C (PreScission) protease have also been constructed and used primarily by the Stroud laboratory.
Because CSMP focuses on the potential value of the MISTIC fusion protein as a tool for expression of membrane proteins in E. coli, we made LIC constructs (termed HMisT) bearing, in series, His6, MISTIC, and the TEV site. We have also generated HMT LIC and HMisT LIC vectors that have an N-terminal signal peptide for use on target proteins where the N-terminal domain is known to be extracellular (ssHMT and ssHMisT).
Development of a rapid expression assay based on GFP for proper expression and membrane insertion (Daniel Minor)
In an effort to facilitate the screening and evaluation of target proteins, we have developed an LIC expression vector that uses green fluorescent protein (GFP) to monitor target protein expression levels in E. coli directly. This vector bears an LIC site followed by GFP and a His8 affinity tag. The premise is that only proteins that are expressed and properly folded will give robust fluorescence signals, as it is known that inclusion body formation suppresses the formation of the GFP chromaphore. Our initial experiments confirmed that well-expressed proteins turn the cells visibly green.
In the past year of effort we have used our GFP vector format to screen the expression of 314 membrane proteins from extremophilic organisms. We found that 67/314 (21%) of our extremeophile targets expressed as well or better than the benchmark proteins in the GFP assay. To investigate whether there was a correlation between GFP fluorescence values (FSUs) and the amount of protein produced in the membrane fraction, we took 36 targets (29 from the >60,000 FSU pool and seven from the <60,000 FSU pool) and subjected them to an expression optimization protocol. We evaluated expression levels for each construct (108 total) at three expression temperatures (18, 25, and 37°C). 17 of our targets, once optimized, express at >5 mg/l in the membrane fraction. Thus, the GFP screen appears to be a reliable way to identify targets that have good potential to express at high levels in the membrane. Based on our results, there is a 90% probability that a target with a fluorescence value >60,000 FSU can be expressed at 0.5 mg/l or better, levels that are sufficient for further characterization and crystallization trials.
We pursued purification and characterization of 24 of the targets that expressed at 0.5 mg/l or better. We are actively pursuing purification optimization for each candidate as well as crystallization trials. Our plans for FY04 are to pursue crystallization and structure determination of our cohort of well-expressed extremeophile membrane proteins. Additionally, we plan to work with Dr. Greg Hura at ALS beamline 12.3.1 to collect Small Angle X-ray Scattering (SAXS) data on all well-behaved samples. Recent implementation of robotic sample handling on the 12.3.1 SAXS end station together with our cohort of well-expressed, readily purified targets provides an excellent opportunity to investigate the potential of a yet underutilized way to obtain medium resolution information (particularly regarding multimerization state) for membrane proteins. It is anticipated that such efforts will also aid in sample improvements for our efforts on structure determination by crystallization and X-ray diffraction methods.
Protein expression core (Witek Kwiatkowski, Casey Johnson, Luis Esquivies, Christopher Dickson, Inno Maslennikov, SALK)
We have standardized the steps of vector construction, detergent screening, mini-scale expression tests, and protein scale-up and subsequently trouble-shooting. Using these standardized protocols, we plan to process 4 vectors made per week, and 2 protein expression and scale-up per week, on average. Once the expression vectors are made and confirmed, mini-scale protein expression tests are performed. Experiments to determine the optimal detergent type, most effective solubilization time and detergent concentration will be performed and the data entered into CSMP Central Database. Once the detergent is selected from the screening and expression is confirmed, the expression vectors are tested for scale-up protein preparation. As a general guideline, we evaluate by testing detergent exchange and concentration by NMR, testing by HPLC sizing, ion exchange chromatography, testing oligomeric state of the protein in solution by Analytical Ultracentrifuge, choosing purification protocol, setting up crystallization screens. If necessary, we change detergent (decyl-d-maltopyranoside (DM), dodecyl-d-maltopyranoside (DDM), octyl l-d-glucopyranoside (OG), nonyl-d-glucopyranoside (NG) are the choices) and repeat. This is performed by the Protein Analysis Core. A typical target protein will take 7 working days to pass through the detergent screening with day one and two being used to solubilize membrane preparations prepared before-hand, days 3 and 7 to run the samples on size exclusion chromatography and run signature SDS–PAGE gels and western blots and change and test different buffer conditions. Most of this work is not particularly suited to automation except through the use of auto-injectors on the HPLC systems.
Purification and characterization: high throughput characterization of membrane protein detergent complex (Kwiatkowski, SALK)
The identification and characterization of integral membrane proteins (IMPs) require stable protein-detergent complexes (PDCs) to be formed. We used a combination of light scattering, refractive index measurement, mass spectroscopy, NMR spectroscopy, and ultracentrifugation to carefully characterize detergents and proteins and their relative compositions in the PDCs. This information was essential to evaluate oligomeric states of samples, and thus the feasibility of crystallization.
We use 1D-NMR spectroscopy to quantitatively monitor detergent concentration during sample purification. These data provide a useful gauge of the detergent’s extraction potential for a given protein, detergent exchange efficiency, and detergent concentration after final ultra-filtration. To assess the homogeneity and oligomeric state of PDCs we used HPLC in line with light scattering, UV and refractive index instruments or, alternatively, the sedimentation velocity measurement by analytical centrifugation. These methods provide us with the quantified detergent-to-protein ratio and the mass of the entire PDC, and thus with the precise PDC composition. They also allow us to assess the capability of detergent additives to influence these important parameters.
Crystallization and structure determination: automation of crystallization trials (Robert Stroud)
In the process of 3D structure determination of biological macromolecules, crystallization constitutes one of the major bottlenecks. The scarcity of sample for crystal trials constitutes a limiting step in particular in the case of membrane proteins. At the CSMP, we have implemented a ‘high-throughput’ crystallization strategy that covers two aspects: Robotic screening and optimization of crystallization conditions. A nanoliter scale crystallization robot (Mosquito© from TTP LabTech) is used to perform crystallization trials in 96 well-format plates. It is designed to prepare drops in both hanging- or sitting-drop mode using the vapor diffusion method. Drop size can be reduced to 200 nl (100 nl + 100 nl) without loss of accuracy and efficiency. This robot is easy to use, and it is a positive displacement pipetting robot that uses disposable tips. It therefore eliminates the risk of cross-contamination, and it eliminates the need for cleaning the tips.
This system is used for screening initial crystallization conditions and also for optimization using a set of additive screens. We keep approximately 20 commercial crystallization screens of 96 conditions on-hand giving researchers 1,920 unique crystallization conditions in addition to custom chemical components. Working with 100 nl of a protein solution and 100 nl of the crystallization screen requires <10 μl of protein solution instead of the 192 μl needed for manual drop setting methods (2 μl + 2 μl drops). This represents a 20× reduction in the protein needed for each screen and thus a 20× increase in the number of screens that we can perform with a given volume of protein preparation. This improvement in efficiency permits effective screening with proteins that express 20× less that before, putting our minimum useful expression level around 0.1 mg/l. This crystallization robot was purchased by CSMP, and is shared, (as are costs) by all members of CSMP, and also by the Macromolecular Structure Group (msg.ucsf.edu), the Gladstone Research Institute, and the QB3 Institute. About 20 different groups currently use this robot.
Optimizing X-ray structure determination (James Holton, Robert Stroud, ALS/UCSF)
Work continues on narrowing the large numerical gap between collected data sets and published structures. World wide, this gap is roughly 50 to 1, and our analysis of the problem has demonstrated that this gap is due to a lack of understanding of the data quality requirements of modern structure-solving algorithms. Clearly, it is always best to have high quality data, but optimizing throughput, efficiency and success rates requires that the minimum data quality requirements be known.
Establishing these limits empirically would require collecting data sets from a wide range of proteins and systematically varying different parameters such as crystal size, mosaic spread, exposure time and others. This is a very large parameter space. To aid this search, a quantitative model of the diffraction experiment and all its various sources of error was constructed. This diffraction experiment simulator (MLFSOM) is now a mature system and has begun to produce exciting results. Nearly all important physical phenomena in the diffraction experiment, including radiation damage, spot shape, anomalous differences, diffuse scattering, shutter jitter and beam flicker are all modeled from first principles. The simulation is performed on an absolute scale, so once the correct real-world values of flux, crystal volume and other physical parameters are entered the simulated images are not only remarkably realistic, but on the same scale as measured diffraction patterns. The simulated images are output in the .SMV file format so they can then be processed with modern data-processing software. In this way, the effect of varying experimental parameters (such as exposure time in seconds or crystal size in microns) can be precisely correlated to data quality and the “threshold of solvability” established for each. The advantage of absolute scale is that the optimal parameters derived from the simulation have the same units as the optimal parameters for data collection. For example, it has been found that the read-out noise of a modern detector has almost no impact on the accuracy of anomalous difference measurements, which implies that MAD/SAD data sets can be divided over many more images than previously thought. This prediction was experimentally tested, and it was found that higher redundancy with the same total exposure actually improves overall data quality. This new data collection strategy is now strongly encouraged at ALS 8.3.1.
In May 2007, the ADSC Q210 detector in ALS 8.3.1 was replaced with a new, larger, ADSC Q315r detector. The increased surface area and lower intrinsic noise have enabled larger unit cell work, improved signal-to-noise (longer detector distances) and a 10-fold increase in the allowable data redundancy over the old detector. It is now clear from our studies [1] and others that radiation damage is fundamentally unpredictable. We have shown that heavy atom sites decay exponentially with dose, and the decay constants are reproducible for a given sample type. We have established that the best strategy for MAD data collection is to begin with a complete data set of very short exposures (1 s or less) and then repeat the complete data set with 2× the exposure time, then 4× the exposure time and continue until the crystal shows signs of decay. This decay can either be evaluated by monitoring the XANES spectrum of the metal [1] or by visual inspection of the decaying diffraction pattern. If the sites are robust, then all the data sets can be merged together, and if the sites are found to decay rapidly, then the radiation induced phasing (RIP) technique can be applied between the first and last data sets. Thus the same set of data is applicable to several structure solution pathways.
Of course, once the requirements of a crystal are known, one must find a crystal that meets them. Two novel screening technologies are in development at ALS 8.3.1: the Offline Target Indication System (OTIS) and the “plate goniometer”. OTIS circumvents the crystal-centering problem by using optical microscope images of a hand-centered crystal to re-center it at a later time. This effectively enables unattended data collection, which is done late at night. Screening is then done during the daylight hours, when users are more alert. Delayed data collection has now become a popular feature for users with overnight shifts.
The “plate goniometer” is an in situ screening system developed in collaboration with the SIBYLS beamline 12.3.1 and Fluidigm Corporation. It is now operational and screening crystals at a rate of one per minute. With this system, crystals can be probed in their growth chamber, which eliminates the caveats and the labor of harvesting, mounting and cryoprotecting each crystal, all important considerations in crystal optimization. We have recently begun applying this technology to novel structural projects, including a membrane protein from CSMP.
Describe plans for FY04 research activities
The initial goals and principal persons proposed continue along the lines of their initial goals. We need to make a more seamless transfer of membrane proteins to electron microscopic analysis, which is underutilized due to supply at present. As new proteins are now being produced the equation has evolved and demands this streamlined transfer of material to UC Davis.
Bioinformatic methods will be applied increasingly here to help in pinpointing structures of highest impact on the membrane proteome. Increased focus will be added to the human membrane protein genes being assembled in GATEWAY vectors by Dr. Choe. These will be expressed as MISTIC fusions at CSMP.
We have set-up a multi-center crystallization method comparison with several PSI centers (Hauptman–Woodward Institute, Ismagilov Laboratory at the University of Chicago, and the Kenis Laboratory at the University of Illinois Champaign-Urbana, Fluidigm Corporation) that specialize in high throughput crystallization methods. We will be able to assess the success of each method via their results with a standardized protein preparation that we provide. CSMP is producing sufficient DjkA protein in a PHS state to provide all of these groups with enough protein to run a selected number of our favorite commercial crystallization screens.
At the end of year 3, the Stroud laboratory has 735 prokaryotic membranes cloned, 240 proteins demonstrating significant expression, 85 that have been solubilized, 36 in membrane purification; 11 entered crystallization, 11 crystallized, 6 diffracted and have 3 structures. We can expect a dramatic ramp up as we enter more proteins into purification.
All the CSMP centers combined have 1072 proteins listed on our website database CSMP Central. To date, 771 have been cloned, 564 have tested positive for useful expression, 62 have been solubilized, 38 were PHS, 13 produced crystals, 11 diffracted, and 11 structures were determined.
Progress on production and structural determination of non-redundant proteins from classes of challenging proteins: introduction and background
Many eukaryotic membrane protein families have close homologs in bacteria whose structures provide insight into the structures of eukaryotic family members. Therefore a major focus is to determine the structures of E. coli membrane proteins. E. coli however, does not contain representatives of many families found in mammals, hence the focus on archaea and human membrane proteins. The major subclass in human is those membrane proteins that are localized in functional form in the plasma membrane of the cell. These include many drug targets.
Describe your plans and progress in selecting targets for production and structure determination and structural coverage at granularity consistent with the PSI-2 policies and appropriate for your class of challenging proteins
Target selection
Objective: Develop a target selection strategy to sample a broad range of membrane proteins from set of selected prokaryotic and eukaryotic organisms.
Targets of special interest
(a) Extensive pipeline of E. coli and prokaryotic membrane proteome proteins (Stroud), (b) Membrane Protein Complexes from Prokaryotes (Stroud), (c) E. coli membrane proteins expressed from the in vitro/in vivo comparison (Stroud), (d) ABC and SLC transporters (Giacomini), (e) Bacterial receptor kinases (Choe), (f) Lactate symporter and H+/nucleobase-ascorbate symporter YgfO (Kaback), (g) Transporters and channels (Stroud). Ammonia channels, Mercury transporters, aquaporins (Stroud), and (h) extremeophile membrane proteins (Minor).
Target selection (Andrej Sali, UCSF)
As part of the CSMP effort, we selected a set of target proteins for structural studies that maximizes coverage of integral α-helical membrane proteins in the S. cerevisiae while at the same time minimizing the number of sequences.
We used our analysis to guide target selection for the structural genomics of membrane proteins in yeast, aiming for complete coverage of integral membrane proteins predicted to have three or more alpha helices. We found that complete profile-based sequence coverage of the yeast genome required 361 total targets of the 622 total.
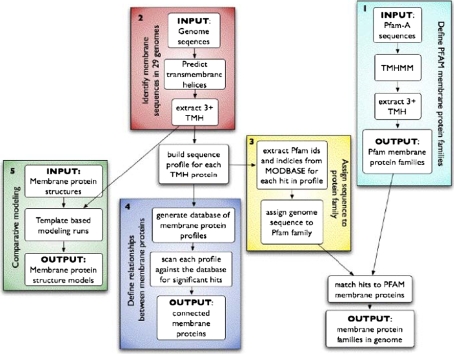
The overall membrane protein annotation process consisted of the following five steps detailed in Fig. 1: Membrane protein annotation pipeline. First, we identified integral membrane protein families in the Pfam database of protein families. Second, we collected protein sequences encoded by a diverse set of 34 genomes of interest to CSMP and predicted α-helical transmembrane proteins. Third, we generated sequence profiles for the identified membrane proteins in each genome and annotated the sequences in each sequence profile with organism and protein family identifiers. Fourth, we enumerated membrane protein families in each genome and connected significantly related membrane proteins both within organisms and across organisms. Finally, we constructed comparative protein structure models for all known sequences from Uniprot based on known atomic structures of membrane proteins.
Fig. 1.
Bioinformatic flow chart for CSMP protein target selection
Complete profile-based sequence coverage of the yeast genome therefore requires 361 total targets of the 622 total. These sequences were added to the target selection tool and entered in to the experimental structural characterization pipeline in the Stroud group.
One goal of structural genomics is to improve comparative modeling of sequence space. Finding homologs that can be modeled based on a template structure is the first step in the modeling process. We use our sequence profiles for each target sequence as a proxy for how many sequences could be modeled if we had a structure of the target. In total, there are 415,983 sequences in Uniprot that are detectably homologous to one or more of the CSMP targets. We find that our target selection methodology yields more unique modelable sequences and better profile-based sequence coverage than randomly selected sequences at 30% sequence identity.
Focus on human ABC and SLC transporter homologs in yeast
Two selected targets, the yeast genes STE6 and YN_99, code for ABC transporters that are homologous to human multidrug transporters in the B and G families, respectively. There are 48 characterized ABC transporters in the human genome and 18 are disease-associated. Additional structural data these transporter families will be invaluable for interpreting the results of functional studies and suggesting mechanisms for clinical phenotypes.
Expression of membrane proteins
Extensive pipeline of E. coli and prokaryotic membrane proteome proteins (Robert Stroud). In the Stroud laboratory, we have a three phase organization to our work. First, we initially started on a 120 protein project investigating in vitro translation/expression. The results of this work were published in 2007 and the target proteins that expressed successfully moved into the purification pathway, this work is continuing. The second phase was our “408” protein project that was headed by Melissa Del Rosario. The proteins in this project were selected with the aid of the bioinformatics group in the Andrei Sali laboratory to provide extensive coverage of the Pfam classes in E. coli. Phase 2 has provided a large number of proteins that are ready for expression and solubility testing. From prior experience it was decided to test for solubility in octyl glucopyranoside (OG) detergent only. With this operational filter, we still had enough proteins that solubilized in OG to saturate our purification resources. In Phase three, since we had covered a significant part of the E. coli genome already in the first two phases, it was decided to complete the entire E. coli membrane protein proteome by selecting the remaining unselected proteins. All three phases are continuing presently.
E. coli membrane proteins expressed from the in vitro/in vivo comparison (Robert Stroud)
Of the 120 proteins that were subject to expression testing in both the E. coli, and by E. coli cell-free methods, 12 of the membrane proteins [2] expressed at greater than one milligram per liter of culture and were able to be solubilized in OG and were therefore selected as targets for X-ray crystallography. Four have gone forward to crystal trials, and have been crystallized. CcmG and YijD crystals have gone to the ALS and diffract to 2.3 and 7.5 Å, respectively. The structure of CcmG is currently being solved by molecular replacement (a structure of the soluble component of CcmG has been published, but not the transmembrane segment). Crystals of YijD are being further optimized with grid and additive screens.
Protein production and structure determination (UCSF-Harries, Miercke, Stroud)
Prokaryotic membrane proteins
We currently have 735 prokaryotic membrane proteins in our protein purification pipeline. So far, 155 proteins express at levels high enough to proceed to solubility testing using a detergent screen with six different detergents representing three detergent chemical families. 85 of the solubilized proteins have been successfully purified on Ni IMAC. Once the Ni immobilized metal affinity chromatography (IMAC) purification efficiency has been assured by SDS–PAGE and western blots, the histidine tags are cleaved for preparative SEC runs. If the SEC produces a pure homogenous single peak, the sample is tested for stability; at this time, 34 of the post-SEC proteins are pure, homogenous, and stable (PHS). If a protein preparation can not be adequately purified by SEC, ion-exchange chromatography using methyl sulphonate (S) or quaternary ammonium (Q) matrices are used for further purification. Size exclusion chromatography is routinely employed to test the PHS condition of all preparations. Once a protein preparation is PHS it is ready for crystal trials. At the end of year 3, the Stroud laboratory has 735 prokaryotic membranes cloned, 240 proteins demonstrating significant expression, 55 that have been solubilized, 36 in membrane purification; 11 entered crystallization, 11 crystallized, six diffracted and have four structures. One structure is of a primitive Rh factor from Nitrosomonas (PDB: 3bhs, [3]), two are published structures of AQPM solved at 1.67 Å and 2.3 Å (PDB: 2evu and 2f2b, [4]), and the other is an AmtB-GlnK complex refined to 1.95 Å (PDB: 2ns1, [3]) (Table 2). We can expect a dramatic ramp up as we enter more proteins into purification. However, the bottleneck for us is protein purification. We therefore seek to augment the effort there with increased efficiency, additional chromatography stations, and possibly additional personnel if we can appropriately refocus less productive prototype experiments.
Table 2.
STROUD prokaryotic membrane protein progress
| Progress step | Step description | No. in group | Total no. through this level |
|---|---|---|---|
| 12 | Structure | 5 | 5 |
| 11 | Diffraction | 2 | 8 |
| 10 | Crystals | 0 | 8 |
| 9 | PHS | 25 | 33 |
| 8 | S-Chrom | 0 | 33 |
| 7 | Q-Chrom | 0 | 33 |
| 6 | Tag Cleaved | 0 | 33 |
| 5 | SEC | 2 | 35 |
| 4 | Aff Chrom | 0 | 35 |
| 3 | Soluble | 22 | 55 |
| 2 | Significant exp | 240 | 240 |
| 1 | Selection | 735 | 735 |
All the CSMP centers combined have 1072 proteins listed on our website database CSMP Central. To date, 771 have been cloned, 564 have tested positive for useful expression, 62 have been solubilized, 38 were PHS, 13 produced crystals, 11 diffracted, and 11 structures were determined (Table 3).
Table 3.
CSMP membrane protein progress
| Progress step | Step description | No. in group | Total no. through this level |
|---|---|---|---|
| 12 | Structure | 11 | 11 |
| 11 | Diffraction | 11 | 11 |
| 10 | Crystals | 2 | 13 |
| 9 | PHS | 25 | 38 |
| 8 | S-Chrom | 0 | 38 |
| 7 | Q-Chrom | 0 | 38 |
| 6 | Tag Cleaved | 0 | 38 |
| 5 | SEC | 2 | 40 |
| 4 | Aff Chrom | 0 | 40 |
| 3 | Soluble | 22 | 62 |
| 2 | Initial exp | 628 | 628 |
| 1 | Selection | 1072 | 1072 |
Human membrane proteins
Thirty-six human membrane protein targets have entered the Protein Expression and Production Pipeline. Ten of these express adequately to continue through the pipeline. So far, nine of these have been solubilized and moved on to the purification process. Purification protocols for all 9 are in the process of being fine-tuned. One human protein has been crystallized.
Membrane protein complexes from prokaryotes (Robert Stroud)
We have initiated collaboration with David Eisenberg’s group at UCLA. He searched the four bacterial genomes to identify genes encoding putative protein-protein complexes where one member is a predicted to be a membrane protein. We obtained a total of 42 membrane protein-containing complexes (86 proteins) from Acinetobacter baylyi ADP1, Escherichia coli K12, Mycobacterium tuberculosis H37RV, and Pseudomonas aeruginosa PAO1, were chosen as targets for CSMP. All have two subunits with the exception of one complex from E. coli, which has four protein components.
Our aim is to co-express members of each complex in the same E. coli cell host since co-expression may allow association during synthesis, and bring advantages of solubility, activity and stability compared to proteins expressed separately. We initially attempted an LIC Duet cloning strategy marketed by Novagen; however this method has been successful for only one complex.
We have cloned the all of the genes on separate compatible plasmids that can be co-transformed. Expression testing is now ongoing.
Membrane transporters (Kathy Giacomini)
Membrane transporters play a critical role in drug disposition and response. The Pharmacogenomics of Membrane Transporters project (PMT) is focused on discovering and functionally evaluating genetic polymorphisms in 50 membrane transporters in the human genome. These transporters are in two major super families, ATP Binding Cassette (ABC) Superfamily and Solute Carrier Superfamily (SLC). The goal of our studies in the CSMP project is to develop expression constructs of human membrane transporters in appropriate expression vectors and functional assays so that the transporters can be expressed in abundance to facilitate the determination of structures. Our laboratory clones the transporters from human and mammalian tissues and functionally characterizes the transporters in mammalian expression systems. To date, 20 transporters have been cloned and functional assays performed.
The goal of our studies in the CSMP is to develop expression and purification protocols of functional human membrane transporters for downstream structure determination and characterization. Another goal for the coming year is to develop a collaboration with Volker Dotsch in the expression and purification of membrane transporters in the Solute Carrier Superfamily (SLC). Volker has developed a non-cellular system of expression of large quantities of membrane proteins.
Table 4 below describes progress made in our project over the first 3 years.
Table 4.
Progress of transporter cloning and functional characterization in the Giacomini Laboratory
| Transporter | MW | Functional clone | Yeast expression | Solubilized | Purified | |
|---|---|---|---|---|---|---|
| 1 | hMATE1 | 61919 | X | ~0.8 mg/l | No | No |
| 2 | rMATE1 | 61436 | X | ~0.5 mg/l | Yes | No |
| 3 | hENT1 | 50216 | X | ~2 mg/l | Yes | Yes |
| 4 | hENT2 | 50110 | X | ~1.2 mg/l | ~50% | Yes |
| 5 | hENT3 | 51712 | X | ~0.8 mg/l | ~60% | No |
| 6 | hCNT1 | 71580 | X | ~1.0 mg/l | Yes | Yes |
| 7 | hCNT2 | 71991 | X | POOR | Yes | Yes |
| 8 | hCNT3 | 76925 | X | ~0.5 mg/l | Yes | No |
| 9 | hOCTN1 | 62151 | X | ~1.0 mg/l | ~50% | No |
| 10 | hOCT1 | 61150 | X | ~1.0 mg/l | Yes | No |
| 11 | hOCT2 | 62560 | X | ~0.8 mg/l | Yes | No |
| 12 | hOCT3 | 61276 | X | ~0.5 mg/l | ~60% | No |
| 13 | hABCG2 | 72309 | X | 1.0 mg/l | No | No |
| 14 | hMATE2 | – | X | Not tested | Not tested | Not tested |
| 15 | hOAT1 | 61812 | X | Not tested | Not tested | Not tested |
| 16 | hOAT2 | 59807 | X | Not tested | Not tested | Not tested |
| 17 | hOAT3 | 59853 | X | Not tested | Not tested | Not tested |
| 18 | hOAT4 | 62166 | X | Not tested | Not tested | Not tested |
| 19 | hOCTN2 | 62747 | X | Not tested | Not tested | Not tested |
| 20 | CTR1 | – | X | Not tested | Not tested | Not tested |
Bacterial receptor kinases (SALK-Choe, Kefala)—bacterial histidine kinase receptors (SALK-Choe, Kefala)
Crystal structures of bacterial histidine kinase receptors are the main targets of our efforts. To facilitate the structure determination, we have carried out a systematic approach in selecting and constructing expression vectors of these family members. Based on the complete analysis of E. coli genome, we identified 25 such E. coli’s kinase receptors, 23 of which are histidine kinase receptors and the remaining two being tyrosine kinase receptors. All these receptors are part of two-component transmembrane signaling systems.
We have prepared a two-prong approach in making expression constructs in order to analyze the effectiveness of Mistic fusion on protein expression. Our laboratory uses Gateway expression vector technology to construct these vectors so it is very easy of put these vectors together. One is fused with Mistic (Misticated), and the other without Mistic (non-Misticated). Both carry an octahistidine tag as an affinity tag to facilitate purification. These are further expanded by removing the putative signal sequence present in the gene, also by the addition of response regulators. This combinatorial mix of about 100 protein constructs are at various stages of the pipeline for overproduction. About 12 of those have reached the stage of well-behaving protein (grade A) purified at sufficient quantity for further work. The crystallization efforts are being made with various combinations of additives and detergents. The project utilizes significantly a high-throughput crystallization using Honeybee 96 robot in the laboratory. For human targets, we continue to optimize the high-throughput tools and vectors as well as control vectors to benchmark the progress in order to process 35 plates currently in possession, each containing 96 human targets. Lately, we have initiated a new approach using Mistic as a signal sequence of yeast expression system. All these expression vector systems rely on the Gateway technology to ease the exchange of expression vectors for a target gene.
Bacterial secondary transporters (UCLA—Kaback): Nucleobase Cation Symporter 2 (NCS2) family
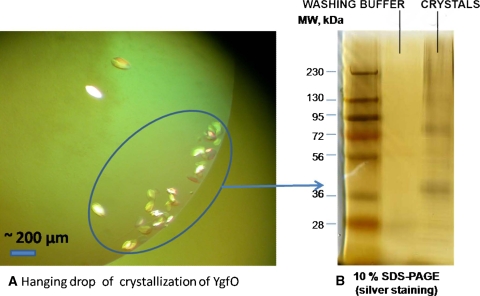
The H+/nucleobase-ascorbate symporter YgfO, which belongs to the NCS2 family of transport proteins, is ubiquitously found in all cells from archaea to eukaryotes. This highly conserved family consists of purine, pyrimidine and l-ascorbate transporters, with members specific for the cellular uptake of uracil, xanthine or uric acid (microbial and plant genomes) or vitamin C (mammalian genomes), as well as several important purine-related drugs (5-fluorouracil, allopurinol, oxypurinol). Thus, a crystal structure of YgfO would be relevant to the development of anti-cancer drugs, drugs to treat gout and anti- oxidants. We cloned four members of NCS2 family from E. coli, T. thermophilus, P. furiosus as well as T. maritimus, and expressed them in E. coli DE3 strain. Well-expressed YgfO from E. coli and T. thermophilus are solubilized with Fos-choline (FC)-14, purified by IMAC (Talon resin), yielding 1–2 mg/l culture. Both proteins are stable on ice for at least 2–4 weeks. Recently, initial crystallization screening by microbatch and hanging drop vapor diffusion yielded a few ‘hits’ for YgfO purified from E. coli HRK43. After brief optimization, relatively large crystals (50 × 200 μm) were obtained from salt-based crystallization conditions and xanthine (Fig. 2).
Fig. 2.
a Photomicrograph of crystals for YgfO obtained from salt-based crystallization conditions and xanthine. b Photograph of a silver stained SDS-PAGE gel showing the protein composition of the crystals harvested from the salt-based crystallization conditions and xanthine
The ion transporter superfamily (ITS)
Lactate permease (LctP) belongs to the ITS family found in Gram-negative and Gram-positive bacteria, as well as Archaea. All LctP members are proposed to have 14–16 transmembrane -helices with both N- and C-terminal exposed to the periplasm. We used the PhoA fusion approach where the expression construct is made with an embedded alkaline phosphatase sequence, permitting the gene expression to be easily monitored by a simple alkaline phosphatase assay. LctP appears to contain 14 transmembrane domains. E. coli LctP transports both l- and d-lactate and is a lactate/H+ symporter. It is also highly interesting that LctP is one of a group of acidic substrates that seems to be coupled to the pH gradient (interior alkaline) at ambient acidic pH values, but coupled to the membrane potential (interior negative) at more alkaline pH values. Recently, it was reported that LctP from Neisseria gonorrhea is important for pathogenesis. Crystals of LctP were obtained with PEG400/(NH4)2SO4 (pH 8.6) after several months incubation. At the ALS beamline 8.2.1, anisotropic diffraction is observed to 3.2/6.0 Å and a data set was completed to 7.0 Å.
Topology of MISTIC
MISTIC is a protein from Bacillus subtilis that is reported to facilitate expression of eukaryotic membrane proteins in E. coli. We have utilized the alkaline phosphatase (phoA) fusion approach to study the topology of MISTIC in the membrane of E. coli. The following PhoA fusions have been constructed: MISTIC-K28, -Y58, P63, Q66, K86, G115 and T139. The letters and numbers refer to positions in MISTIC where PhoA (alkaline phosphatase) fusions were constructed. For example, K28 means that the fusion construct is MISTIC from position 1 to 28, and the rest of the fusion protein is alkaline phosphatase. In order to be active alkaline phosphatase must be translocated into the periplasm. Since none of the fusions that Lan Guan constructed—which run the length of MISTIC—have alkaline phosphatase activity, the observations would be interpreted to mean that MISTIC has no transmembrane domains. Incidentally, this is a technique that was developed originally by Manoil and Beckwith in the early ′90s, and it has been used to test the hydropathy models of many polytopic membrane proteins in addition to LacY.
Transporters and channels (Stroud)
Ammonia channels
Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å [3].
Ammonia conductance is highly regulated. A PII signal transduction protein GlnK is the final regulator of transmembrane ammonia conductance by the ammonia channel AmtB in E. coli. The complex formed between AmtB and inhibitory GlnK at 1.96 Å resolution shows that the trimeric channel is blocked directly by GlnK, and how in response to intracellular nitrogen status, the ability of GlnK to block the channel is regulated by uridylylation/deuridylylation at Y51. ATP and Mg2+ augment the interaction of GlnK. The hydrolyzed product, adenosine 5′-diphosphate (ADP) orients the surface of GlnK for AmtB blockade. 2-oxoglutarate diminishes AmtB/GlnK association and plausible sites for 2-oxoglutarate binding were identified.
Rh Sub-family of Ammonia Channels: Rh50 of Nitrosonomas europea. (Stroud) Nitrosonomas europaea is an obligate lithoautotrophic ammonia-oxidizing gram-negative bacterium. NeRh is similar to evolutionary precursors of the Rh family found in eukaryotes. It is 38% identical and has strong similarity to human Rh factors. It has been demonstrated to conduct ammonia without pH dependence seen in the human Rh channels and the mechanism is remarkably conserved to that of the Amt subfamily. Gas conduction is not yet demonstrated in our functional testing. Unlike many Rh ammonia channels, NeRh is not glycosylated. It is a trimer of 42 kDa monomers. NeRh has been expressed in E. coli obtained in pure, homogeneous and stable form. Robotic crystal trials and optimization has yielded crystals that diffracted to 2.0 Å and the structure solved by molecular replacement. The refinement is complete for this structure and is a significantly better model for the structure and functional mechanism of the human channels.
The Kell/KX complex is responsible for the second most common erythrocyte antigenic phenotype and we have initial over-expression in S. cerevisiae.
Human RhCG has also been successfully expressed in the membrane fraction of S. cerevisiae and HEK293 cells, and it is soluble in OG and DDM.
Mercury transporters (Stroud)
In order to survive in mercury-rich environments, bacterial mercury resistance operons utilize mercury transporting membrane proteins, specifically MerT and MerC. In these Mer systems the toxic mercury ion, Hg+2, is imported through these transporters into the cell, where it is then reduced from Hg+2 to Hg, a relatively inert elemental form of mercury. These proteins are mechanistically interesting, and they represent a novel structural family.
Two bacterial mercury transporters, MerT and MerC, have been successfully cloned from Shigella flexneri into bacterial over-expression plasmid vectors as His-affinity tagged fusion proteins. Upon induction of expression in E. coli, both MerT and MerC are highly over-expressed and localized to the cell membrane, yielding approximately 4 mg of protein per liter of culture. We are near to finalizing this purification protocol and scaling up efforts to product quantities of MerT of suitable purity and low-detergent concentration for crystallization screening.
Aquaporins (Stroud)
Aquaporins that also function in H2S transport are found in archaea. One structure has been solved for methanobacter AQPM, to 1.65 Å. A second for A. fulgidus has been crystallized. The glycerol channel from the malaria parasite PfAQP has been subjected to gene design and synthesis, and has been crystallized with diffraction to 2.04 Å. Human AQP4 has also been successfully been crystallized after 2 years of work and a 1.8 Å dataset has been obtained. This diffraction data permitted the determination of the structure to a new higher level of resolution than before. All of these successful structure determinations are in the process of being written up for publication.
Structure determination by Cryo EM (UCDavis—Stahlberg)
The Stahlberg lab will continue developing the 2D membrane crystallization technology, the STEM imaging possibilities, and the maximum likelihood software tools. STEM imaging will advance due to the installation in the summer 2008 of an aberration-corrected and energy filtered 200 kV FEG STEM/TEM instrument at UC Davis, for which the Stahlberg lab is designing a new electron detector system and protocol to optimally harvest the phase contrast signal from that instrument. The Stahlberg lab is also hosting the next international workshop on electron crystallography of membrane proteins at UC Davis, which will take place Sept. 7–13, 2008 (http://2dx.org/workshop/2008). The Stahlberg lab continues structural investigations of a number of membrane protein systems by lipid membrane reconstitution, 2D crystallization and electron microscopy imaging trials. We have successfully reconstituted cell-free expressed CCR5 (from the Choe lab) and see first signs of structure formation within the membranes. UhpB (Choe) is reconstituted and forms first small 2D crystals. Reconstitution was successful, but so far no crystals were obtained for the sensor protein PhoQ (Choe), the lactate permease LctP (Kaback), and ABC-R (from the lab of Krys Palczewski). The Stahlberg lab also has obtained 2D crystals of a KcsA-MloK1 chimera protein, in collaboration with Crina Nimigean, NY. These crystals might reveal the KcsA gating mechanism, since the MloK1 cyclic nucleotide binding domains can activate or block the KcsA channel function. 2D crystals of the membrane embedded KscA-MloK1 chimera are obtained at different pH values, and structural analysis is ongoing. We are streamlining the process of membrane protein structure determination by electron crystallography of 2D crystals.
2D Crystal trial volume reduction
Current 2D crystallization trials are done by dialyzing a protein-detergent-lipid mixture of a volume of ~100 μl against detergent-free buffer. The Stahlberg lab has developed devices and a protocol for dialyzing much smaller 5 μl volumes. First results show that 2D crystallization does not scale linearly. Crystallization conditions for small and large volumes are different, and better crystals are generally obtained with larger volumes. Nevertheless, the smaller volumes allow screening a larger number of conditions with available protein material, after which the best conditions can be refined with larger volumes.
STEM imaging
Cryo-EM imaging of frozen hydrated and tilted specimen in the electron microscope suffers from beam-induced specimen movement or specimen charging, both of which severely limit the resolution of the structure in the direction perpendicular to the membrane plane. The Stahlberg group has compiled data that show that Scanning Transmission Electron Microscopy (STEM) is not affected by this problem, thereby bypassing the major bottleneck of electron crystallography. However, STEM imaging exposes the samples to very high dose rates of electron beam irradiation, and STEM imaging conventionally does not give phase contrast images, so that the resulting images have little contrast at lethal electron doses for the proteins. The Stahlberg lab now has developed an aberration-corrected STEM imaging protocol that allows to operate the instrument at membrane-protein compatible electron doses (5 electrons per square angstrom), and also allows access to the phase contrast signal of the electron-beam/sample interaction. The Stahlberg lab also has obtained proof of a strong phase contrast signal under the developed conditions. While it is not yet clear if the phase contrast signal is comparable to the phase contrast signal in conventional bright-field cryo-EM imaging, this development nevertheless appears likely to allow membrane protein structure analysis by cryo-EM, without the limitation from beam-induced specimen movement or charging.
Image processing
The Stahlberg lab continues developing new algorithms for membrane protein structure reconstruction from cryo-EM images of badly ordered 2D crystals. Realizing that the majority of the 2D crystals are not perfectly ordered, or that quasi-native membrane environments provide no crystallinity at all, such image processing will enable structure determination also from reconstituted but not crystalline membrane proteins. The Stahlberg lab is developing a single-particle 3D maximum likelihood software solution for such images, which is integrated into the user friendly 2dx software package that the Stahlberg lab is distributing (http://2dx.org).
Structure determination of integral membrane proteins by NMR (Salk, Riek)—small bacterial targets
Based on our structural investigations with MISTIC, we aim to determine several 3D structures of small integral membrane proteins of E. coli. E. coli has at least 52 integral membrane proteins with size smaller than 150 amino acids. Cloning and expression of all of these proteins have been initiated using MISTIC-technology in collaboration with S. Choe.
In parallel, cloning and expression of them without MISTIC has been established in collaboration with R. Stroud. In an initial screening, four of these proteins have been selected for expression and purification. Two of them (YidH and YgaP) were expressed at 10 mg/l level, were purified by Ni IMAC and show a dimeric size based on SDS-gels and size exclusion chromatography. Extensive detergent screening was applied to both proteins. For YidH a good quality TROSY NMR spectrum was obtained and sequential assignment is in progress. For this purpose TROSY-based HNCA, HNCACB and NOESY experiments were recorded. The TROSY NMR spectrum of YgaP is not yet of good quality and further detergent screening is initiated.
3D structure determination of corticotropin releasing factor receptor, a GPCR of family B: We aim to determine the 3D structure of corticotropin releasing factor receptor (CRF-R), a G-protein coupled receptor of family B1, by solution NMR.
In collaboration with the Choe lab we initiated the in vitro expression of the five somatostatin receptors sst1-sst5, which belong to the family A GPCRs. The yield for ss2 and sst5 is about 1 mg/ml reaction mixture. Binding studies with its ligand somatostatin are positive and indicate that both receptors are well folded. A 13C, 15N-labeled somatostatin was synthesized in collaboration with the Rivier group (The Salk Institute) and the complex formation between sst2 and 13C, 15N-labeled somatostatin is initiated to determine the 3D structure of the ligand in complex with its receptor.
Structural studies on KcsA potassium channel: The core of the KcsA potassium channel in its closed conformation has been determined by the MacKinnon group. We studied in detail the secondary structure as well as the dynamics of KcsA between open and closed state. It is our finding that conformational exchange dynamics between locally open and closed configurations in the filter as well as at the C-terminal end of transmembrane helix 2 are governing channel gating elucidating the mechanism of channel conductance and gating [5]. To our knowledge, this is the first study to correlate dynamics with function of a membrane protein. In addition, novel techniques are developed to study the structures and dynamics of membrane proteins.
Summary of all structures determined
The CSMP provided support during the resolution extension phase of the membrane protein Aquaporin M from Methanococcus marbergensis during Year 1, and supported the X-ray structure of the AmtB/GlnK complex (2NS1) during Year 2. The CcmG structure has been determined. It shows the ecto domain that reduces cytochromes clearly, but the transmembrane domain remains poorly determined. Refinement should define the domain A structure of Nitrosomonas europaea Rh50 has also been determined to a resolution of 1.99 Å in the Stroud laboratory. In addition, we determined the structure of AcrB—an already determined structure since it was purified in its endogenous form on the Ni column due to presence of two histidines. This is a case where we determined the entire structure, in two hours after data collection, albeit only to realize that it was already determined. It was the minor contaminant of the preparation of another membrane protein of similar size! Human AQP4 is being refined currently; the dataset is of high quality, defracting to ~1.80 Å. The Stahlberg laboratory obtained a 16 Å structure of the bacterial membrane protein MloK1 and the structure has been deposited at the EMBD Electron Microscopy Database (5548). The Minor laboratory obtained a 2.07 Å structure or the human membrane protein Kv7.4 (2OVC). The Kaback laboratory has continued to elucidate the various conditional structures of lactose permease, determining the wild type and the acidic and neutral forms (2V8 N/3.60, 2CFP/3.30, and 2CFQ/3.60, respectively.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Robert M. Stroud, Email: stroud@msg.ucsf.edu
William Harries, Email: harries@msg.ucsf.edu.
References
- 1.Holton JM (2007) XANES measurements of the rate of radiation damage to selenomethionine side chains. J Synchrotron Radiat 14(1):51–72. doi:10.1107/S0909049506048898 [DOI] [PMC free article] [PubMed]
- 2.Savage DF, Stroud RM (2007) Structural basis of aquaporin inhibition by mercury. J Mol Biol 368:607–617. doi:10.1016/j.jmb.2007.02.070 [DOI] [PMC free article] [PubMed]
- 3.Gruswitz F, O’Connell J III, Stroud RM (2007) Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein GlnK, at 1.96 angstroms. Proc Natl Acad Sci USA 104(1):42–47. doi:10.1073/pnas.0609796104 [DOI] [PMC free article] [PubMed]
- 4.Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM (2005) Structural basis for conductance by the archeal aquaporin AqpM at 1.68 angstroms. Proc Natl Acad Sci USA 102(52):18932–18937. doi:10.1073/pnas.0509469102 [DOI] [PMC free article] [PubMed]
- 5.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R (2007) Conformational dynamics of the Kcsa potassium channel governs gating properties. Nat Struct Mol Biol 14(11):1089–1095. doi:10.1038/nsmb1311 [DOI] [PMC free article] [PubMed]