Abstract
The development of forward genetics as a functional system in Toxoplasma gondii spanned more than three decades from the mid-1970s until now. The initial demonstration of experimental genetics relied on chemically-induced drug resistant mutants that were crossed by co-infecting cats, collecting oocysts, sporulating and hatching progeny in vitro. To capitalize on this, genetic markers were employed to develop linkage maps by tracking inheritance through experimental crosses. In all, three generations of genetic maps were developed to define the chromosomes, estimate recombination rates, and provide a system for linkage analysis. Ultimately this genetic map would become the foundation for the assembly of the T. gondii genome, which was derived from whole genome shotgun sequencing, into a chromosome-centric view. Finally, application of forward genetics to multigenic biological traits showed the potential to map and identify specific genes that control complex phenotypes including virulence.
Keywords: Genetic mapping, Virulence, Phylogeny, Population genetics
1. Introduction
Toxoplasma gondii is a widespread protozoan parasite of animals that also infects humans. While only an occasional cause of human disease, it has emerged as a model for studying the complex biology of apicomplexan parasites. This is due in large part to the ease of propagation and experimental manipulation both in vitro using a variety of cell lines and in the laboratory mouse. The complex life cycle would seem to preclude development of genetics, and yet these features have been exploited to develop powerful genetic tools. This advance required unusual cooperation between many individuals across different groups and spanning different technologies. This is my personal recollection of their efforts and the often serendipitous events that influenced key advances along the way. Italics have been added to emphasize my recollection of material that was paraphrased from writings or comments made by others, and which I found to be particularly influential.
2. Early development
2.1. A sage in the north woods
The development of experimental genetics in T. gondii was founded by pioneering efforts of Elmer Pfefferkorn; and yet he got a late start, having already made significant contributions over several decades to the development of sindbis virus as a model. As Elmer tells it, he had decided that virology was becoming too crowded and he felt his best course would be to change fields. Having moved north from Harvard Medical School (USA) to Dartmouth Medical School in New Hampshire (USA), he began looking for a new project to carry him into retirement. Elmer was always modest about his contributions, but in fact his influence in virology was considerable. For example, when I arrived at Washington University in the early 1990s, several of my virologist colleagues including Sonya Schlessinger, Milton Schlessinger and Charlie Rice, knew Elmer from his Sindbis days almost 20 years earlier. They held great admiration for his work, which was known for its clarity, precision and insight into areas of molecular biology that were not discovered until years after his initial work. The same features would become true of his work on Toxoplasma.
During the last cycle of his National Institutes of Health (NIH, USA) grant on Sindbis, Elmer went to the Dartmouth library, read up on potential topics and decided to move into Toxoplasma genetics. The life cycle had been discovered in the early 1970s (see contributions by J.P. Dubey (2009) and D.J.P. Ferguson (2009)) and presumably Elmer had read the recent accounts of how meiosis only occurs in the cat gut, and that it is only efficiently initiated after feeding of bradyzoites from tissue cysts that develop in mice (Dubey and Frenkel, 1972). Despite this somewhat daunting life cycle, he decided to try to employ tools that had proven useful in virology and to see if forward genetics could be developed in Toxoplasma. It took remarkable perception to foresee the potential of genetics in this system despite its biological complexity. Apparently even Elmer had some reservations as he briefly considered working on lichens or moss, but decided in the end that parasites were more interesting. Perhaps he was influenced by the fact that around the same time, Richard Carter and David Walliker had initiated experiments on recombination in Plasmodium spp. (Walliker et al., 1971, 1973, 1975). Elmer, and members of his laboratory including his wife Lorraine who played a critical role in these studies, launched a series of studies in Toxoplasma that would establish it as a model for biochemistry, cell biology, and lay the ground work for genetics (Pfefferkorn, 1981, 1988, 1990).
Elmer’s approach was straightforward and in typical Yankee fashion, wasted no effort: they would develop tools in T. gondii similar to those he had pioneered in viruses. They began by defining culture conditions to monitor growth and viability by plaque formation on monolayers of human foreskin fibroblasts. They established methods for chemical mutagenesis (Pfefferkorn and Pfefferkorn, 1979) and isolated temperature sensitive mutants (Pfefferkorn and Pfefferkorn, 1979), similar to his efforts to first produce these in mammalian cells years earlier. They then used these tools to generate mutants resistant to agents that targeted pyrimidine and purine salvage pathways (Pfefferkorn and Pfefferkorn, 1976, 1977a, 1977b, 1978; Pfefferkorn, 1978), and thereby defined their contributions to parasite biochemistry (Schwartzman and Pfefferkorn, 1980, 1981, 1982). Similar approaches proved useful for characterizing the sensitivity and resistance mechanisms to antibiotics that inhibit the parasite (Pfefferkorn et al., 1988, 1992a, 1992b, 1993). Others would later copy these strategies to explore basic areas of biology such as invasion (Dobrowolski and Sibley, 1996), egress (Black et al., 2000), the cytoskeleton (Morrissette et al., 2004) and the cell cycle (Gubbels et al., 2008).
In addition, Elmer and members of his laboratory established conditions for performing genetic crosses with T. gondii. A key step was the demonstration that a single cloned organism was capable of generating tissue cysts and of initiating the complete life cycle in the cat (Pfefferkorn et al., 1977). This established that there were no predefined mating types, but rather micro and macrogametes differentiate from a common progenitor. They performed the first genetic crosses in T. gondii by co-feeding to cats, the homogenates of mouse brains containing tissue cysts bearing different parental lines marked with distinct drug resistance traits (Pfefferkorn and Pfefferkorn, 1980). Isolation of progeny was then used to test inheritance of the markers, which could be accurately counted by plaque formation in media containing single or double drug combinations. By careful extrapolation he was able to show that sexual development results in an equal probability of self-fertilization and out-crossing (Pfefferkorn and Pfefferkorn, 1980). Deductive reasoning based on these ratios allowed him to conclude that tachyzoites and bradyzoites were haploid except for the fused zygote, which must be the only diploid stage. This was confirmed by cytological studies some years later (Cornelissen et al., 1984). Analyses of several genetic crosses demonstrated independent segregation of markers, a hallmark of Mendelian genetics (Pfefferkorn and Pfefferkorn, 1980; Pfefferkorn and Kasper, 1983). At the time, so few markers existed that it was not possible to determine whether this represented segregation of independent chromosomes or recombination within a single chromosome due to crossing over, although clearly the odds would favor the former. The absence of cytologically visible chaismata in studies on Eimeria was used to suggest that crossovers within the chromosomes may not occur in Coccidia (Pfefferkorn 1990). While other predictions of work from Elmer’s laboratory were borne out by further testing, it is fortunate that this latter idea proved incorrect, as otherwise the utility of genetics would have been limited.
2.2. Genetics from the ground up
When I joined John Boothroyd’s laboratory at Stanford University (USA) to begin a postdoctoral fellowship in 1987, I felt fortunate to be joining one of the best groups in molecular parasitology. Primarily known for his work on trans-splicing in trypanosomes, John had decided to take up Toxoplasma as a second research area. When I arrived, the laboratory was busy generating cDNA libraries, cloning genes and examining the basic molecular components of this parasite. John was an enthusiastic convert with a knack for motivating people to tackle tough but interesting problems. He launched the idea of bringing genetics into the molecular era. He convinced Elmer to assist us with genetic crosses, and talked Ron Davis and Gil Chu, also at Stanford University, into helping with chromosome separation. John’s enthusiasm convinced me that it would be a great project to generate a genetic map using molecular probes, something I knew nothing about but was happy to jump into. Combined with Elmer’s measured counsel, we set out to develop genetics as an experimental tool.
Elmer’s original markers had all been drug resistance traits and while they had served their purpose, it seemed unlikely that we could generate a sufficient number of these to provide a system for genetic mapping. Instead, we turned to restriction fragment length polymorphisms (RFLPs), which had been used to generate linkage maps in a variety of organisms. Our strategy was to analyze polymorphisms between different laboratory strains by Southern blotting with DNA probes. We included the RH strain used by most groups, Elmer’s CTG line that he had performed the original crosses with, and a clone of the ME49 strain called P strain, isolated by Lloyd Kasper, a former postdoctoral fellow of Elmer’s, also now on the faculty at Dartmouth. We had no idea at the time, but this choice spanned 95% of the genetic diversity present in most isolates from North America. We performed Southern blots of these isolates using as probes cDNA clones from others in the laboratory and genomic clones randomly chosen from small insert libraries. Progress was slow as most probes identified the same RFLP pattern in all three strains and it took on average 12 restriction digests to identify a single polymorphism. This implied a high degree of similarity between the strains, yet the significance of this would not be known for many years to come. Biallelism was also apparent at this early stage, but we also did not appreciate it until much later. In the end, the level of polymorphism was so low, we turned to using cosmids as probes, since they spanned much larger regions (35–40 kb) and hence captured greater polymorphism. Nylon filters, containing replicate sets of genomic DNAs that had been digested with a bank of restriction enzymes, were repeatedly probed, striped and reprobed until we built up a collection of polymorphic markers. Many of these were also mapped by Southern blotting to chromosomes separated by pulsed-field gel electrophoresis (PFGE), thus providing a molecular karyotype (Sibley and Boothroyd, 1992a).
Once we had an adequate collection of markers, we proceeded to genotype 19 recombinant progeny sent to us by Elmer, who had performed a cross between a clone of the type II ME49 strain (B7) and the doubly resistant CTG ARAR-SYNR line that he had isolated several years before. (A more complete summary of the genetic crosses that have been performed to date can be found at: http://toxomap.wustl.edu/Experimental_crosses.htm). Elmer isolated progeny by segregation of the two drug markers found in the CTG parent, such that they formed singly resistant clones. Genotyping of 64 distinct markers across all 19 progeny was used to generate the first linkage maps for T. gondii (Sibley et al., 1992). The chromosome maps were assembled manually by comparing genotypes for each new marker with a posted wall-chart that graphically depicted parental and recombinant genotypes for each clone. These were cross-checked against physical linkage groups assigned by PFGE, and ordered using the frequency of single and double crossovers. These efforts were greatly aided by a number of Stanford undergraduates involved in research, most notably Allen LeBlanc who performed a staggering number of Southern blots.
The first generation RFLP map defined 11 distinct linkage groups, yet only comprised ~150 cM in total genetic distance. Mapping resolution was limited, but we were able to link one of the two drug resistance markers to a specific chromosome. Despite earlier predictions, we found that intrachromosomal crossovers did occur; yet they were relatively rare. This was partly due to low density of markers but also an early indication that the map unit was large. Later refinements proved that it was in the order of 100 kb (Khan et al., 2005), almost six-fold larger than what would be found in malaria (Su et al., 1999). In practical terms, this meant that recombinant progeny, rather than the density of markers, would become the most limiting aspect of linkage mapping in T. gondii. One unfortunate oversight was that we failed to save the pool of recombinants from that cross, and would henceforth be limited to only the initial 19 progeny Elmer had isolated. In the future, we would preserve the recombinant pools by cryopreserving uncloned populations and this would prove to be invaluable.
2.3. The link with clinical toxoplasmosis and discovery of clonality
The other great influence in Toxoplasma at Stanford was Jack Remington, who was part of the Department of Medicine and ran a research laboratory at the Palo Alto Medical Foundation. Jack was always tremendously supportive of the work we were doing, even though his focus was primarily on research that was directly relevant to the patient. Despite the different styles of the two laboratories, we met for joint meetings and had great interactions over the years. One of Jack’s Infectious Disease fellows Brian Danneman, can be credited with our venture into analyzing genetic diversity of T. gondii samples from clinical samples, something that led to an important and unanticipated discovery. As a clinician, Brian attended many patients with advanced HIV (this was before highly active antiretroviral therapy (HAART)) who had severe toxoplasmic encephalitis (Dannemann et al., 1992). At this time, CNS biopsies and CFS were still being obtained to confirm the diagnosis of toxoplasmosis. Brian had collected a set of T. gondii strains from such patients and added these to an already diverse collection of strains housed in the Remington laboratory. To my knowledge, Brian was the first to suggest using molecular markers to analyze these strains for genetic diversity. His idea was met with a great deal of skepticism, as the prevailing wisdom often recounted by experts was that Toxoplasma strains are all pretty much the same, so why bother analyzing them? Despite this sentiment, we decided to look at his strains with some of our markers, which had been converted to PCR-based analysis to provide faster genotyping.
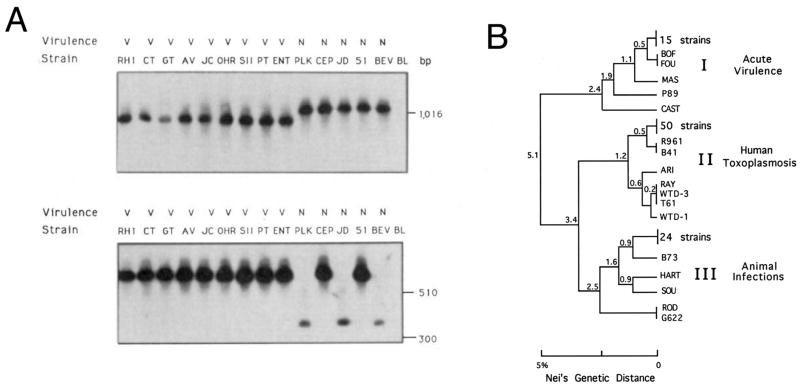
Near the end of my time in John’s laboratory, I finally got around to running the PCR analysis of Brian’s strains. At that time, PCR products were typically digested with restriction enzymes to reveal polymorphisms, resolved by gel electrophoresis and then Southern blotted to detect specific banding patterns. At the end of one particularly long day, I developed the film of Brain’s clones. One glance told me something was very unusual: all of the clones Brian had defined as virulent in the mouse model shared the exact same genotype, while those that were less virulent in mice were genetically variable. I mulled over the result for a few days, considering that it was possibly an artifact of PCR contamination. When I took it to show John, he agreed with typical cautious enthusiasm that there were several potential explanations; but if true, it would be important. I decided to regrow the strains, isolate new DNA and test them more carefully. Follow-up experiments confirmed that the original blot was correct and it became central in the publication showing that virulent strains of T. gondii belong to a single clonal genotype (Sibley and Boothroyd, 1992b) (Fig. 1A).
Fig. 1.
Demonstration of clonality in Toxoplasma gondii. A) Analysis of T. gondii strains for polymorphisms using two genetic markers SAG1 (top) and 850 (bottom). All of the mouse-virulent clones shared the same genotype at both marker (and subsequently many others that were tested) while non-virulent clones differed. Note that biallelism was evident in these early markers. Reproduced with permission from Sibley and Boothroyd (1992b). B) Phylogenetic tree of 106 strains analyzed by Dane Howe using six polymorphic markers. Three predominant clones typified the population structure. This pattern was later shown to be characteristic of North America and Europe. While this pattern has largely held up over the years, not all strains shown here are in fact members of the clonal lineages, or have the precise phenotypes shown to the right. For details on a more current analysis of these and others strains see Khan et al. (2007) and Sibley and Ajioka (2008). Phenotypic correlations of these lineages are shown at the right. Genetic distance was extrapolated from restriction fragment length polymorphism differences according to Nei’s genetic distance. Reproduced with permission from Howe and Sibley (© 1995 by the Infectious Diseases Society of America).
2.4. Three clones, a tree and a pony
We expanded our analysis of strains of T. gondii once I had moved to Washington University in St. Louis (USA) to start my own group. Dan Howe, a quiet self-assured Midwesterner and the first postdoctoral fellow to join the laboratory, genotyped more than 100 strains that had been generously sent to us by many colleagues. Most remained skeptical about our studies, remarking that strains of Toxoplasma were not really different, so why spend time genotyping them? We had heard this comment before and so persisted in our requests and eventually most agreed to send us strains for what seemed like a harmless, if misguided, study. Armed with this new collection of strains and some improved markers, Dan Howe made the remarkable discovery that not only were virulent lineages clonal, nearly all strains could be grouped into one of three highly similar yet distinct clonal lineages. We were surprised by this finding since it did not necessarily follow that because one clonal group existed, that this feature would define the entire population. Dan Howe was responsible for generating the tree shown in Fig. 1B, which has now been reproduced many times in reviews and borrowed by various colleagues. We were very excited by this finding and tried hard to convince a member of the National Academy of Sciences (USA) to sponsor it for the ‘Proceedings’. He sent the paper out for review, but ultimately the reviewers were unenthused with our discovery and thought it was fairly predictable. This perplexed us since the experts had previously predicted there would be no meaningful differences between strains. I telephoned the academy member to argue our case, saying how remarkable it was that the strains grouped into only three highly similar lineages, as shown by our phylogenetic tree. Somewhat exasperated by my persistence, I recall he blurted out …a Shetland pony could have drawn that tree! We thought that was exactly the point, the population was so highly structured that this implied something really noteworthy. The editor was unpersuaded and we ultimately had to settle on publishing the paper elsewhere (Howe and Sibley, 1995). Over the years, it has been one of the most cited papers from my laboratory, eclipsing the original ‘Nature’ paper describing clonality.
Spanning the same time frame, Marie Laure Darde and her colleagues examined a collection of clinical T. gondii isolates from France, also concluding that the population was highly structured (Dardé et al., 1992; Ajzenberg et al., 2002a, 2002b). Comparison of our studies revealed that the same three lineages predominate in North America and Europe. Importantly, Dan’s tree provided the framework for many subsequent findings including: i) strains differ in virulence and this can be mapped genetically (Su et al., 2002; Saeij et al., 2006; Taylor et al., 2006), ii) strains that did not fit the clonal groups are in fact members of distinct lineages that are common in other geographic regions (Lehmann et al., 2006; Khan et al., 2007), iii) the close genetic distance between the lineages reflects a recent origin (Su et al., 2003; Khan et al., 2006), and iv) the clonal lineages were generated by only a few genetic crosses in the wild, from highly similar parental strains (Grigg et al., 2001; Boyle et al., 2006).
While it might be true that a Shetland pony could have drawn that original tree, it would take many years of significant effort by more than a few good minds to unravel the mysteries underlying that simple tripartite arrangement.
3. Genetic mapping version 2
3.1. Despite and by the odds
Along with Dan Howe’s finding of the three major lineages, we also recognized that the different clonal groups had very different phenotypic correlates (Howe and Sibley, 1995). Notably type I strains were acutely virulent in the mouse model. However, such strains were rare in the wild and most clinical cases of toxoplasmosis were caused by type II strains, which were relatively avirulent in mice. Type III strains were even less virulent in mice and only rarely isolated from humans. Combined with Elmer’s earlier work, it suggested that genetic approaches might be useful for defining the components that contribute to such phenotypes. We choose “acute virulence” in the mouse model as the trait we would map in a genetic cross between type I and type III. Tachyzoites of type I strains kill outbred mice with a single infectious organism delivered by i.p. needle inoculation (this is extrapolated from the fact that infection at low doses always leads to death with no evidence of serological survivors). In contrast, type III strains are avirulent even at doses 5–6 logs higher than this. Regardless of its clinical relevance, we recognized that acute virulence in mice was extremely stable, the magnitude was sufficiently different between lineages, and the variability low enough that it would be amenable to mapping. Dana Mordue, a postdoctoral fellow who had arrived in the laboratory after graduate work in immunology at the University of Iowa (USA), examined the basis of acute virulence in mice more closely. She found that type I strains outgrew and out-disseminated the slower type II and III strains, yet somewhat paradoxically, type I strains elicit a strong Th1 response, eventually leading to death by cytokine shock (Mordue et al., 2001). Antonio Barragan, who had received his medical degree and doctoral training in malaria at the Karolinska Institute (Sweden), went on to show that enhanced motility and migration are also characteristic of type I strains (Barragan and Sibley, 2002). It seemed logical that such properties could be related to virulence and these rapidly became part of our genetic analysis. Other more specific attributes of the host response to infection have since come to light and these are discussed in more detail by Jon Boyle and Jay Radke (2009).
In pursuing the mouse model of toxoplasmosis, we reasoned that even if the differences were specific to mice, identifying the gene(s) involved might still be informative about disease in other hosts including humans. Others differed in this view indicating that the study of mouse virulence was medically unimportant given that mice are not taxpayers (Frenkel and Ambroise-Thomas, 1997). Likewise, the fruit flies of Morgan and peas of Mendel were not taxpayers, yet study of these model genetic systems has advanced our knowledge of human physiology and medicine in many ways that could not have been anticipated. Of course model organisms will not always yield relevant answers, but they allow us to probe the important questions in ways that cannot be addressed directly in humans.
The idea that specific genes would mediate differences in virulence and that one could map these genetically was met by more than a little criticism by experts in the field. First of all, they countered that passage history can affect virulence and the enhanced virulence of type 1 strains was likely to be just an artifact of laboratory passage (Frenkel and Ambroise-Thomas, 1997). However, many type I isolates presented with acute virulence in early passage, as found by Brian Danneman, and later confirmed by us (Sibley and Boothroyd, 1992b). Moreover, Alfred Sabin’s original report on congenital toxoplasmosis contains a description that mice died on primary isolation of the RH strain (Sabin, 1941). There was however a legitimate concern that the RH strain was not suitable for genetic crosses, since it has been passed extensively and has lost the ability to undergo differentiation in the cat. This property is not unique to type I strains, as it had been shown that as few as 30 passages in mice can give rise to loss of sexual development (Frenkel et al., 1976). For this reason, we chose the GT1 stain, isolated from a goat, found to be virulent on early passage and able to complete the life cycle (Dubey, 1980).
Other objections were also raised to our model for explaining phenotypic differences based on virulence genes. It was questioned whether a single gene would have a measurable contribution to a complex trait like virulence outside the context of the genetic background. This view espoused the idea that the interaction between genes was more important than the intrinsic nature of the genes themselves. Support came from analysis of the progeny from a cross between types II and III, both of which are relatively avirulent. As a result of reassortment of genes, several of the progeny showed enhanced virulence, albeit tested in inbred mice, which are inherently more susceptible (Grigg et al., 2001). This suggested that perhaps type I strains were only virulent because they contained a suitable combination of genes, which under different circumstances would not mediate pathology. While clearly important, the idea of genetic interaction is not mutually exclusive to the possibility that some genes would work in a variety of different genetic backgrounds or even irrespective of genetic background. Such topics made for heated discussions between proponents of various models, despite the fact that evidence at this stage was largely anecdotal.
We eventually realized that the real challenge was not predicting which model was correct, but devising a system to define the number of different contributing genes and estimate the magnitude to which they contribute to a complex trait. Fortunately others, working in model systems, had paved the way once again. Quantitative trait locus (QTL) mapping provides a powerful means of assessing such relationships and had been used for complex traits in animals and plants (Lander and Botstein, 1989; Churchill and Doerge, 1994; Lander and Kruglyak, 1995; Lynch and Walsh, 1998). We reasoned that it might be possible to use a similar approach to define virulence in T. gondii. The beauty of this approach is that it can parse the contribution of different genes, define whether they have small or large effects, and even predict how they interact. We were so excited to try QTL mapping, we paid little attention to advice from colleagues working on mouse genetics. They assured us that there would likely be dozens of genes involved, each with a small effect, and that mapping such traits would require hundreds of F1 progeny; in short a nearly impossible task. Had we been more cognizant that this scenario was actually the probable one, it is unlikely that we would have pursued the strategy with the eagerness that we did.
3.2. Rebuilding the genetic map and the heritability of virulence
The original cross between the type I GT1 strain and the type III CTG strain was performed in collaboration with J.P. Dubey at the United States Department of Agriculture (USA) (Fig. 2A). Following in Elmer’s footsteps, Dan Howe first generated mutants that were resistant to fluorodeoxyuridine following chemical mutagenesis with ethyl nitrosourea (Pfefferkorn and Pfefferkorn, 1979). Because the GT1 strain is highly virulent and kills mice, chronic infections were obtained by infecting outbred mice and treating with sulfadiazine to allow for survival. The other parent for the cross was a double mutant of the CTG stain that was resistant to arabinoside A and sinefungin (referred to as ARAR/SYNR), as previously isolated by Elmer in one of his early crosses (Pfefferkorn and Pfefferkorn, 1980). Following feeding of tissues cysts from the two parental strains to kittens, oocysts were collected, sporulated in vitro, and used to isolate progeny by in vitro culture in monolayers of human foreskin fibroblasts. We isolated the first set of doubly resistant mutants by co-culture in FUDR/ARA or FUDR/SYN, verifying that these were in fact recombinants. Dan had moved on to a position as Assistant Professor at the University of Kentucky (USA) before it came time to analyze the clones in detail, and this task was taken up by Chunlie Su, a new postdoctoral fellow who had recently arrived in the laboratory after completing his Ph.D. in Pathobiology at Pennsylvania State University (USA).
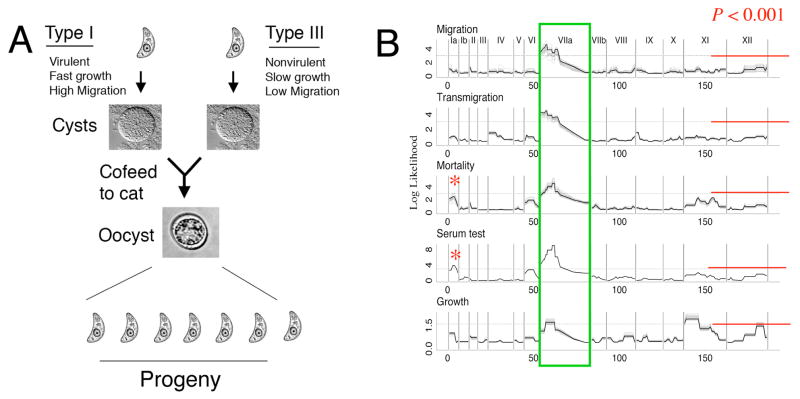
Fig. 2.
Diagram of genetic cross and outcome of mapping studies. A) Diagram of the genetic cross performed between types I and III parasites, highlighting key phenotypic differences between those. A complete listing of the genetic crosses performed to date can be found at http://toxomap.wustl.edu/Experimental_crosses.htm. B) Remarkably, a number of important phenotypes relating to pathogenesis converged on the central region of chromosome (Chr.) VIIa (boxed). Included in this were migration, transmigration, mortality and serum response. A second minor quantitative trait locus (QTL) for serum and mortality was detected on Chr. 1a. In contrast growth was mapped to several different chromosomes, marked by distinct QTLs that had statistical significance. The solid line indicates the association between phenotypes and markers listed along the X axis, and grouped by chromosome (numbers listed at top). Shaded lines represent 95% confidence intervals as estimated by permutation tests. Reproduced with permission from Taylor et al. (2006).
By this time PCR had become much more routine and less costly, and it offered the potential to accelerate the analysis if we could amplify regions flanking the polymorphic restriction sites, digest those with enzymes and resolve the fragment on ethidium bromide-stained gels. Chunlei set about converting the Stanford era Southern blotting markers to PCR-based analysis. This required that the DNA segments be separately PCR-amplified, sequenced, aligned and analyzed for unique restriction sites. For some markers, Chunlei could identify the exact site that had been previously mapped by Southern blot. In other cases, the polymorphic site lay outside the boundaries of the DNA fragments used for probing. In such cases, sometimes Chunlei could identify new polymorphisms that could be distinguished by restriction digests within the original probe, but in other cases these markers were retired in favor of new ones that had to be developed from scratch. It is important to keep in mind that there was no whole genome sequence available at the time, and only a limited set of expressed sequence tags (ESTs) from which to derive new single nucleotide polymorphisms (SNPs). Chunlei combed through these data to generate some new markers, sequenced genes from other strains based on GenBank entries, and also obtained some markers from Tovi Lehmann at the NIH, who was investigating microsatelite markers for typing of T. gondii (Lehmann et al., 2000).
Overall, Chuneli converted 53 markers to PCR format and typed 26 progeny from the types IxIII cross. He also put each of the progeny clones through outbred mice to derive mortality data based on inoculation with different dilutions of tachyzoites, thus estimating their virulence. The linkage mapping data files quickly outgrew the hand-drawn genotype maps posted on the wall, that we had used previously. Instead, Chunlei turned to MapMaker (Lander et al., 1987) to generate maps of the chromosomes defining 13 linkage groups. Mapping of acute virulence using MapManager QTX (Manly et al., 2001) revealed this trait was associated with a strong QTL on chromosome (Chr.) VII and a minor peak on Chr. IV (Su et al., 2002). The QTL analysis suggested a single locus on Chr.VII was predominately involved; however, there was no physical underpinning for the linage map and no indication of the distance spanned by the flanking markers. While these data provided us with the first solid evidence that it might actually be possible to map the genetic basis of virulence, we were still a long way from finding a gene.
While various doubts were raised about the feasibility of mapping virulence genes, there were nonetheless several bright lights along the way that kept us on course. One particularly stabilizing influence was Jim Ajioka, whose background in fly genetics gave him an expertise that we lacked and whose opinion was consequently quite influential. Jim was enthusiastic about our approach, and many times reassured with comments such as genetics would show the way. Other insights came from key experiments along the way, even when they did not lead to publication. For example, Dan Howe reasoned that if the phenotype induced by type I strains was dependent on the host response during infection (i.e. either under- or over-response of the immune system), then co-infection with both type I and type III strains might result in a permissive host where both strains would grow equally well. This proved not to be the case; rather type I would consistently outgrow type III in a mixed infection. We interpreted this to mean that virulence was intrinsic to the genotype and hence mediated by specific genes. Of course other interpretations were possible, but this belief kept us going when progress seemed to move glacially.
3.3. Assembling the genome: another late night with Dr. Roos
When we arrived at the VIIth International Toxoplasma meeting in Tarrytown, New York (USA) in 2003, we came with the sequence files of Chunlie’s markers. David Roos and his team had been working with Ian Paulsen at The Institute for Genomic Research (TIGR, USA) to assemble the whole genome reads from the type II ME49 strain of T. gondii. The assemblies represented about six-fold coverage at the time, and had been grouped into several hundred scaffolds ranging in size from a few kb to several mb. However, without a physical map, there was no way to assemble the genome. Unlike the Stanford era probes, which were simply anonymous pieces of DNA, Chunlei’s markers were sequence-based. Thus, we recognized that the genetic markers and linkage map might be used to assemble the scaffolds into an ordered genome. David Roos, Bindu Gajria, Chunlei and I gathered around David’s laptop computer to try this the first night. Typical of David’s relaxed schedule, things didn’t really get going until late in the evening, but it soon became evident that this process was not only possible but that it was going to go remarkably quickly. That is, other than frequent interruptions for David to espouse at length on various tangential, albeit interesting side topics. As the night stretched on into morning, I retired and left things for David and the others to complete. In the morning, David presented a preliminary alignment to the T. gondii genome that showed more than half of the large scaffolds could be pinned to the genetic map, even at this early stage. We knew then that genetic mapping and whole genome sequencing were going to converge into a single unified map. They would buttress each others shortcomings and allow us to tie linkage analysis to a physical framework and hence map genes. We always believed this would work, yet it might have failed if the genome assemblies were inaccurate due to repeats, if the genetic linkage map was biased due to strong epistasis, or if coverage of either was too scant to make meaningful connections. Some of these problems did and do exist on a small scale, but this did not stop us from proceeding.
As new genome assemblies became available from TIGR and the linkage map improved, we updated the genome map through repeated iterations of this process. The rapid progress in assembling the genome map would not have been possible without the complete and open cooperation between the groups. Later efforts to add sequences derived from the ends of bacterial artificial chromosomes and cosmid clones that contained large genomic inserts provided greater closure and refinement of the map, and these efforts were contributed to selflessly by many participants. Of all the participants, Chunlei Su deserves a great deal of credit for advancing things to the point where this integration became a reality. Without his efforts to convert the markers, the Stanford map would have remained disconnected from the genome sequence, which would have remained unassembled in many fragments.
3.4. Minding the gaps
While we could now account for large blocks of the genome, it was clear we lacked markers for many scaffolds, others were not linked yet to the genome, and it still remained possible there were whole chromosomes we were missing. Additionally, the region on Chr. VII that was associated with virulence contained a huge gap of more than 2 mb that was beneath the peak QTL. In short, while we had come a long way, there was still no way to link phenotype to gene. Around this time, Chunlei moved to the University of Tennessee (USA) as an Assistant Professor, and the task of completing the genetic map fell to two new postdoctoral fellows, Asis Khan and Sonya Taylor. Asis had done his graduate work at Jadavpur University (India) on molecular epidemiology of shiga toxin producing Escherichia coli and wanted to pursue experimental genetics. Sonya had trained with Andy Tait and Mike Turner at Glasgow University (United Kingdom), mapping arsenical resistance in trypanosomes using high throughput genotyping of genetic crosses. Neither was averse to hard work and together they teamed up to complete the genetic map.
We faced two familiar problems: not enough markers and not enough progeny. Fortunately, with the increased coverage of the genome available from TIGR, the major scaffolds were coming into focus. We could now see the distribution of markers across large physical regions for the first time. This enabled us to set a course of placing a genetic marker every 300 kb across the genome, or what would turn out to be approximately every 3 cM. Asis Khan took up this challenge. We had previously generated a large set of ESTs for T. gondii and fortunately these were split over different strains (Ajioka et al., 1998; Li et al., 2003), allowing us to mine them for polymorphisms. Asis mapped ESTs to the genome scaffolds provided by TIGR, identified polymorphisms and converted these into PCR-based markers that were analyzed by restriction digestion and gel electrophoresis. In total, we generated ~ 250 markers that could be typed over the combined crosses. Meanwhile, Sonya took up the parallel challenge of developing more progeny. As fortune would have it, we had one remaining vial of a separate genetic cross between types II and III that was conducted by Elmer and still stored in our liquid nitrogen tank after nearly 10 years. Elmer had since closed his laboratory and all remaining stocks had been lost or disposed of. We thawed this vial from the 1996 cross, which became known as c96, and Sonya generated a series of unique clones and genotyped those using the set of informative markers.
More than 25,000 PCR-amplifications, digestions, gel lanes, and countless hours of analyses later, we repeated the process of linking the genetic map to sequenced scaffolds for the final time. Collectively, the data included ~ 250 markers analyzed across 71 progeny from three separate crosses, including both types II×III and I×III combinations (Khan et al., 2005). We generated a composite genome map that now defined 14 separate linkage groups, stretched the total genetic map to almost 600 cM, and defined the parameters of recombination among the three major lineages: it became the foundation for the chromosome-centric view of the genome that is housed in ToxoDB (http://ToxoDB.org) (Kissinger et al., 2003; Gajria et al., 2007).
4. Finding genes that control important phenotypes
4.1. The hunt for virulence
In combining our data to complete a combined genetic map we enlisted the help of John Wootton at National Center for Biotechnology Information (USA), who had also had a hand in the genetic linkage map of Plasmodium falciparum (Su et al., 1999). John helped us refine our mapping programs and provided key insight into the statistical methods. Working with John was a treat because of his critical eye for detail and intuitive sense for biology. When I updated him with our progress at a meeting we attended, he astutely remarked to me that now that we had our map, Sonya was likely to be avidly tracking the virulence gene. John also has a keen sense of scientific adventure.
Although we now had a complete genome map, including a physical scaffold underlying the virulence region on Chr. VIIa, the recombination parameters of T. gondii were such that our search for a gene was still in jeopardy. The area between the closest two markers flanking this QTL remained more than 2 mb in size, containing hundreds of candidate genes. Getting closer could only be achieved by finding progeny with crossovers within this region. Sonya returned to the freezer and starting isolating new progeny. In total she genotyped ~ 1,000 clones, in the end finding three clones that had recombinations across the central region of Chr. VIIa. During this several month-long process, Sonya rarely strayed far from either the bench doing PCR or the tissue culture hood cloning progeny. These new clones were also tested for acute virulence and enhanced migration, in collaboration with Antonio Barragan who was now back at the Karolinska Institute (Fig. 2A). Remarkably, these phenotypes converged on a single region on Chr. VIIa (Fig. 2B), although fine mapping later revealed that migration was controlled by a separate locus from acute virulence (Taylor et al., 2006). Sonya completed genotyping of these new clones, narrowed the virulence region to 140 kb, and appeared in my office with a list containing 23 genes. As we considered the candidate list, the gene at the right end of the region caught our eye: annotated as a rhoptry-like protein, it was extremely polymorphic and characterized by a high degree of type I SNPs. It quickly became our top candidate. While progress up until this point had taken many years of slow accrual, things were about to accelerate dramatically.
I attended the Biology of Parasitism at the Marine Biological Laboratory (MBL) in Woods Hole (USA) in July 2005 and gave a lecture about our efforts to map and analyze virulence in Toxoplasma. Hiba El Hajj, a student in Jean Francois Dubremetz’ laboratory at the Université de Montpellier (France) was enrolled for the summer course at the MBL. Knowing that she worked on rhoptry proteins, I told her that one of our best candidates for virulence was a potential rhoptry protein, and I wondered if she knew anything about it. It turned out it was one of several proteins she had been working on, and indeed one they were quite interested in. After the course, I wrote to Jean Francois and asked if we could obtain an expression construct that Hiba had generated, as it might be useful in testing the role of this gene in the phenotype we had been mapping. That fall they generously agreed to send the plasmid, we generated transgenes in the type III background using reverse genetic approaches that had been developed over the years (see the contribution by John Boothroyd (2009) for more details), and began testing them in animals. The first mice began dying the following winter-spring, confirming that we had our gene at last. Fortuitously, this candidate, now called ROP18 by the Dubremetz group, contained a putative serine threonine kinase domain. We began working furiously to localize it to rhoptries, examine secretion into host cells and test the role of its kinase activity in virulence. Notably, with the help of Michael White’s laboratory at the University of Montana (USA), we also analyzed differences in growth rate, which turned out to be multigenic and contribute only a small effect to acute virulence (Fig. 2). As the mouse geneticists had predicted, some complex traits are truly multigenic, lacking contribution from one or two major loci, and hence could be extremely difficult to map to specific genes.
That spring I also got an email from John Boothroyd, indicating that they had their eye on the region of Chr. VIIa that Chunlei had mapped some years ago and thought it might contribute to a phenotype they had been mapping. We were aware of their efforts as Jeroen Saeij had presented his work at the Molecular Parasitology Meeting in Woods Hole the previous fall, and we had provided them with the mapping data and progeny from the new crosses during the development of the genome map. Their approach was novel; they were mapping genes in the parasite that affected host gene transcription using microarrays. This ultimately led to the discovery of ROP16, another rhoptry protein that is secreted into the host cell and affects STAT3/6 phosphorylation (Saeij et al., 2007). With the help of Jon Boyle, they were also analyzing differences in virulence among the progeny of the types IIxIII crosses. Among four to five loci that they identified by QTL analysis, one coincided with the virulence locus we had previously mapped on Chr. VII. Again, reverse genetic approaches quickly confirmed that expression of the type II ROP18 gene in the type III background could increase virulence. Jon Boyle was also the first to recognize that differences in the upstream regions were responsible for the under-expression of this gene in the type III lineage (Boyle et al., 2008). More recently, we have shown this is due to a loss of the upstream region in types I and II that was associated with increased expression and enhanced virulence (Khan et al., 2009).
Our convergence on ROP18 seemed like a remarkable coincidence at the time, since we thought we were mapping much different phenotypes in entirely different genetic backgrounds. The outcome also supported both previous models that had been suggested for explaining the genetic basis of differences in virulence: small contributions from a large number of loci as in the case of types IIxIII differences, and a single predominant locus as in the case of types IxIII differences. John and I presented our work at the Gordon Conference on Parasitism in June of 2006, concluding that ROP proteins were potent mediators of virulence and they operated by secretion into the host cell following parasite invasion. I recall that Piet Borst who was in attendance summed up with the comment the awesome power of genetics. Over the next few months, we communicated quite often with the Stanford group and worked collectively to submit back-to-back papers that were ultimately published in ‘Science’ (Saeij et al., 2006; Taylor et al., 2006). Hiba and Jean Francois would add their report indicating that over-expression of ROP18 enhanced parasite growth and that it was an active kinase that decorated the parasitophorous vacuole membrane (El Hajj et al., 2007). While there was some overlap between our studies, this process was marked by a friendly level of competition that ended up bringing out the best in each group. It was a fitting end that after 20 years we had finally demonstrated the potential for forward genetics by arriving at the same family of genes, yet by very different routes. Of course, this is not so much the end as the beginning and there remain many challenges to figuring out the various functions of ROPs. While the role of two active ROP kinases was shown conclusively by genetics, their biochemical functions remain elusive. The recent publication of several structures for ROP family members and a model for regulation of ROP18 activity will aid in further defining these biochemical functions (Qiu et al., 2008). Moreover, phylogenetic and hidden Markov model analyses conducted by Lucia Peixoto in David Roos’ laboratory, have shown there are more than 30 ROP kinase family members in Toxoplasma. Further studies will be needed to explore the extent to which they contribute to infection in different hosts including humans, and how widely distributed and variable they are among natural isolates of the parasite.
4.2. Rhoptries and déjà vu all over again
Science often converges on similar topics and we were certainly not the first to recognize the importance of rhoptries. Indeed, Jean Francois has devoted considerable effort in his laboratory to elucidating their functions (Dubremetz, 2007). Additionally Peter Bradley, while a postdoctoral fellow in John Boothoyrd’s laboratory, had generated an enormously useful proteome of rhoptries (Bradley et al., 2005). This and other similar efforts greatly accelerated progress in the genetic mapping of virulence factors due to the availability of reagents and awareness of the potential importance of rhoptry proteins in the biology. Additionally, other events critically shaped our thinking and yet are not fully documented in the published literature. One key early contribution was made by Sebastian Hakannson, a postdoctoral fellow who had a background in Yersina protein secretion, when he joined our laboratory in St. Louis. Seb described the process by which rhoptries discharge part of their contents directly into the host cell cytosol to form small, empty vesicles that he termed “evacuoles” (Håkansson et al., 2001). He went on to show that evacuoles take on the fate of the parasitophorus vacuole, avoiding fusion with lysosomes and endosomes, and recruiting endoplasmic reticulum and mitochondria (Håkansson et al., 2001). He also speculated that rhoptry secretion might be important for delivering important effector proteins into the host cell, analogous to type III secretion in bacteria. Prior to returning to Umea University (Sweden) in 2000, Seb had also discovered in the early ESTs from T. gondii a putative tyrosine kinase with homology to a rhoptry protein. He surmised that it would likely be a key effector released from rhoptries and altering host cell function. Since we had no intention to work on rhoptries, I readily agreed that he was free to take this project with him. For various reasons, Seb’s project never came to fruition, although his insight proved prescient. When we confirmed the role of ROP18 in virulence, I went to his old notebook to check what gene he had found. It was not ROP18, the gene associated with virulence, or ROP16 that had been implicated in altering host cell transcription, but rather ROP17, a related rhoptry protein that is also predicted to be an active kinase of yet unknown function. More than being a good genome sleuth, Sebastian deserves the credit for correctly predicting the role of ROP proteins as important effectors, years before genetics and the rest of us caught up.
5. Perspectives for the future
As aptly said by Donald Rumsfeld “there are also unknown unknowns. There are things we don’t know we don’t know”. (Press Conference at NATO Headquarters, Brussels, Belgium, June 6, 2002; http://www.defenselink.mil/transcripts/transcript.aspx?transcriptid=3490)
While not sophisticated, our approach to genetic mapping had been reliable, scalable and sufficient for the job at hand. However, with the advent of the genome and post-genomics, it has rapidly become obsolete. In planning the Affymetrix microarrays for T. gondii, Amit Bahl in David Roos’ laboratory had the foresight to include the genetic markers as well as SNPs defined by the EST database in the design of the chip. It didn’t take long to show that hybridization of genomic DNA to the array can provide the genotype for many markers across the genome simultaneously in a single experiment. Mike Behnke in Michael White’s laboratory, along with Amit Bahl in David Roos’ laboratory have gone on to show, together with John Wootton, that these array-based markers provide an enhanced resolution genetic map that will replace the PCR-based typing method we had worked so hard to achieve. As a result of this leap, the next round of mapping will be far easier and have greater precision, and is sure to reveal new discoveries not foreseen by prior technology.
While building the genetic maps and generating the progeny was a difficult task, it should have compounding effects for mapping future phenotypes. First of all, all of the data is publicly housed, allowing ready access (http://www.toxodb.org/; http://toxomap.wustl.edu/). Moreover, the clones have been deposited in the Biodefense and Emerging Infections Research Resources Repository (BEIR) repository (www.beiresources.org/), making them available to other investigators. Unlike model organisms such as mice, the F1 generation of T. gondii can be propagated indefinitely, cryopreserved and reanalyzed at a later date to explore additional phenotypes. Some caution needs to be observed however, as phenotypes may change with repeated passage. The limited phenotypes explored to date have relied on mouse virulence using needle inoculation of tachyzoites, an unnatural route of infection. Significant differences may also occur following oral infection with bradyzoites (or sporozoites), in either intestinal pathology, induction of immune responses, or in chronicity. Exploitation of these more natural routes of infection and pathogenesis models may reveal genes that are of importance to other hosts, including humans. While transcriptional responses have been explored in some detail, additional, signaling pathways in host cells may be differentially affected by strains of T. gondii and these could be mapped genetically. As a general guide, phenotypes that can easily be mapped by QTL analysis need to have a mean difference between groups that is greater than the within group variance. Many potential phenotypes could be piloted in the parental clones and then typed on the progeny.
While Elmer’s original methods for performing genetic crosses have been tremendously useful, they remain difficult and require the use of animals that ultimately must be sacrificed. Thus far, the available crosses are limited and it would be very beneficial to add a types IxII cross and perform crosses with other more diverse groups such as those from South America. Development of an in vitro system would both speed this process and reduce the need to use animals. Significant barriers remain to this goal as T. gondii only undergoes sexual development in intestinal epithelial cells of the cat. Suitable cell lines do not exist and their isolation should be one primary initial goal. Additionally, it may be possible to coax development in other cell types, under appropriate growth conditions. Attempts to develop in vitro genetics should be a high priority. These and other important technical developments will drive the future of genetics in T. gondii.
Acknowledgments
I am extremely indebted to many members of my laboratory and to many colleagues who have contributed generously to the events described here. I am also grateful for the funding my laboratory has received over the years from the National Institutes of Health, the United States Department of Agriculture, the American Foundation for AIDS Research, the American Heart Association, Merck Research Laboratories, and the Burroughs Wellcome Fund. Our approaches were often not hypothesis driven, pushed the limits of feasibility and were exceedingly overly ambitious. Only the support of unusually open-minded peer reviewers made it possible to pursue these projects.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajioka JA, Boothroyd JC, Brunk BP, Hehl A, Hillier L, Manger ID, Overton GC, Marra M, Roos D, Wan KL, Waterston RH, Sibley LD. Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Gen Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Bañuls AL, Tibayrenc M, Dardé ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Intl J Parasitol. 2002a;32:27–38. doi: 10.1016/s0020-7519(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Cogné N, Paris L, Bessieres MH, Thulliez P, Fillisetti D, Pelloux H, Marty P, Dardé ML. Genotype of 86 Toxoplasma gondii isolates associated with human congenital toxoplasmosis and correlation with clinical findings. J Infect Dis. 2002b;186:684–689. doi: 10.1086/342663. [DOI] [PubMed] [Google Scholar]
- Barragan A, Sibley LD. Transepithelial migration of Toxoplasma gondii is linked to parasite motility and virulence. J Exp Med. 2002;195:1625–1633. doi: 10.1084/jem.20020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MW, Arrizabalaga G, Boothroyd JC. Ionophore-resistant mutants of Toxoplasma gondii reveal host cell permeabilization as an early event in egress. Mol Cell Biol. 2000;20:9399–9408. doi: 10.1128/mcb.20.24.9399-9408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd JC. Toxoplasma gondii: 25 years and 25 major advances for the field. Int J Parasitol. 2009;39 doi: 10.1016/j.ijpara.2009.02.003. [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Radke JR. A history of studies that examine the interactions of Toxoplasma with its host cell: Emphasis on in vitro models. Int J Parasitol. 2009;39 doi: 10.1016/j.ijpara.2009.01.008. [in this issue] [DOI] [PubMed] [Google Scholar]
- Boyle JP, Rajasekar B, Saeij JPJ, Ajioka JW, Berriman M, Paulsen I, Sibley LD, White M, Boothroyd JC. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc Natl Acad Sci (USA) 2006;103:10514–10519. doi: 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Saeij JP, Hrada SY, Ajioka JW, Boothroyd JC. Expression QTL mapping of Toxoplasma genes reveals multiple mechanisms for strain-specific differences in gene expression. Euk Cell. 2008;7:1403–1414. doi: 10.1128/EC.00073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in T. gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen AWCA, Overdulve JP, Van der Ploeg M. Determination of nuclear DNA of five eucoccidian parasites, Isospora (Toxoplasma) gondii, Sarcocystis cruzi, Eimeria tenella, E. acervulina, and Plasmodium berghei, with special reference to gametogenesis and meiosis in I. (T.) gondii. Parasitol. 1984;88:531–553. doi: 10.1017/s0031182000054792. [DOI] [PubMed] [Google Scholar]
- Dannemann B, McCutchan JA, Israelski D, et al. Treatment of toxoplasmic encephalitis in patients with AIDS: a randomized trial comparing pyrimethamine plus clindamycin to pyrimethamine plus sulfonamides. Ann Intern Med. 1992;116:33–43. doi: 10.7326/0003-4819-116-1-33. [DOI] [PubMed] [Google Scholar]
- Dardé ML, Bouteille B, Pestre-Alexandre M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiological implications. J Parasitol. 1992;78:786–794. [PubMed] [Google Scholar]
- Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- Dubey J. Mouse pathogenicity of Toxoplasma gondii isolated from a goat. Am J Vet Res. 1980;41:427–429. [PubMed] [Google Scholar]
- Dubey JP, Frenkel JF. Cyst-induced toxoplasmosis in cats. J Protozool. 1972;19:155–177. doi: 10.1111/j.1550-7408.1972.tb03431.x. [DOI] [PubMed] [Google Scholar]
- Dubey JP. History of the discovery of the life cycle of Toxoplasma gondii. Int J Parasitol. 2009;39 doi: 10.1016/j.ijpara.2009.01.005. [in this issue] [DOI] [PubMed] [Google Scholar]
- Dubremetz JF. Rhoptries are major players in Toxoplasma gondii invasion and host cell interaction. Cell Microbiol. 2007;9:841–848. doi: 10.1111/j.1462-5822.2007.00909.x. [DOI] [PubMed] [Google Scholar]
- El Hajj H, Lebrun M, Arold ST, Vial H, Labesse G, Dubremetz JF. ROP18 is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathogens. 2007;3:e14. doi: 10.1371/journal.ppat.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DJP. Identification of faecal transmission of Toxoplasma gondii: Small science, large characters. Int J Parasitol. 2009;39 doi: 10.1016/j.ijpara.2009.01.003. [in this issue] [DOI] [PubMed] [Google Scholar]
- Frenkel JK, Ambroise-Thomas P. Genomic drift of Toxoplasma gondii. Parasitol Res. 1997;83:1–5. doi: 10.1007/s004360050197. [DOI] [PubMed] [Google Scholar]
- Frenkel JK, Dubey JP, Hoff RL. Loss of stages after continuous passage of Toxoplasma gondii and Besnoitia jellisoni. J Protozool. 1976;23:421–424. doi: 10.1111/j.1550-7408.1976.tb03799.x. [DOI] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, AJM, Pinney DF, Roos DS, Stoeckert CJ, Wang J, Brunk BP. ToxoDB: an integrated Toxoplasma gondii database resource. Nucl Acids Res. 2007;36:D553–556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science. 2001;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- Gubbels JM, Lehmann M, Muthalgi M, MEJ, Brooks CF, Szatanek T, Flynn J, Parrot B, Radke J, Streipen R, White MW. Forward genetic analysis of the apicomplexan cell division cycle in Toxoplasma gondii. PLoS Path. 2008;4:e36. doi: 10.1371/journal.ppat.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson S, Charron AJ, Sibley LD. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 2001;20:3132–3144. doi: 10.1093/emboj/20.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- Khan A, Bohme U, Kelly KA, Adlem E, Brooks K, Simmonds M, Mungall K, Quail MA, CA, Chillingworth T, Churcher C, Harris D, Collins M, Fosker N, Fraser A, Hance Z, Jagels K, Moule S, Murphy L, O’Neil S, Rajandream MA, Saunders D, Seeger K, Whitehead S, Mayr T, Xuan X, Watanabe J, Suzuki Y, Wakaguri H, Sugano S, Sugimoto C, Paulsen I, Mackey AJ, Roos DS, Hall N, Berriman M, Barell B, Sibley LD, Ajioka JW. Common inheritance of chromosome Ia associated with clonal expansion of Toxoplasma gondii. Gen Res. 2006;16:1119–1125. doi: 10.1101/gr.5318106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM, Sibley LD. Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc Natl Acad Sci (USA) 2007;104:14872–14877. doi: 10.1073/pnas.0702356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Taylor S, Ajioka JW, Rosenthal BM, Sibley LD. Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are viruelnce in mice. PLoS Genetics. 2009 doi: 10.1371/journal.pgen.1000404. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Taylor S, Su C, Mackey AJ, Boyle J, Cole RH, Glover D, Tang K, Paulsen I, Berriman M, Boothroyd JC, Pfefferkorn ER, Dubey JP, Roos DS, Ajioka JW, Wootton JC, Sibley LD. Composite genome map and recombination parameters derived from three archetypal lineages of Toxoplasma gondii. Nuc Acids Res. 2005;33:2980–2992. doi: 10.1093/nar/gki604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger JC, Gajria B, Li L, Paulsen IT, Roos DS. ToxoDB: Accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Blackstone CR, Parmley SF, Remington JS, Dubey JP. Strain typing of Toxoplasma gondii: comparison of antigen-coding and housekeeping genes. J Parasitol. 2000;86:960–971. doi: 10.1645/0022-3395(2000)086[0960:STOTGC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Marcet PL, Graham DH, Dahl ER, Dubey JP. Globalization and the population structure of Toxoplasma gondii. Proc Natl Acad Sci (USA) 2006;103:11423–11428. doi: 10.1073/pnas.0601438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Brunk BP, Kissinger JC, Pape D, Tang K, Cole RH, Martin J, Wylie T, Dante M, Fogarty SJ, Howe DK, Liberator PA, Diaz C, Anderson J, White M, Jerome ME, Johnson EA, Radke JA, Stoeckert CJ, Jr, Waterston RH, Clifton SW, Roos DS, Sibley LD. Gene discovery in the Apicomplexa as revealed by EST sequencing and assembly of a comparative gene database. Gen Res. 2003;13:443–454. doi: 10.1101/gr.693203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, Sinauer Associataes, Inc; Sunderland MA: 1998. [Google Scholar]
- Manly KF, Cudmore J, RH, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- Mordue DG, Monroy F, La Regina M, Dinarello CA, Sibley LD. Acute toxoplasmosis leads to lethal overproduction of Th1 cytokines. J Immunol. 2001;167:4574–84. doi: 10.4049/jimmunol.167.8.4574. [DOI] [PubMed] [Google Scholar]
- Morrissette NS, Mitra A, Sept D, Sibley LD. Dinitroanalines bind alpha-tubulin to disrupt microtubules. Molec Bio Cell. 2004;15:1960–1968. doi: 10.1091/mbc.E03-07-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER. Toxoplasma gondii: The enzymatic defect of a mutant resistant to 5-fluorodeoxyuridine. Exp Parasitol. 1978;44:26–35. doi: 10.1016/0014-4894(78)90077-2. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER. Toxoplasma gondii and the biochemistry of intracellular parasitism. Trends Biochem Sci. 1981;6:311–313. [Google Scholar]
- Pfefferkorn ER. Toxoplasma gondii viewed from a virological perspective. In: Englund PT, Sher A, editors. The Biology of Parasitism. Alan R, Liss, Inc; NY: 1988. pp. 479–501. [Google Scholar]
- Pfefferkorn ER. Cell biology of Toxoplasma gondii. In: Wyler DJ, editor. Modern parasite biology. New York, NY: W.H. Freeman; 1990. pp. 26–50. [Google Scholar]
- Pfefferkorn ER, Borotz SE, Nothnagel RF. Toxoplasma gondii: Characterization of a mutant resistant to sulfonamides. Exp Parasitol. 1992a;74:261–270. doi: 10.1016/0014-4894(92)90149-5. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Borotz SE, Nothnagel RF. Mutants of Toxoplasma gondii resistant to atovaquone (566C80) or decoquinate. J Parasitol. 1993;79:559–564. [PubMed] [Google Scholar]
- Pfefferkorn ER, Kasper LH. Toxoplasma gondii: Genetic crosses reveal phenotypic suppression of hydroxyurea resistance by fluorodeoxyuridine resistance. Exp Parasitol. 1983;55:207–218. doi: 10.1016/0014-4894(83)90015-2. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, MEE, McAdams E. Toxoplasma gondii: In vivo and in vitro studies of a mutant resistant to arpinocid-N-oxide. Exp Parasitol. 1988;65:282–289. doi: 10.1016/0014-4894(88)90133-6. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Nothnagel RF, Borotz SE. Parasiticidal effect of clindamycin on Toxoplasma gondii grown in cultured cells and selection of a drug-resistant mutant. Antimicrob Agents Chem. 1992b;36:1091–1096. doi: 10.1128/aac.36.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC. Arabinosyl nucleosides inhibit Toxoplasma gondii and allow the selection of resistant mutants. J Parasitol. 1976;62:993–999. [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC. Specific labeling of intracellular Toxoplasma gondii with uracil. J Protozool. 1977a;24:449–453. doi: 10.1111/j.1550-7408.1977.tb04774.x. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC. Toxoplasma gondii: Characterization of a mutant resistant to 5-fluorodeoxyuridine. Exp Parasitol. 1977b;42:44–55. doi: 10.1016/0014-4894(77)90060-1. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC. The biochemical basis for resistance to adenine arabinoside in a mutant of Toxoplasma gondii. J Parasitol. 1978;64:486–492. [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC. Quantitative studies of the mutagenesis of Toxoplasma gondii. J Parasitol. 1979;65:363–370. [PubMed] [Google Scholar]
- Pfefferkorn ER, Pfefferkorn LC, Colby ED. Development of gametes and oocysts in cats fed cysts derived from cloned trophozoites of Toxoplasma gondii. J Parasitol. 1977;63:158–159. [PubMed] [Google Scholar]
- Pfefferkorn LC, Pfefferkorn ER. Toxoplasma gondii: Genetic recombination between drug resistant mutants. Exp Parasitol. 1980;50:305–316. doi: 10.1016/0014-4894(80)90034-x. [DOI] [PubMed] [Google Scholar]
- Qiu W, Wernimont A, Tang K, Taylor S, Lunin V, Schapira M, Fentress SJ, Hui R, Sibley LD. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 2009 doi: 10.1038/emboj.2009.24. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin AB. Toxoplasmic encephalitis in children. J A Med Assoc. 1941;116:801–807. [Google Scholar]
- Saeij JPJ, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Coller S, Boyle JP, Jerome ME, White ME, Boothroyd JC. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman JD, Pfefferkorn ER. Toxoplasma gondii: Studies of purine synthesis and salvage using mutant host cells and parasites. Exp Parasitol. 1980;53:77–86. doi: 10.1016/0014-4894(82)90094-7. [DOI] [PubMed] [Google Scholar]
- Schwartzman JD, Pfefferkorn ER. Pyrimidine synthesis by intracellular Toxoplasma gondii. Exp Parasitol. 1981;67:150–158. [PubMed] [Google Scholar]
- Schwartzman JD, Pfefferkorn ER. Toxoplasma gondii: purine synthesis and salvage in mutant host cells and parasites. Exp Parasitol. 1982;53:77–86. doi: 10.1016/0014-4894(82)90094-7. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: Clonal expansion driven by infrequent recombination and selective sweeps. Ann Rev Microbiol. 2008;62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Boothroyd JC. Construction of a molecular karyotype for Toxoplasma gondii. Mol Biochem Parasitol. 1992a;51:291–300. doi: 10.1016/0166-6851(92)90079-y. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature (Lond) 1992b;359:82–85. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- Sibley LD, LeBlanc AJ, Pfefferkorn ER, Boothroyd JC. Generation of a restriction fragment length polymorphism linkage map for Toxoplasma gondii. Genetics. 1992;132:1003–1015. doi: 10.1093/genetics/132.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Evans D, Cole RH, Kissinger JC, Ajioka JW, Sibley LD. Recent expansion of Toxoplasma through enhanced oral transmission. Science. 2003;299:414–416. doi: 10.1126/science.1078035. [DOI] [PubMed] [Google Scholar]
- Su C, Howe DK, Dubey JP, Ajioka JW, Sibley LD. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc Natl Acad Sci (USA) 2002;99:10753–10758. doi: 10.1073/pnas.172117099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Haijj EL, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- Walliker D, Carter R, Morgan S. Genetic recombination in malaria parasites. Nature. 1971;232:561–562. doi: 10.1038/232561a0. [DOI] [PubMed] [Google Scholar]
- Walliker D, Carter R, Morgan S. Genetic recombination in Plasmodium berghei. Parasitol. 1973;66:309–320. doi: 10.1017/s0031182000045248. [DOI] [PubMed] [Google Scholar]
- Walliker D, Carter R, Sanderson A. Genetic studies on Plasmodium chabaudi: recombination between enzyme markers. Parasitol. 1975;70:19–24. doi: 10.1017/s0031182000048824. [DOI] [PubMed] [Google Scholar]