Abstract
Aims
We report a comparative study on the mRNA expression of ErbB receptor tyrosine kinases, and in particular ERBB4 transcript variants, in two common paediatric brain tumours: medulloblastoma (MB) and pilocytic astrocytoma (PA).
Methods
While the conventional real-time quantitative PCR was used to measure the expression of ERRBs and ErbB4-processing proteases genes, the LightCycler FRET probes were specifically designed to investigate all of the known ERBB4 juxtamembrane (JM) and cytoplasmic (CYT) transcript variants.
Results
The overall expression of ERBBs suggests that ErbB2/ErbB4 heterodimers and ErbB4 homodimers may be major functional units of the ErbBs in MB, while ErbB2/ErbB3 heterodimers may play a more prominent role in addition to ErbB4-containing dimmers in PA. Different expression patterns of ERBB4 JM transcripts in MB, PA and normal brain were observed. The JM-d variant was only detected in MBs, while JM-c was present in MB and PA but was not identified in normal brain. The expression of cleavable ERBB4 transcript variants was elevated in PAs and MBs compared to normal brain, while mRNA levels of ErbB4-processing proteases were similar in both tumour types and normal brain. This suggests that proteolytic cleavage of ErbB4 may be more common in MB and PA, which leads to signaling events divergent from those in normal brain.
Conclusion
Taken together, these results suggest that ErbB4 processing and function may be altered in brain tumours such as MB and PA via differential expression of JM transcript variants.
Keywords: ErbB4, transcript variants, real-time quantitative PCR, LightCycler hybridization probe, medulloblastoma, pilocytic astrocytoma
Introduction
The ErbB subfamily of receptor tyrosine kinases (RTKs) consists of four receptors: epidermal growth factor receptor (EGFR/ErbB1/Her1), ErbB2 (Neu/Her2), ErbB3 (Her3), and ErbB4 (Her4). Alterations of ErbB1 and ErbB2 can result in oncogenic activity, and they are currently being targeted in cancer therapy [1]. However, the role in tumourigenesis of the most recently characterized ErbB family member, ErbB4, is less clear. For example, an oncogenic role for ErbB4 in breast cancer has been suggested in both cell biological [2-4] and clinical [5, 6] studies, while other findings indicate that ErbB4 can mediate anti-proliferative and differentiation responses in breast cancer cells [7-9] and that its expression is associated with a favorable prognosis [10-12].
Recent studies suggest that these opposing roles may be explained by proteolytic processing of the ErbB4 proteins encoded by alternatively spliced transcript variants. Several alternatively spliced isoforms of ErbB4 have been identified, which vary either in the extracellular juxtamembrane region (four JM isoforms) or the cytoplasmic region (two CYT isoforms).
The JM isoforms differ in their sensitivity to proteolytic cleavage by tumour necrosis factor-α-converting enzyme (TACE). It has been shown that the JM-a isoform with a 23 amino-acid cassette encoded by exon 16 (exon 16a, Fig. 1) represents a cleavable form of ErbB4, whereas the JM-b isoform with a unique 13 amino-acid cassette encoded by an alternatively spliced exon (exon 16b, Fig. 1) is protease resistant [13, 14]. In addition, two rare ErbB4 isoforms, JM-c (with neither the 23 amino acids from JM-a nor the 13 amino acids from JM-b) and JM-d (with both cassettes) were identified in a study of medulloblastomas (MB) [15]. While the JM-c isoform would be expected to be resistant to cleavage and the JM-d isoform cleavable, this has not been demonstrated as yet. The cleavage of ErbB4 by TACE causes receptor ecto-domain shedding from the cell surface [14] and triggers a secondary cleavage by γ-secretase within the membrane region of ErbB4, releasing a soluble intracellular domain (ICD) [16, 17]. The ICD of ErbB4 can have several fates. It may either translocate into the nucleus [16] to regulate gene transcription [18-21], be sequestered and retained in the cytoplasm [22], or function as a BH3-only protein to promote mitochondria-regulated apoptosis [21, 23].
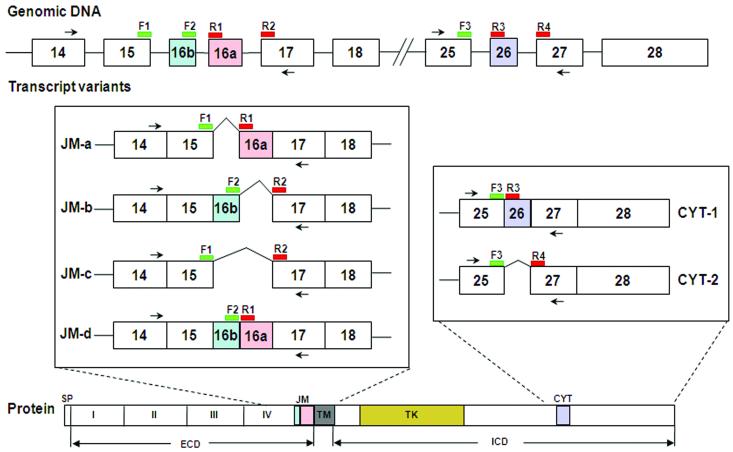
FIGURE 1.
Schematic representation of part of the exon structure of ERBB4 mRNA around the JM and CYT regions, and the design of LightCycler hybridization probes for detecting the expression of alternatively spliced variants. Two primer pairs, PC2338-PC5221 and PC5268-PC5269 (shown by black arrows in the figure) that flank the JM and CYT variable regions, respectively, were used to amplify all the JM and CYT related variants of ERBB4. For quantitative analysis of each JM variant, two donor probes, F1 and F2, were designed according to the 3′end sequences of exon 15 and exon 16b respectively and two acceptor probes, R1 and R2, were designed to match the 5′end sequences of exon 16a and exon 17 respectively. Thus, the dual-labelled probe pair F1-R1 can specifically detect the JM-a variant, while F2-R2 detects JM-b, F1-R2 detects JM-c and F2-R1 detects JM-d. For CYT variant analysis, one donor probe (F3) homologous to the 3′ end sequence of exon 25 and two acceptor probes (R3 and R4) matching 5′end sequences of exons 26 and 27 were used to generate probe pair F3-R3 for CYT-1 and F3-R4 for CYT-2. The donor probes (F1, F2 and F3) were labelled with 3′ Fluorescein. The acceptor probes (R1, R2, R3 and R4) were labelled with 5′ LightCycler Red 610 or 640 fluorescent dyes and the 3′ hydroxyl groups were blocked with a phosphate to prevent Taq DNA polymerase extension. To avoid any steric problems between the donor and acceptor fluorophores on a probe pair, a gap of 1 to 5 nucleotides (4 to 25Å distance) was designed to separate the two probes from each other. During the annealing step of real-time PCR, the PCR primers and the LightCycler probes hybridize to their specific target regions causing the donor dye (on the F probe) to come into close proximity to the acceptor dye (on the R probe). When the donor dye is excited by light from the LightCycler instrument, energy is transferred by Fluorescence Resonance Energy Transfer (FRET) from the donor to the acceptor dye. The energy transfer causes the acceptor dye to emit light at a longer wavelength which can be detected by the LightCycler instrument’s optical unit. The increase in measured fluorescent signal is directly proportional to the amount of accumulating target DNA and can therefore be used for real-time quantitative PCR analysis. SP: signal peptide; I-IV: extracellular domains I-IV; TM: transmembrane domain; TK: tyrosine kinase domain; CYT: cytoplasmic region; ECD: extracellular domain; ICD: intracellular domain.
Alternative splicing also affects the primary structure of the cytoplasmic domain of ErbB4 proteins. The CYT-1 isoform has a docking site for the SH2-domain of phosphatidyl inositol-3 kinase (PI3K) and a putative WW-domain binding motif, which is encoded by an additional exon (exon 26, Fig. 1) not included in the CYT-2 transcript [18, 24, 25]. Thus, the alternatively spliced variants of ERBB4 can potentially generate receptors with eight different primary structures, each providing different signaling capabilities and leading to diverse cellular responses: proliferation, differentiation, apoptosis or survival.
Most of the above findings are based on studies with breast cancers or breast cancer cell lines. The brain is an organ with naturally high expression of ErbB4 [26], and it has been shown that nuclear signaling by proteolytically cleaved ErbB4 directly regulates neural precursor cell fate during development [27]. However, only a few studies have investigated the roles of ErbB4 in brain tumours. A relationship between elevated co-expression of ErbB4 and ErbB2 in MB and increased metastatic disease with reduced survival has been reported [28-30]. One further study suggests that the ErbB4 CYT-1:CYT-2 ratio positively correlates with ErbB2 levels and may be an indicator of MB aggressiveness [31]. Despite the fact that the two rare ERBB4 variants, JM-c and JM-d, were first identified in MB [15], no quantitative analysis of all the known ERBB4 transcript variants has been conducted to date.
Here we present a comprehensive expression analysis of all ErbB family receptors together with a detailed quantitative study of all the known ERBB4 JM and CYT transcript variants in two common paediatric tumour types with very different biological characteristics, the highly malignant medulloblastoma and the relatively benign pilocytic astrocytoma (PA). In addition, this is the first study to examine in parallel the expression of genes whose products are involved in the proteolytic cleavage of ErbB4 isoforms.
Materials and methods
Clinical tissue samples, RNA isolation and cDNA synthesis
RNA from a total of 31 medulloblastoma (MB) samples and 35 pilocytic astrocytoma (PA) samples were included in the analysis. The tumours were resected at the Karolinska Hospital, Stockholm and the Sahlgrenska University Hospital, Gothenburg, Sweden, between 1987-1997. The study was approved by the Ethical Committee of the Karolinska Hospital (No. 91:16) and Cambridge Local Research Ethics Committee, Cambridge, UK (Ref. LREC 03/115). Histopathological classification was revised according to the most recent World Health Organization (WHO) recommendations [32]. All MB cases in the analysis were classic medulloblastomas and there were no desmoplastic or large-cell variants; none of the PA cases showed features consistent with a diagnosis of pilomyxoid astrocytoma. Detailed clinical characteristics and genomic analysis of the MB and PA series used in the study were described previously by M McCabe [33] and D Jones [34] respectively. All tumour pieces were selected for nucleic acid extraction after histological examination to ensure a minimum composition of 70% tumour cells.
Total RNA was extracted from human tissue as described previously [35]. Normal cerebellar RNA was a mixture of equal amounts of RNA extracted from the cerebellum of two individuals who died from non brain-related disease (obtained from the Cambridge University Hospitals Human Tissue Bank, Cambridge, UK). Total brain RNA (FirstChoice Human Brain Reference RNA, Ambion, Austin, TX) was pooled from multiple donors and several brain regions as described by the manufacturer.
RNA quantity and purity was checked by absorbance measurements at A260/280 using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Two micrograms of total RNA were used for each cDNA synthesis reaction. To ensure no genomic DNA contamination, DNase I treatment was performed (amplification grade deoxyribonuclease I, Invitrogen, Paisley, UK) before reverse transcription by SuperScript III Reverse Transcriptase (Invitrogen, Paisley, UK), followed by ribonuclease H (Invitrogen, Paisley, UK) treatment, according to the manufacturer’s protocol. Each cDNA sample was then diluted to 100 μl before being used as a template for quantitative PCR.
Real-time quantitative PCR using SYBR Green I
All the PCR primers used in quantitative PCR are listed in Table 1. Primers for ERBB1, ERBB2 and ERBB3 were as previously described by Junttila et al [36]. All other primers in the study were designed by the authors. Real-time quantitative PCR was carried out using a LightCycler Instrument (Roche Diagnostics, Mannheim, Germany) and LightCycler FastStart DNA Master SYBR Green I (Roche Diagnostics, Burgess Hill, UK), following manufacturer’s recommendations. Briefly, 1 μl of the cDNA solution described above was used in a 10 μl PCR reaction containing 1x SYBR Green I master mix with an appropriate concentration of MgCl2 and primers. All samples were run in duplicate. Each PCR run also included a no-template control as well as a consistent triplicate calibrator comprising a mixture of multiple cDNAs as template, to normalize the data from several PCR runs for the same target cDNA. 18S ribosomal RNA expression was used as an internal reference for relative quantification, as described elsewhere [37].
TABLE 1.
Sequences of gene specific primers and hybridization probes used in quantitative PCR analysis
| Target mRNA | Assay Format | Primer/Probe | Sequences (5′ to 3′) |
|---|---|---|---|
| ErbB1 (EGFR) | SYBR Green I | Forward primer (PC4886) | CCACCTGTGCCATCCAAACT * |
| Reverse primer (PC4887) | GGCGATGGACGGGATCTT * | ||
| ErbB2 | SYBR Green I | Forward primer (PC4888) | AGCCTTGCCCCATCAACTG * |
| Reverse primer (PC4889) | AATGCCAACCACCGCAGA * | ||
| ErbB3 | SYBR Green I | Forward primer (PC4890) | CCCTGCCATGAGAACTGCAC * |
| Reverse primer (PC4891) | TCACTGTCAAAGCCATTGTCAGAT * | ||
| ErbB4 | SYBR Green I | Forward primer in TK domain (PC1542) | CCTGGAAGAAAGACGACTCGTTC |
| Reverse primer in TK domain (PC1541) | CGTCACTCTGATGGGTGAATTTCC | ||
| ErbB4 JM region | Hybridization Probe | Forward primer flanking JM region (PC2338) | CAGTGTGAGAAGATGGAAGATG |
| Reverse primer flanking JM region (PC5221) | CCACAATGACCAGAATGAAGAG | ||
| JM-a | Hybridization Probe | Donor-probe F1 (PC5213) | GCCATCCAAACTGCACCCAA-Flu |
| Acceptor-probe R1 (PC5215) | Red610-GTAACGGTCCCACTAGTCATGACTGCATTTA-p | ||
| JM-b | Hybridization Probe | Donor-probe F2 (PC5214) | AAGACTGCATCGGCCTGATGGATA-Flu |
| Acceptor-probe R2 (PC5216) | Red640-AACTCCCCTGATTGCAGCTGGA-p | ||
| JM-c | Hybridization Probe | Donor-probe F1 (PC5213) | GCCATCCAAACTGCACCCAA-Flu |
| Acceptor-probe R2 (PC5216) | Red640-AACTCCCCTGATTGCAGCTGGA-p | ||
| JM-d | Hybridization Probe | Donor-probe F2 (PC5214) | AAGACTGCATCGGCCTGATGGATA-Flu |
| Acceptor-probe R1 (PC5215) | Red610-GTAACGGTCCCACTAGTCATGACTGCATTTA-p | ||
| ErbB4 CYT region | Hybridization Probe | Forward primer flanking CYT region (PC5268) | CCAGTCCAAATGACAGCAAG |
| Reverse primer flanking CYT region (PC5269) | CCTTGTTCAGCAGCAAAACC | ||
| CYT-1 | Hybridization Probe | Donor-probe F3 (PC5217) | CTTCCAGAGCAAGAATTGACTCGAATAGG-Flu |
| Acceptor-probe R3 (PC5218) | Red610-GAAATTGGACACAGCCCTCCTCC-p | ||
| CYT-2 | Hybridization Probe | Donor-probe F3 (PC5217) | CTTCCAGAGCAAGAATTGACTCGAATAGG-Flu |
| Acceptor-probe R4 (PC5219) | Red640-CCAGTTTGTATACCGAGATGGAGGTTTTG-p | ||
| TACE | SYBR Green I | Forward primer (PC5128) | GGATGTGAAGATGTTGCTAGAG |
| Reverse primer (PC5129) | CTGGGCTATAATAAGCCTTTG | ||
| Presenilin-1 | SYBR Green I | Forward primer (PC5140) | GCCCTGCACTCAATTCTG |
| Reverse primer (PC5141) | ACACTTCCCCCAAGTAAATG | ||
| Presenilin-2 | SYBR Green I | Forward primer (PC5142) | TCCTCTTCACCTATATCTACCTTG |
| Reverse primer (PC5144) | CACAGCCACGAGATCATAC | ||
| Nicastrin | SYBR Green I | Forward primer (PC5134) | AGCCTCTCCCACCATCTTC |
| Reverse primer (PC5135) | CATACAGAGCACGTCCCAG | ||
| Pen-2 | SYBR Green I | Forward primer (PC5145) | TTCCTTGTCCCAGCCTAC |
| Reverse primer (PC5146) | GGTGAGCACTATCACCCAG | ||
| Aph-1a | SYBR Green I | Forward primer (PC5151) | GGTTAGCATCGCTGAGTGAG |
| Reverse primer (PC5152) | GAAAGGCTGAAGTCAGGAAG | ||
| H6PD | SYBR Green I | Forward primer (PC5242) | GATCCTGCCTTTCCGAGAC |
| Reverse primer (PC5244) | CCGCACTGCTGACATTG | ||
| 18S rRNA | SYBR Green I | Forward primer (PC1745) | GTGGAGCGATTTGTCTGGTT |
| Reverse primer (PC1746) | CGCTGAGCCAGTCAGTGTAG |
The donor probes were labelled with Fluorescein (Flu) dye at the 3′end. The acceptor probes were labelled with LightCycler Red610 (R610) or Red640 (R640) at the 5′ end, and the 3′ hydroxyl group of the probe was blocked with a phosphate (p). TACE: tumor necrosis factor-α-converting enzyme; H6PD: hexose 6-phosphate dehydrogenase.
Primers indicated are as designed by Junttila et al (36).
Real-time quantitative PCR using LightCycler Hybridization Probes
Two sets of LightCycler hybridization probes were used to specifically detect the expression of all the ERBB4 JM and CYT related transcript variants identified to date. The design of the primers and probes used in the assay are shown in Figure 1, and the sequences are listed in Table 1. Probes were synthesized by Metabion (Martinsried, Germany). Real-time PCR experiments were carried out using a LightCycler 480 system (Roche Diagnostics, Burgess Hill, UK). To measure the expression of each JM and CYT variant, 1 μl of cDNA solution was used in a 10 μl PCR reaction containing 1x LightCycler 480 Probes Master (Roche Diagnostics, Burgess Hill, UK) with appropriate primers and probes (Fig. 1). The LC480 instrument was employed in Multi Color HybProbe detection format with a filter combination of Fluos (483-533 nm), Red 610 (483-610 nm) and Red 640 (483-640 nm), according to the manufacturer’s protocol. Each sample was run in duplicate, and each run included a no-template negative control as well as the consistent calibrator (mixture of various cDNA as template) in triplicate. Parallel experiments were done for 18S rRNA and hexose 6-phosphate dehydrogenase (H6PD) using LightCycler 480 SYBR Green I Master (Roche Diagnostics, Burgess Hill, UK) and running with SYBR Green I (483-533 nm) detection format. The expression levels of the two genes were used as internal references to normalize cDNA input quantity across samples when doing relative quantification analysis. Both absolute and relative quantification analyses were performed using LC480 software version 1.2, following manufacturer’s instructions.
Statistical methods
Results are expressed as means ± s.d. from an appropriate number of experiments as indicated in figure legends. Statistical differences were analysed by a Student’s t-test with a significance level set at p<0.05.
Results
Relative mRNA expression of ErbB family RTKs in MB, PA and normal brain
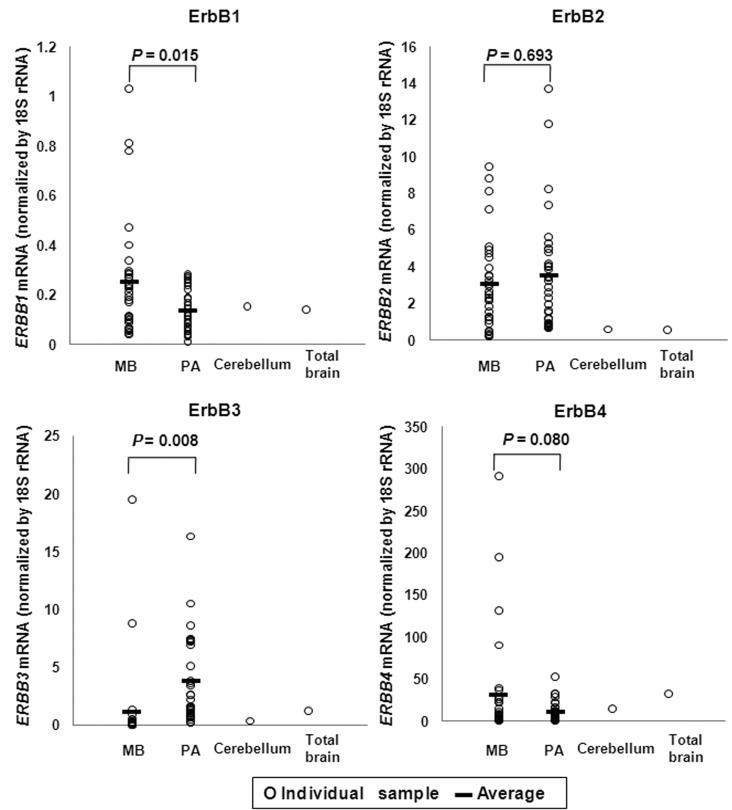
The overall transcript expression pattern for the four ErbB family RTKs, ErbB1-4, was compared for the two series of common childhood tumours (31 MB and 35 PA cases) and normal brain tissues (cerebellum and total brain). Transcripts encoding transmembrane regions of ErbB1, ErbB2 and ErbB3 [36], and the tyrosine kinase (TK) domain of ErbB4 were analyzed by real-time RT-PCR using a SYBR Green I system. This analysis showed that ERBB2 was highly expressed in both MBs and PAs when compared with the normal brain samples (Fig. 2). ERBB1 expression was higher in MB than in PA (P<0.05) and normal brain, ERBB3 mRNA was expressed at significantly higher levels in most PAs when compared with MB (P<0.05) and normal brain. This is consistent with previous microarray data reporting the paired over-expression of ERBB3 and SOX10 in PA [38]. Both tumour types had ERBB4 expression levels that were close to that of normal brain tissues, but the average mRNA levels of MB showed a trend slightly above those of PA (P=0.08).
FIGURE 2.
Relative mRNA expression of ErbB family receptors in MB, PA and normal brain. RNA samples representing normal brain tissues (cerebellum and total brain), 31 MB tumours and 35 PA tumours were analyzed for transcripts encoding transmembrane regions of ErbB1, ErbB2 and ErbB3, and the tyrosine kinase domain of ErbB4, by real-time RT-PCR. Expression of 18S rRNA was used to normalize ErbB expression. SDs of two parallel analyses were < 5% of the means. MB, medulloblastoma; PA, pilocytic astrocytoma.
We have previously reported high-resolution whole-genome array-CGH analyses of these tumours [33, 34]. In no cases were the genes encoding the ErbB receptors involved in small, focal deletions or amplifications. However, the genes were frequently encompassed by large-scale genomic gains or losses, usually extending to involve the majority of a chromosome arm or whole chromosome. There were single copy gains of a large region encompassing ERBB1 in 10 of the 31 MB cases and 10 of the 35 PA cases; single copy gains of ERBB2 in 14 MBs but not in any PAs; and single copy gains of ERBB3 in two MBs and two PAs. ERBB4 was encompassed by a region of single copy gain in one MB and single copy loss in four MBs while no copy number changes in the ERBB4 region was found in PAs. We then compared the expression data with the array-CGH data. However, no correlation between the copy number changes and the expression levels of ERBBs was found in the series of MBs or PAs (data not shown).
Relative expression of ERBB4 transcript variants between MB, PA and normal brain
Differences in the splice variants of ERBB4 were investigated in order to determine whether ErbB4 might function differently in MB, PA and normal brain even though mRNA expression levels are similar, as shown in Figure 2.
To specifically measure the mRNA level of all the known ERBB4 transcript variants, and especially to detect the JM-c and JM-d transcripts, LightCycler hybridization probes were used in real-time RT-PCR analysis. Figure 1 shows the design of this experiment. To measure the expression of the four ERBB4 JM variants, for example, donor probes (F1 and F2) homologous to the 3′ ends of exons 15 and 16b and acceptor probes (R1 and R2) matching the 5′ ends of exons 16a and 17 were generated. By employing FRET technology in real-time PCR, different combinations of the above probe pairs can specifically recognize the alternative splice junctions in each of the four known JM transcript variants. Thus, F1 and R1 probes can specifically detect JM-a, F2 and R2 detect JM-b, F1 and R2 detect JM-c, and F2 and R1 detect JM-d (Fig. 1). Theoretically, there would be no cross-reaction caused by non-specific binding during probe hybridization and signal detection procedures. We proved the specificity of the hybridization probes by performing control experiments using plasmid DNA with cloned ERBB4 transcript variants as templates prior to detecting the transcripts in tumours and normal brain tissues.
The relative expression of each ERBB4 transcript variant in MB and PA cases, using 18S rRNA as a normalization reference, is shown in Figure 3. Since the expression of ERBB4 transcript variants are much lower than that of 18S, another reference (H6PD) known to have low but even expression in MB, PA and normal brain was also used in this study, to ensure no inaccuracies were generated by using an inappropriate reference gene for relative quantification. Analysis using H6PD (data not shown) showed a similar pattern of results to those generated by 18S (Fig. 3). So, to be consistent with the other qPCR experiments using 18S as a reference, only the 18S results are presented here.
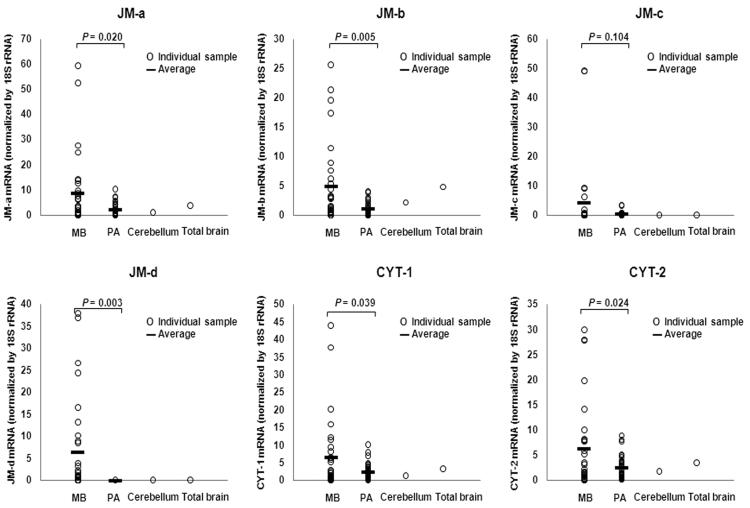
FIGURE 3.
Relative expression of ERBB4 transcript variants in MB, PA and normal brain. Real-time RT-PCR with LightCycler Hybridization Probes was used to specifically detect the junctions between alternatively spliced exons representing the ERBB4 transcript variants JM-a, JM-b, JM-c, JM-d, CYT-1 and CYT-2. The relative expression of each variant was compared between 31 MB tumours, 35 PA tumours and normal brain tissues (cerebellum and total brain) using 18S rRNA as a reference for normalization. SDs of two parallel analyses were < 5% of the means. MB, medulloblastoma; PA, pilocytic astrocytoma.
The JM-a, JM-b, CYT-1 and CYT-2 variants of ERBB4 are differentially expressed in MBs and PAs. The expression levels of those ERBB4 transcripts are generally higher in MBs than in PAs (P<0.05). The JM-d transcript variant was only found to be expressed in MB tumours, while JM-c was expressed in both MBs and PAs at similar levels (P=0.1). However, neither JM-c nor JM-d could be detected in normal brain tissues (cerebellum or total brain) (Fig. 3).
Composition of ERBB4 transcript variants in individual tumours and normal brain
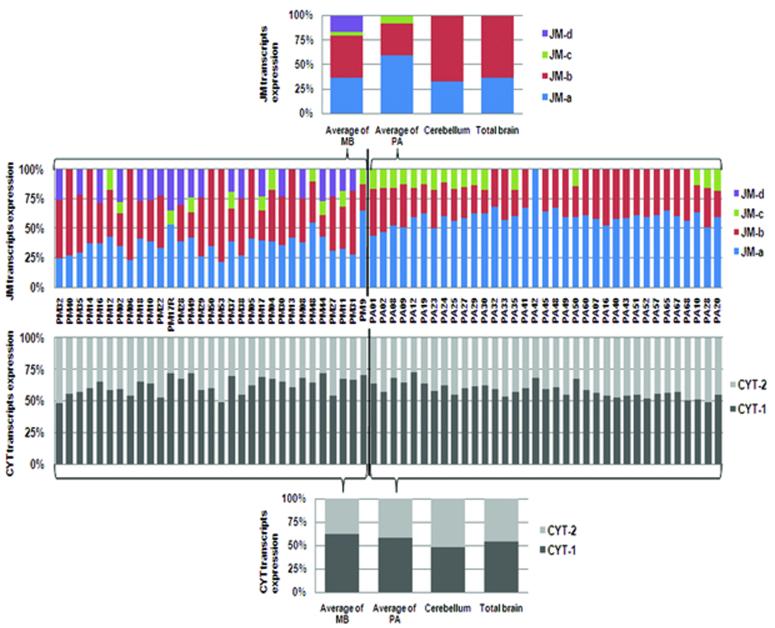
To demonstrate the expression pattern of ERBB4 transcript variants in individual MB and PA tumours and in normal brain, the percentage of each variant as a proportion of total JM or CYT transcripts was calculated for each sample by absolute quantification analysis of the real-time PCR data generated with hybridization probes.
There were clear differences in the expression patterns of ERBB4 JM transcripts between the two tumour types (MB and PA), and also between the tumours and normal brain tissues (Fig. 4, upper panel). In cerebellum and total brain, ERBB4 JM transcripts consist solely of the JM-a and JM-b variants. JM-b is the predominant transcript in normal brain tissues (around 65%), while JM-a was expressed at a lower level (around 35%). This is consistent with previous real-time RT-PCR data on alternative splicing of ERBB4 in normal human tissues [39], which found that brain, cerebellum, skeletal muscle and heart expressed predominantly the JM-b transcript. In most PA tumours, however, JM-a was expressed at higher levels than JM-b. On average, around 60% of JM transcripts in PA tumours were JM-a, while only around 30% were JM-b. Furthermore, the JM-c variant of ERBB4 was detected in 17 out of 35 PA tumours (49%), and comprised on average about 7.4% of JM transcripts in PAs. The transcript for the JM-c isoform was identified in 2/5 optic PAs, 2/4 hypothalmic PAs, 11/21 cerebellar PAs and 2 of the 4 PAs at other sites. Thus it appears to occur in PAs at all common sites. Medulloblastomas showed a more complicated alternative splicing pattern of ERBB4 JM transcripts than PAs. JM-a and JM-b comprised the major types of JM domain in MB (around 70% of total JM expression) and were expressed in almost every MB tumour sample in the series (except PM17R, which lacked JM-b expression). In addition, 11 out of 31 tumours expressed JM-c (at around 5% of total JM expression), and 20 of 31 expressed JM-d (at around 16% of total JM variants). Seven MB tumours expressed all four of the JM transcript variants. Correlations between the expression pattern of JM variants and the clinical data of the two series of tumours were analysed. No association was found between the expression of any transcript variants of ERBB4 (or the expression of cleavable isoforms) and survival (data not shown).
FIGURE 4.
Composition of ERBB4 transcript variants in individual tumour samples and normal brain. LightCycler Hybridization Probes were used to specifically detect the junctions between alternatively spliced exons representing the ERBB4 transcript variants JM-a, JM-b, JM-c, JM-d, CYT-1 and CYT-2. The composition of ERBB4 variants in individual tumours was calculated as the percentage of the total JM or CYT expression of each transcript variant. The upper panel shows the composition of JM-region transcript variants and the lower panel shows the CYT variants. The average expression pattern of 31 MB tumours and 35 PA tumours was also calculated and used in comparison with normal brain tissues (cerebellum and total brain). SDs of two parallel analyses were < 5% of the means. MB, medulloblastoma; PA, pilocytic astrocytoma.
If the MB and PA cases studied are divided into childhood and adult using the cutoff age of 15 years, the expression of JM-c was more frequently found in adult cases of either MB (63%) or PA (67%) than in childhood cases (26% of childhood MB and 42% of childhood PA, Table 2). There was no relationship between the age of the patients and the expression of JM-d transcripts in MBs
TABLE 2.
Summary of the age of MB and PA cases with expression of ERBB4 JM-c or JM-d variants
| Tumor type | Group of cases* | Median age at operation (yrs) | Range of age at operation (yrs) | No. of cases | No. of cases expressing JM-c | No. of cases expressing JM-d |
|---|---|---|---|---|---|---|
| MB | Children | 7 | 0.6 - 15 | 23 | 6 (26%) | 15 (65%) |
| Adults | 22 | 18 - 41 | 8 | 5 (63%) | 5 (63%) | |
| PA | Children | 6 | 0.5 - 14 | 26 | 11 (42%) | 0 |
| Adults | 24 | 17 - 33 | 9 | 6 (67%) | 0 |
A cut-off age of 15 years was used to divide the cases into childhood and adult groups because this age is used by the American Cancer Society (www.cancer.org) and the United Kingdom Office for National Statistics (www.statistics.gov.uk) to define childhood cancer incidence.
The expression pattern of CYT-region transcript variants was very similar among the individual tumours of both types (Fig. 4, lower panel). The CYT-1:CYT-2 ratio in the PAs and MBs was roughly 60:40 - slightly different from the pattern in normal brain tissues, where CYT-1 and CYT-2 were equally expressed.
Relative expression of proteases responsible for ErbB4 cleavage in MB and PA
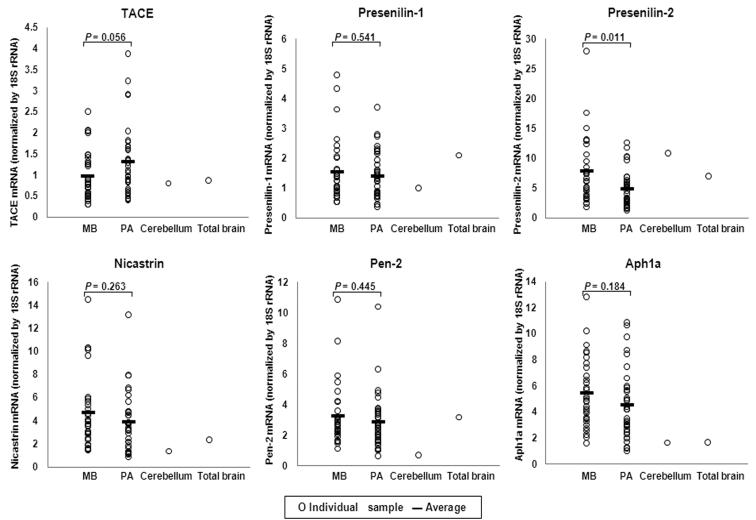
To test whether proteolytic processing of ErbB4 in MB and PA tumours is likely, the expression level of transcripts coding for the proteases known to be involved in ErbB4 cleavage were assessed using real-time RT-PCR with gene-specific primers (Table 1) and the SYBR Green I system. The results are shown in Figure 5.
FIGURE 5.
Relative mRNA expression of ErbB4-processing proteases in MB, PA and normal brain. RNA samples representing normal brain tissues (cerebellum and total brain), 31 MB tumours and 35 PA tumours were analyzed for transcripts encoding TACE and components of the γ-secretase complex (Presenilin-1, Presenilin-2, Nicastrin, Pen-2 and Aph1a). Expression of 18S rRNA was used to normalize protease expression. SDs of two parallel analyses were < 5% of the means. MB, medulloblastoma; PA, pilocytic astrocytoma; TACE, tumour necrosis factor-α-converting enzyme.
The mRNA of TACE, the protease believed to first cut the JM-a isoform of ErbB4 at the juxtamembrane region [14], showed similar expression levels in MBs to those of normal brain tissues; while a higher average expression was found in the PAs (Fig. 5).
Presenilin is the catalytic subunit of the γ-secretase complex [40]. This complex cleaves ErbB4 within its transmembrane domain after the ecto-domain has been removed by TACE, releasing a soluble intracellular domain of ErbB4 [16, 17]. The expression levels of transcripts for both homologs of Presenilin, Presenilin-1 (PS1) and Presenilin-2 (PS2), were assessed. Similar levels of Presenilin mRNA were expressed in MB, PA, and in normal brain (Fig. 5). Besides the Presenilins, three other proteins (Nicastrin, Pen-2 and Aph-1) are necessary for γ-secretase complex function [41]. The expression of transcripts for these proteins was also investigated. The data showed that substantial levels of mRNA for each of these proteins were expressed in both MBs and PAs, at a slightly higher level (Nicastrin, Aph1a) or at similar levels (Pen-2) to those seen in normal brain (Fig. 5).
Discussion
MB and PA are two of the most frequently occurring childhood brain tumours. MB is a highly malignant, invasive embryonal tumour of the cerebellum, while PA is relatively benign. This study compares the mRNA expression of the ErbB RTKs, and in particular the ERBB4 transcript variants in these two tumour types that represent opposite ends of the malignancy spectrum of childhood brain tumours.
Dimerized receptors are the basic functional unit of ErbB signaling, with each partner contributing unique features. In this study, we first measured the overall mRNA expression patterns of ErbB family RTKs in the two series of MB and PA tumours by real-time RT-PCR. ERBB2 was found to be over-expressed in both MBs and PAs when compared with normal brain tissues (Fig. 2). The present qPCR data also confirmed a significantly higher expression level of transcripts for ERBB3 in our series of PA tumours as compared to MB and normal brain (Fig. 2). This confirms earlier reports using expression microarray analysis [38]. Studies on the structure of the ErbB2 ectodomain have shown that it lacks the capacity for ligand binding and does not readily form homodimers [42, 43]. Rather, it acts as a preferred partner for the other activated ErbBs since its fixed conformation resembles the ligand-activated state of EGFR [42-45]. ErbB3 does not have a functional kinase domain and thus can also not form functional homodimers [46, 47]. However, heterodimers formed by ErbB2 and ErbB3 are the most potent mitogenic and transforming dimers among all the possible combinations of the ErbB receptors [45, 48-50]. The high levels of expression of both ERBB2 and ERBB3 in PAs may therefore indicate a significant role for ErbB2/ErbB3 heterodimers in these tumours. In MB, the relative expression levels of ERBB1 transcripts was higher than in PA and normal brain, but the expression levels of ERBB1 mRNA overall in both tumour types appeared far lower than other ErbB family members (Fig. 2), making implications of this finding unclear. Previous studies have reported that ErbB4 is highly expressed in normal brain compared to many other normal tissues [26], and expression of ErbB4 in association with ErbB2 is associated with poor prognosis in MB [28-30]. Our qPCR data showed that total ERBB4 mRNA (as determined by measurements encompassing the coding region for the TK domain) was expressed at a higher level in MB and normal brain than in PA (Fig. 2). The trend to high expression levels of ERBB4 mRNA in MBs (P=0.08, Fig. 2) was further confirmed by subsequent relative expression data of individual ERBB4 variants (JM-a, JM-b, CYT-1 and CYT-2), as each of them showed significantly higher expression levels in MBs compared to PAs (P<0.05, Fig. 3). In summary, the overall expression of ERBBs suggests that ErbB2/ErbB4 heterodimers and ErbB4 homodimers may be the major functional units of the ErbB family RTKs in MB, while in PA, ErbB2/ErbB3 heterodimers may play a more prominent role in addition to ErbB4-containing dimers.
Some reports indicate that differential expression of specific ErbB4 isoforms, rather than changes in total ErbB4 expression, may be associated with some cancers. For example, a protease-cleavable ErbB4 isoform (JM-a CYT-2) has been shown to promote growth of estrogen receptor (ER) positive breast cancer [39], and an elevated CYT-1:CYT-2 ratio may promote tumour growth and act as a prognostic indicator in MB [31]. We therefore analyzed the expression of all ERBB4 JM and CYT transcript variants in MB, PA and normal brain. Although an analysis of how the JM and CYT variants are combined is very difficult with current technology, the expression levels of all the known ERBB4 JM or CYT transcript variants were investigated specifically and quantitatively in this study by using real-time PCR with FRET probes (Fig. 1). Previous real-time qPCR studies of ERBB4 transcripts could not distinguish JM-d from either JM-a or JM-b and also could not assess JM-c expression [36, 39, 51, 52]. Here we were able to compare the relative expression of each ERBB4 transcript variant across the different tumour samples as well as the composition of ERBB4 transcript variants in individual tumours.
Although the previous study demonstrated that CYT1:CYT2 ratios displayed correlation with MB variants [31], no obvious differences in CYT-1:CYT-2 ratios were detected in our series of MBs or PAs. The absence of differences in CYT1: CYT2 ratio in the present study could be related to absence in the series of desmoplastic as well as large cell MB variants which included in the previous study.
However, the differential expression patterns of JM-related transcript variants between MB, PA and normal brain are very interesting. Firstly, this study detected more alternatively spliced ERBB4 transcript variants in tumour samples than in normal brain tissues. The rare JM-d ERBB4 transcript was only detected in MBs while the other rare variant, JM-c, was found in both MB and PA. Neither of these variants was detected in normal brain tissues (cerebellum or total brain) (Fig. 3 and 4). The detection of both JM-c and JM-d in MBs is consistent with previous findings by Gilbertson et al [15]. They also found the expression of JM-d in developing fetal cerebellum but not in normal human adult cerebellum, so they suggested that the expression of JM-d in MBs reflects the expression patterns seen in primitive neuroectodermal tissue [15]. Thus, the lack of JM-c or JM-d expression in our normal brain samples could be due to the fact that these tissues were of adult origin. It is notable that not all the childhood MB cases showed JM-d expression, while its expression was also detected in the majority of tumours from young adults (Table 2). Prior to the present study, the JM-c variant of ERBB4 was reported only in some MBs [15]. This study is therefore the first to report the existence of JM-c in PAs. Moreover, in both tumour types, JM-c expression was more frequently found in adult cases (Table 2). These findings confirm that in addition to the common JM-a and JM-b isoforms, other ERBB4 JM variants are expressed in brain tumours. In the MBs, all known JM transcript variants are generally expressed.
Another important feature of the pattern of ERBB4 expression in MB and PA is the elevated percentage of cleavable transcript variants when compared with normal brain tissues (Fig. 4). In PAs, the JM-a variant encoding a cleavable isoform of ErbB4 was expressed as a larger fraction of total JM variants than in normal brain. In MBs, although the average expression of JM-a and JM-b is very similar (37% and 43% of total JM expression respectively), the JM-d variant, which also possesses the TACE sensitive sequence and represents another type of cleavable JM isoform, is expressed at around 16% of total JM expression. Thus the total percentage of cleavable ErbB4 isoforms in MB (about 53% on average) was also higher than in normal brain tissues. The expression levels of the transcripts coding for the two proteases responsible for ErbB4 cleavage were relatively constant in MB, PA and normal brain (Fig. 5). Together, these results suggest that proteolytic processing of cleavable ErbB4 isoforms is more likely to occur in MB and PA, and may lead to signaling events divergent from that in normal brain, where the TACE-resistant JM-b isoform is predominantly expressed. Further studies on the protein-level expression of ErbB4 isoforms and ErbB4-processing proteases in MB and PA tumours, and the different subcellular localization of ErbB4 fragments in brain tumours or cell lines, will therefore be required to examine the potential role of proteolytic cleavage of ErbB4 in the behavior of brain tumours such as MB and PA.
Acknowledgements
We would like to thank Dr Michael L. Mimmack from the Institute of Biotechnology, University of Cambridge, for his help in hybridization probe design.
Financial support
This work was supported by grants from Cancer Research UK and the Samantha Dickson Brain Tumour Trust.
Abbreviations
- CYT
cytoplasmic region
- ECD
extracellular domain
- FRET
fluorescence resonance energy transfer
- H6PD
hexose 6-phosphate dehydrogenase
- ICD
intracellular domain
- JM
juxtamembrane region
- MB
medulloblastoma
- PA
pilocytic astrocytoma
- PI3K
phosphatidyl inositol-3 kinase
- PS1
presenilin-1
- PS2
presenilin-2
- qPCR
quantitative polymerase chain reaction
- RTKs
receptor tyrosine kinases
- SP
signal peptide
- TACE
tumour necrosis factor-α-converting enzyme
- TK
tyrosine kinase domain
- TM
transmembrane domain
- WHO
World Health Organization
Footnotes
Conflicts of interest
None
References
- 1.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005 May;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Cohen BD, Kiener PA, Green JM, Foy L, Fell HP, Zhang K. The relationship between human epidermal growth-like factor receptor expression and cellular transformation in NIH3T3 cells. J Biol Chem. 1996 Nov 29;271:30897–903. doi: 10.1074/jbc.271.48.30897. [DOI] [PubMed] [Google Scholar]
- 3.Tang CK, Concepcion XZ, Milan M, Gong X, Montgomery E, Lippman ME. Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor-positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 1999 Oct 15;59:5315–22. [PubMed] [Google Scholar]
- 4.Alaoui-Jamali MA, Song DJ, Benlimame N, Yen L, Deng X, Hernandez-Perez M, Wang T. Regulation of multiple tumour microenvironment markers by overexpression of single or paired combinations of ErbB receptors. Cancer Res. 2003 Jul 1;63:3764–74. [PubMed] [Google Scholar]
- 5.Bieche I, Onody P, Tozlu S, Driouch K, Vidaud M, Lidereau R. Prognostic value of ERBB family mRNA expression in breast carcinomas. Int J Cancer. 2003 Sep 20;106:758–65. doi: 10.1002/ijc.11273. [DOI] [PubMed] [Google Scholar]
- 6.Lodge AJ, Anderson JJ, Gullick WJ, Haugk B, Leonard RC, Angus B. Type 1 growth factor receptor expression in node positive breast cancer: adverse prognostic significance of c-erbB-4. J Clin Pathol. 2003 Apr;56:300–4. doi: 10.1136/jcp.56.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacus SS, Chin D, Yarden Y, Zelnick CR, Stern DF. Type 1 receptor tyrosine kinases are differentially phosphorylated in mammary carcinoma and differentially associated with steroid receptors. Am J Pathol. 1996 Feb;148:549–58. [PMC free article] [PubMed] [Google Scholar]
- 8.Knowlden JM, Gee JM, Seery LT, Farrow L, Gullick WJ, Ellis IO, Blamey RW, Robertson JF, Nicholson RI. c-erbB3 and c-erbB4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene. 1998 Oct 15;17:1949–57. doi: 10.1038/sj.onc.1202107. [DOI] [PubMed] [Google Scholar]
- 9.Sartor CI, Zhou H, Kozlowska E, Guttridge K, Kawata E, Caskey L, Harrelson J, Hynes N, Ethier S, Calvo B, Earp HS., 3rd Her4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol Cell Biol. 2001 Jul;21:4265–75. doi: 10.1128/MCB.21.13.4265-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pawlowski V, Revillion F, Hebbar M, Hornez L, Peyrat JP. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000 Nov;6:4217–25. [PubMed] [Google Scholar]
- 11.Suo Z, Risberg B, Kalsson MG, Willman K, Tierens A, Skovlund E, Nesland JM. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J Pathol. 2002 Jan;196:17–25. doi: 10.1002/path.1003. [DOI] [PubMed] [Google Scholar]
- 12.Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003 Jul;200:290–7. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 13.Elenius K, Corfas G, Paul S, Choi CJ, Rio C, Plowman GD, Klagsbrun M. A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J Biol Chem. 1997 Oct 17;272:26761–8. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- 14.Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumour necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000 Apr 7;275:10379–87. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- 15.Gilbertson R, Hernan R, Pietsch T, Pinto L, Scotting P, Allibone R, Ellison D, Perry R, Pearson A, Lunec J. Novel ERBB4 juxtamembrane splice variants are frequently expressed in childhood medulloblastoma. Genes Chromosomes Cancer. 2001 Jul;31:288–94. doi: 10.1002/gcc.1146. [DOI] [PubMed] [Google Scholar]
- 16.Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001 Dec 7;294:2179–81. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 17.Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, Mei L, Kim TW. Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J Biol Chem. 2002 Feb 22;277:6318–23. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- 18.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003 Aug 29;278:33334–41. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 19.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp Cell Res. 2004 Apr 1;294:469–79. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, Jones FE. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol. 2004 Nov 8;167:469–78. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal GA, Naresh A, Marrero L, Jones FE. Presenilin-dependent gamma-secretase processing regulates multiple ERBB4/HER4 activities. J Biol Chem. 2005 May 20;280:19777–83. doi: 10.1074/jbc.M412457200. [DOI] [PubMed] [Google Scholar]
- 22.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005 Aug 1;65:6764–72. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 23.Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JM, Jones FE. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006 Jun 15;66:6412–20. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- 24.Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M. Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene. 1999 Apr 22;18:2607–15. doi: 10.1038/sj.onc.1202612. [DOI] [PubMed] [Google Scholar]
- 25.Kainulainen V, Sundvall M, Maatta JA, Santiestevan E, Klagsbrun M, Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J Biol Chem. 2000 Mar 24;275:8641–9. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan R, Poulsom R, Hurst HC, Gullick WJ. Expression of the c-erbB-4/HER4 protein and mRNA in normal human fetal and adult tissues and in a survey of nine solid tumour types. J Pathol. 1998 Jul;185:236–45. doi: 10.1002/(SICI)1096-9896(199807)185:3<236::AID-PATH118>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell. 2006 Oct 6;127:185–97. doi: 10.1016/j.cell.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 28.Gilbertson RJ, Perry RH, Kelly PJ, Pearson AD, Lunec J. Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res. 1997 Aug 1;57:3272–80. [PubMed] [Google Scholar]
- 29.Gilbertson RJ, Clifford SC, MacMeekin W, Meekin W, Wright C, Perry RH, Kelly P, Pearson AD, Lunec J. Expression of the ErbB-neuregulin signaling network during human cerebellar development: implications for the biology of medulloblastoma. Cancer Res. 1998 Sep 1;58:3932–41. [PubMed] [Google Scholar]
- 30.Bal MM, Das Radotra B, Srinivasan R, Sharma SC. Expression of c-erbB-4 in medulloblastoma and its correlation with prognosis. Histopathology. 2006 Jul;49:92–3. doi: 10.1111/j.1365-2559.2006.02337.x. [DOI] [PubMed] [Google Scholar]
- 31.Ferretti E, Di Marcotullio L, Gessi M, Mattei T, Greco A, Po A, De Smaele E, Giangaspero F, Riccardi R, Di Rocco C, Pazzaglia S, Maroder M, Alimandi M, Screpanti I, Gulino A. Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene. 2006 Nov 23;25:7267–73. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- 32.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. Lyon, France: IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCabe MG, Ichimura K, Liu L, Plant K, Backlund LM, Pearson DM, Collins VP. High-resolution array-based comparative genomic hybridization of medulloblastomas and supratentorial primitive neuroectodermal tumours. J Neuropathol Exp Neurol. 2006 Jun;65:549–61. doi: 10.1097/00005072-200606000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones DT, Ichimura K, Liu L, Pearson DM, Plant K, Collins VP. Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age. J Neuropathol Exp Neurol. 2006 Nov;65:1049–58. doi: 10.1097/01.jnen.0000240465.33628.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichimura K, Schmidt EE, Goike HM, Collins VP. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996 Sep 5;13:1065–72. [PubMed] [Google Scholar]
- 36.Junttila TT, Laato M, Vahlberg T, Soderstrom KO, Visakorpi T, Isola J, Elenius K. Identification of patients with transitional cell carcinoma of the bladder overexpressing ErbB2, ErbB3, or specific ErbB4 isoforms: real-time reverse transcription-PCR analysis in estimation of ErbB receptor status from cancer patients. Clin Cancer Res. 2003 Nov 1;9:5346–57. [PubMed] [Google Scholar]
- 37.Ichimura K, Vogazianou AP, Liu L, Pearson DM, Backlund LM, Plant K, Baird K, Langford CF, Gregory SG, Collins VP. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene. 2007 Oct 15; doi: 10.1038/sj.onc.1210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Addo-Yobo SO, Straessle J, Anwar A, Donson AM, Kleinschmidt-Demasters BK, Foreman NK. Paired overexpression of ErbB3 and Sox10 in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2006 Aug;65:769–75. doi: 10.1097/01.jnen.0000229989.25171.aa. [DOI] [PubMed] [Google Scholar]
- 39.Junttila TT, Sundvall M, Lundin M, Lundin J, Tanner M, Harkonen P, Joensuu H, Isola J, Elenius K. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 2005 Feb 15;65:1384–93. doi: 10.1158/0008-5472.CAN-04-3150. [DOI] [PubMed] [Google Scholar]
- 40.Steiner H, Haass C. Intramembrane proteolysis by presenilins. Nat Rev Mol Cell Biol. 2000 Dec;1:217–24. doi: 10.1038/35043065. [DOI] [PubMed] [Google Scholar]
- 41.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003 Apr 10;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 42.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr., Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003 Feb 13;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 43.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003 Feb;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 44.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996 Oct;16:5276–87. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997 Apr 1;16:1647–55. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., 3rd Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci U S A. 1994 Aug 16;91:8132–6. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006 Jul;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 48.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996 May 15;15:2452–67. [PMC free article] [PubMed] [Google Scholar]
- 49.Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene. 1995 May 4;10:1813–21. [PubMed] [Google Scholar]
- 50.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995 Sep 1;14:4267–75. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006 Mar 5;141:142–8. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 52.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007 Jan 15;16:129–41. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]