Abstract
The second extracellular loop (EL2) of rhodopsin forms a cap over the binding site of its photoreactive 11-cis retinylidene chromophore. A critical question has been whether EL2 forms a reversible gate that opens upon activation or acts as a rigid barrier. Distance measurements using solid-state 13C NMR spectroscopy between the retinal chromophore and the β4 strand of EL2 show the loop is displaced from the retinal binding site upon activation, and there is a rearrangement in the hydrogen-bonding networks connecting EL2 with the extracellular ends of transmembrane helices H4, H5 and H6. NMR measurements further reveal that structural changes in EL2 are coupled to the motion of helix H5 and breaking of the ionic lock that regulates activation. These results provide a comprehensive view of how retinal isomerization triggers helix motion and activation in this prototypical G protein-coupled receptor.
G protein-coupled receptors (GPCRs) comprise the largest and most diverse superfamily of membrane receptors with a simple architectural core of seven transmembrane (TM) helices (H1 to H7) connected by typically short extracellular and cytoplasmic loops. Sequence variability within the TM helices and extracellular loops allow GPCRs to respond to diverse stimuli including light and a wide variety of ligands. Small molecule ligands can bind within the helical core of the receptor, whereas larger peptide and protein ligands bind at the extracellular loops. The second extracellular loop (EL2) in particular has been the target of a number of functional studies indicating that it plays an integral role in activation of GPCRs that bind either small molecules or large peptide ligands1–4.
The vertebrate visual pigments are unique in the class A GPCRs in that they are activated by photoreaction of an 11-cis retinylidene chromophore. The retinal is covalently attached via a protonated Schiff base (PSB) within the seven TM helix bundle. The crystal structure of rhodopsin indicates that EL2 extends from Trp175 on H4 to Thr198 on H5. The intriguing aspect about the EL2 sequence is that it folds into a highly ordered and stable structure consisting of two short β-strands (β3 and β4) that form a lid over the retinal-binding site5,6. EL2 is constrained by a conserved disulfide bond between Cys110 at the end of H3 and Cys187 on β4 that is critical for the correct folding of rhodopsin7,8. Other than the Cys110-Cys187 disulfide bond, the EL2 sequence is not conserved among the class A GPCRs.
The structure of EL2 in rhodopsin is stabilized by a number of polar residues that form a well-defined H-bonded network (Supplementary Fig. 1a). At the center of this network is Glu181 on the β3 strand. Glu181 is H-bonded to Tyr192 (β4) and Tyr268 (H6), and is connected through water-mediated hydrogen bonds to Ser186 (EL2) and to Glu113 (H3), the counterion to the retinal PSB6. Glu113 is H-bonded to the backbone carbonyl of Cys187 (EL2) through a water molecule and is within H-bonding distance to the hydroxyl group of Thr94 (H2)6. The involvement of Glu113 in this stable H-bonded network is important in raising the pKa of the Schiff base (above 16)9 and ensuring that it remains protonated in the dark state of rhodopsin10,11. Besides the conserved disulfide bond and the H-bonding network involving Glu181, there are a striking number of H-bonding interactions between the β-strands and the ends of the TM helices (e.g. Trp175-Ser202, Ser176-Thr198, Arg177-Asp190, Tyr178-Ala168). Computational studies identified this region as part of a stable folding core of rhodopsin12, suggesting that EL2 is important for maintaining a stable, inactive receptor conformation.
In contrast to the role of EL2 as a stable cap, several studies have suggested that EL2 is dynamic and mediates both receptor activity and ligand binding. Baranski and colleagues proposed that in the C5a receptor, EL2 serves as a negative regulator3 where the loop inserts between the TM helices to block receptor activity, and then is released upon ligand binding. Holst and Schwartz suggested that a short EL2 in the melanocortin receptor, which is unable to insert into the helical TM core, leads to a high level of constitutive activation13. In the recent crystal structure of the β2-adrenergic receptor (β2-AR)14, EL2 is not closely associated with the ligand-binding site. The β2-AR structure, along with the observation that short loops may be correlated with constitutively active GPCRs, raises the question of whether the role of EL2 as a stable cap is unique in rhodopsin due to the crucial requirement that visual pigments must have very low basal activity in the dark.
Here, we use 13C magic angle spinning (MAS) NMR spectroscopy to address the position of EL2 in rhodopsin and in the active metarhodopsin II (meta II) intermediate, and show how motion of EL2 is coupled to motion of TM helix H5 and the insertion of Tyr223 into the region of the “ionic lock” between H3 and H6. Retinal-protein and protein-protein distance constraints from NMR measurements were obtained for rhodopsin and meta II (Supplementary Fig. 1b, Supplementary Table 1) and used to perform restrained molecular dynamic (MD) simulations to obtain an atomistic model of meta II. Chemical shift measurements of the conserved Cys110-Cys187 disulfide bond and distance measurements between the retinal chromophore and the β4 strand of EL2 are consistent with motion of EL2 away from the agonist all-trans retinal Schiff base upon receptor activation. Mutational studies on Glu181 (EL2) and Met288 (H7) show that the H-bonding network on EL2 is coupled to the H-bonding network centered on H5 involving His211, which in turn leads to rearrangement of the intracellular end of H5 in meta II. Together these results explain how EL2 is a pivotal element in locking the extracellular ends of H5, H6 and H7 in inactive conformations in the dark, and how EL2 motion allows the intracellular ends of these helices to shift into active conformations in the light.
RESULTS
Activation of rhodopsin is initiated by photoisomerization of its retinal chromophore within a tightly packed protein environment. Since the all-trans retinal chromophore in the active meta II intermediate does not fit in the retinal binding site of the dark-state of rhodopsin15, conformational changes of a highly strained retinal must induce changes in the structure of the receptor to release the absorbed light energy.
EL2 is displaced from the retinal binding site in meta II
The first indication that the structure or position of EL2 changes in meta II arises from the large chemical shift changes observed for 13Cβ-Ser186, 13Cβ-Cys187 and 13Cα-Gly188 (Fig. 1a). The Cys110-Cys187 disulfide bond is the only conserved feature in EL2. Fig. 1b presents 13C DARR NMR spectra of rhodopsin (black) and meta II (red) labeled with 13Cβ-cysteine. The β-carbon resonances in disulfide bonds occur in a unique chemical shift window (34–50 ppm) and are sensitive to the secondary structure with a range of 34 to 43 ppm for α helices and 36 to 50 ppm for β sheets16. In Fig. 1b, we observe strong crosspeaks between the Cys110-Cys187 β-carbon resonances at 36.4 ppm and 46.8 ppm, respectively. The 46.8 ppm chemical shift of Cys187 is consistent with its location in the β4 strand of EL2. Upon conversion to meta II, the Cys187 resonance shifts to 50.1 ppm due to a change in the conformation of EL2 or a change in the environment around Cys187. The chemical shift of Cys110 does not change appreciably (−0.2 ppm), indicating that the secondary structure of H3 near Cys110 does not change in meta II.
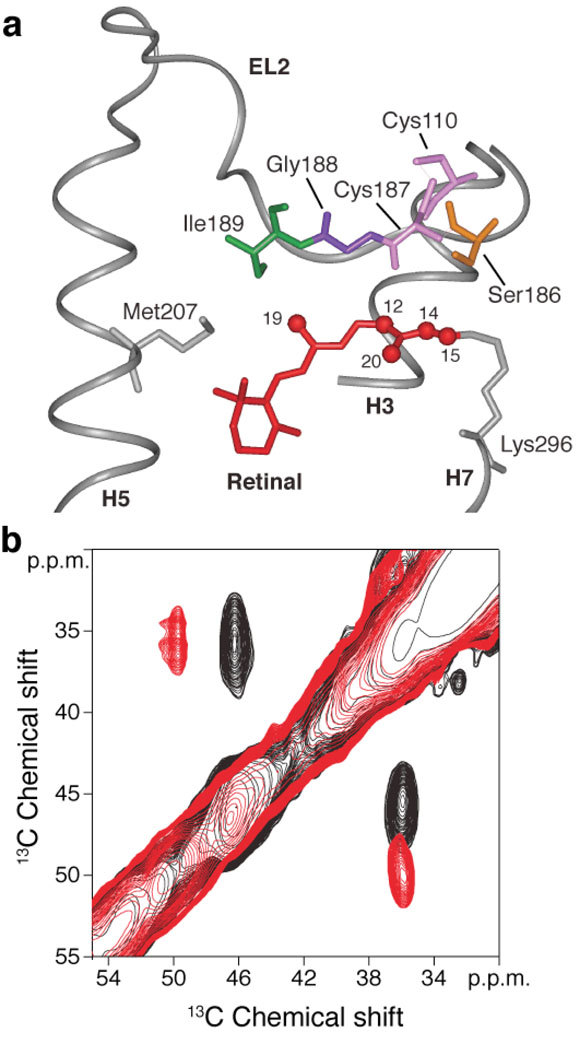
Figure 1.
Structural changes involving the conserved Cys110 - Cys187 disulfide link on activation of rhodopsin. (a) View of the β4 strand of EL2 from the rhodopsin crystal structure6 highlighting the interactions of Ile189, Gly188, Cys187 and Ser186 with the polyene chain of the retinal. Cys110 on the extracellular end of H3 forms a conserved disulfide link with Cys187 in β4. (b) A region from the 2D DARR NMR spectrum of rhodopsin selectively labeled with 13Cβ-cysteine. The figure highlights the crosspeak between Cys187 (46.8 ppm) and Cys110 (36.4 ppm) in rhodopsin (black). On conversion to meta II (red) there is a distinct shift in the crosspeak to 50.1 ppm for Cys187. The 13Cβ chemical shift of Cys110 at ~36 ppm does not change appreciably between rhodopsin and meta II. The eight reduced cysteines in rhodopsin are observed as a broad resonance at ~25 ppm (not shown).
In addition to the chemical shift changes observed in Cys187, we observe a ~1.6 ppm change in the 13Cβ chemical shift of Ser186 and a 2.9 ppm change in the 13Cβ resonance of Gly188 (Supplementary Figs. 2, 3). The 13Cβ-Ser186 chemical shift change may be attributed to a change in the H-bonding interaction of Ser186 with surrounding residues on EL2 and H3, while the 13Cα-Gly188 chemical shift is likely due to changes in backbone torsion angles.
Confirmation of the motion of EL2 away from the retinal binding site in meta II comes from direct distance measurements. The β4 strand of EL2 is aligned almost parallel to the retinal in the binding site with Cys185 close to the PSB end of the retinal and with Ile189 close to the retinal β-ionone ring. We observe close contact between the retinal 13C14 and 13C15 carbons and 13Cβ-Ser186 (Fig. 2a), between the retinal 13C12 and 13C20 carbons and 13C1-Cys187 (Fig. 2b), and between the retinal 13C12 and 13C20 carbons and 13Cα-Gly188 in rhodopsin (Fig. 2c). These contacts are lost in meta II. Moreover, we were not able to observe contacts in rhodopsin or meta II between the retinal 13C9 and 13C12 carbons and U-13C6-Ile189 (Fig. 2d).
Figure 2.
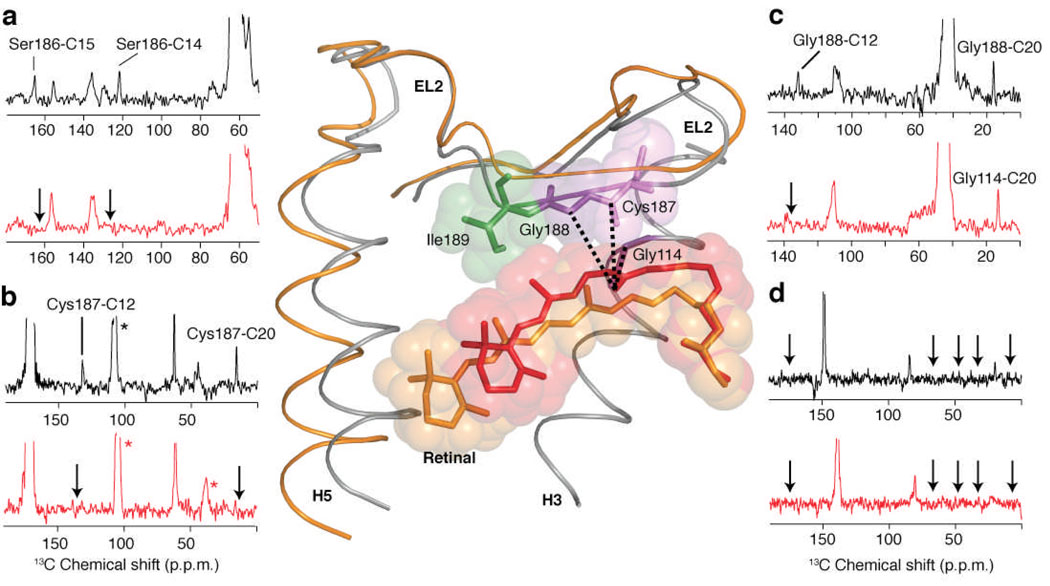
2D 13C DARR NMR spectra of retinal – EL2 interactions. Rows are shown from the 2D 13C DARR NMR spectra of rhodopsin (black) and meta II (red). (a) Rhodopsin labeled with 13Cβ-serine and 13C14,15 retinal. Crosspeaks are observed between Ser186 (63.3 ppm) and the 13C14 and 13C15 resonances in dark rhodopsin, which are lost (arrows) in meta II. (b) Rhodopsin labeled with 13C1-cysteine and 13C12,20 retinal. Crosspeaks are observed between Cys187 (170.8 ppm) and the 13C12 and 13C20 resonances in dark rhodopsin, which are lost (arrows) in meta II. (c) Rhodopsin labeled with 13Cα-glycine and 13C12,20 retinal. Crosspeaks are observed between Gly188 (42.0 ppm) and the 13C12 and 13C20 resonances in dark rhodopsin, which are lost (arrows) in meta II. However, a new Gly-C20 contact is observed in meta II, which is assigned to Gly114 (see text). (d) Rhodopsin labeled with U-13C6-isoleucine and 13C9 retinal. No contacts were observed between Ile189 and C9 on the polyene chain of the retinal in either rhodopsin (black arrows) or meta II (red arrows). The structure of EL2 in rhodopsin is shown (center) indicating the contacts observed between the C20 methyl group and Cys187, Gly188 and Gly114 in rhodopsin. In order to illustrate the displacement of EL2 needed to satisfy the NMR constraints, we have superimposed the rhodopsin crystal structure (grey) with the meta II model (orange) obtained from MD simulations guided by our experimentally determined retinal-protein contacts. Asterisks correspond to MAS sidebands.
As indicated above, the NMR measurements on rhodopsin are compared with the corresponding distances in the crystal structure before converting to meta II. The meta II intermediate we trap at low temperature in n-dodecyl-β-D-maltoside (DDM) is a single well defined state (see Methods). We typically observe strong crosspeaks for 13C…13C distances of ~4.0 Å or less, moderate crosspeaks for distances of up to 5.0 Å and weak crosspeaks for distances up to 6.0 Å. Consequently, the lack of contacts in meta II indicate that retinal – EL2 distances are on the order of 6.0 Å or more. In rhodopsin, we observed strong contacts between the 13C1-Cys187 on EL2 and the retinal 13C12 and 13C20 carbons (Fig. 2b). In the rhodopsin crystal structure6, Cys187 is 4.21 Å and 6.22 Å from the retinal C12 and C20 carbons, respectively. On conversion to meta II, we lose both retinal contacts with Cys187 consistent with an increase in separation between EL2 and the retinal.
Further support for separation between retinal and EL2 in meta II comes from 1) the loss of tyrosine-glycine contacts in meta II and 2) assignment of a crosspeak at 46.5 ppm between the 13C20 methyl carbon on the retinal and a 13Cα-glycine residue. There are two Tyr-Gly contacts that connect EL2 with the extracellular ends of TM helices H3 and H6, namely Tyr268-Gly188 and Tyr178-Gly114. Both contacts are lost in meta II (Supplementary Fig. 3). There are only two glycines in the binding cavity close to the C20 methyl group, i.e. Gly114 on H3 and Gly188 on EL2 (Fig. 3). In meta II, we assign the C20-Gly crosspeak to Gly114 (H3) based on the presence of this resonance in the 2D DARR spectrum of the G188A mutant of meta II. The assignment of a C20-Gly114 contact in meta II indicates that the C20-Gly188 contact is lost despite the large rotation of the C20 methyl group towards EL2 (Supplementary Figs. 4, 5).
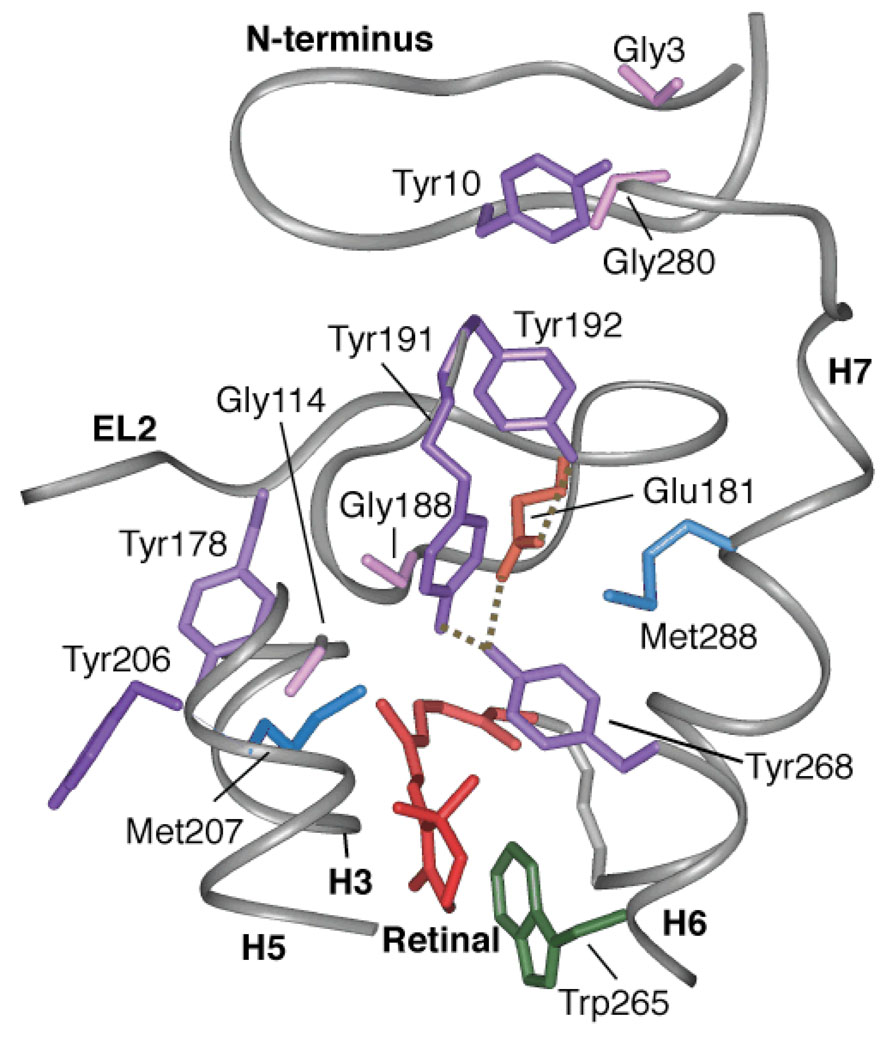
Figure 3.
A view of the extracellular side of the rhodopsin from the crystal structure6. The figure highlights the relative position of six tyrosines: Tyr10, Tyr178, Tyr191, Tyr192, Tyr206 and Tyr268. Of these tyrosines, Tyr191, Tyr192 and Tyr268 are involved in the H-bonding network with Glu181. Tyr268 and Tyr191 are also in close contact with Met288 on H7. Tyr206 on H5 is involved in a second H-bonding network with His211 (H5), Glu122 (H3), Trp126 (H3) and Ala166 (H4) (not shown). Additionally, the figure shows Tyr-Gly interactions on the extracellular side of rhodopsin between Gly188-Tyr268, Gly3-Tyr10-Gly280 and Gly114-Tyr178.
The model in Fig. 2 shows the crystal structure of rhodopsin containing the 11-cis (red) retinal PSB tightly packed against EL2. The distances between the C20 methyl group and the 13C-labeled positions on Gly114, Cys187 and Gly188 are shown. Crosspeaks between the retinal C20 methyl group and each of these amino acids are observed in the dark. We superimpose the position of the all-trans retinal SB (orange) in meta II predicted using restrained MD simulations (Supplementary Table 1). To satisfy distance constraints derived from our NMR measurements, in the MD simulations the retinal shifts slightly toward the cytoplasmic side of the binding site and EL2 moves toward the extracellular surface.
Rearrangement of H-bonding networks involving EL2 and H5
The loss of EL2-retinal contacts in meta II and the changes observed in the chemical shifts for β4 strand indicate that EL2 changes position upon receptor activation. The next question is whether the H-bonding network involving EL2 remains intact or is disrupted in meta II?
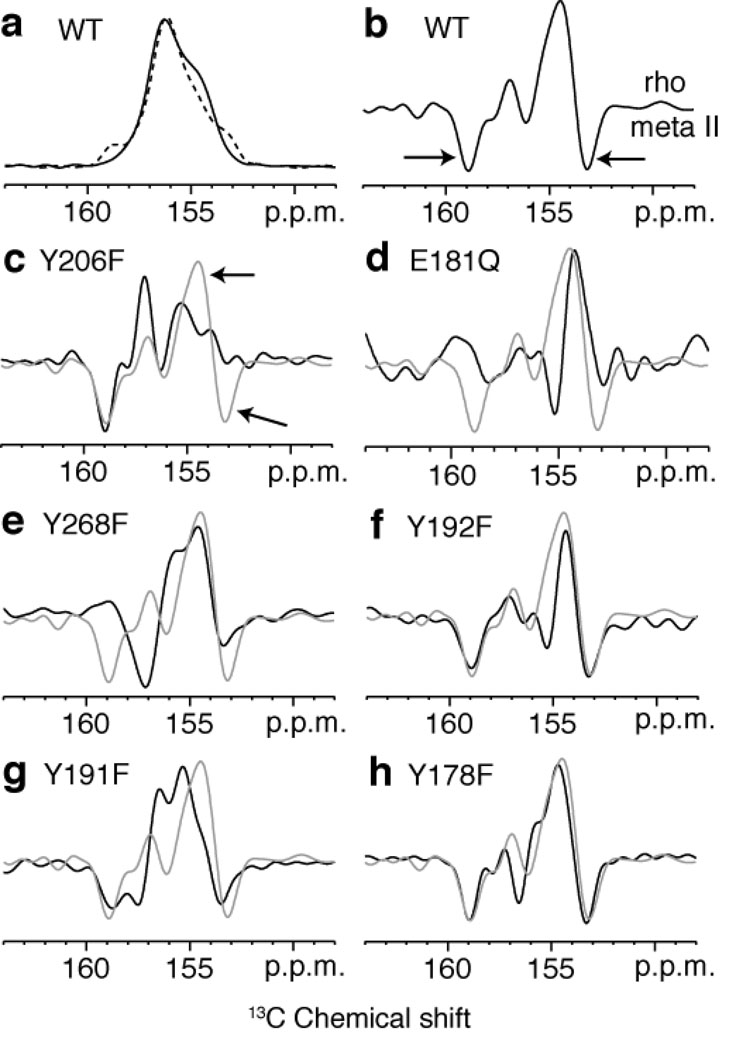
Tyrosines are an integral part of the EL2 H-bonding network6 (Fig. 3). The 13Cζ resonances of the 18 tyrosines in rhodopsin are not resolved (see Fig. 4a, black). However, the difference spectrum between rhodopsin and meta II highlights the 13Cζ-tyrosine resonances that change upon rhodopsin activation (Fig. 4b). There are two well-resolved shoulders in the meta II portion of the difference spectrum (Fig. 4b). The distinct meta II resonance at 153.6 ppm is readily assigned to Tyr206 on H5 on the basis of the loss of a Cζ-Tyr resonance at 153.6 ppm in the meta II component of the Y206F difference spectrum (Fig. 4c). Additional support for this assignment is provided in Supplementary Fig. 6. The upfield shift of 13Cζ-Tyr206 resonance is consistent with a weaker Cζ-OH hydrogen bond in meta II.
Figure 4.
One dimensional (1D) 13C CP-MAS spectra of rhodopsin and meta II labeled with 13Cζ-tyrosine. (a) Overlap of the 13C 1D CP-MAS spectra of the 13Cζ-tyrosine resonance in rhodopsin (black) and meta II (dashed black). Difference spectra for wild-type rhodopsin (b) and several rhodopsin mutants, Y206F (c), E181Q (d), Y268F (e), Y192F (f), Y191F (g), Y178F (h). The difference spectrum in gray corresponds to the wild-type protein.
The downfield resonance at 159.3 ppm is reflective of a more strongly H-bonded tyrosine17. Both tyrosines with unusual chemical shifts must be coupled to the H-bonding network involving Glu181 on EL2 since both resonances are lost in the tyrosine difference spectrum of the E181Q mutant (Fig. 4d). There is no evidence for a tyrosinate anion18, which would have a chemical shift closer to 165 ppm17.
To assign the tyrosine resonance at 159.3 ppm in meta II, difference spectra were collected for a series of rhodopsin mutants (Y268F, Y192F, Y191F and Y178F), where tyrosines in the retinal binding cavity near Glu181 were mutated one at a time to phenylalanine (Figs. 4eh). None of the 13Cζ-tyrosine difference spectra exhibit a complete loss of the negative peak at 159.3 ppm, except the Y268F mutant where the negative peak at 159.3 ppm appears to shift to 157.5 ppm. We are not able to assign the 159.3 ppm resonance to Tyr268 because of the appearance of a positive peak at 159.3 ppm in the dark spectrum of the Y268F mutant, which suggests that the mutation of Tyr268 is causing another tyrosine in the vicinity to become more strongly H-bonded. In the difference spectrum of the Y191F mutant, the negative peak at ~159 ppm is split into two components as compared to the wild-type difference spectrum.
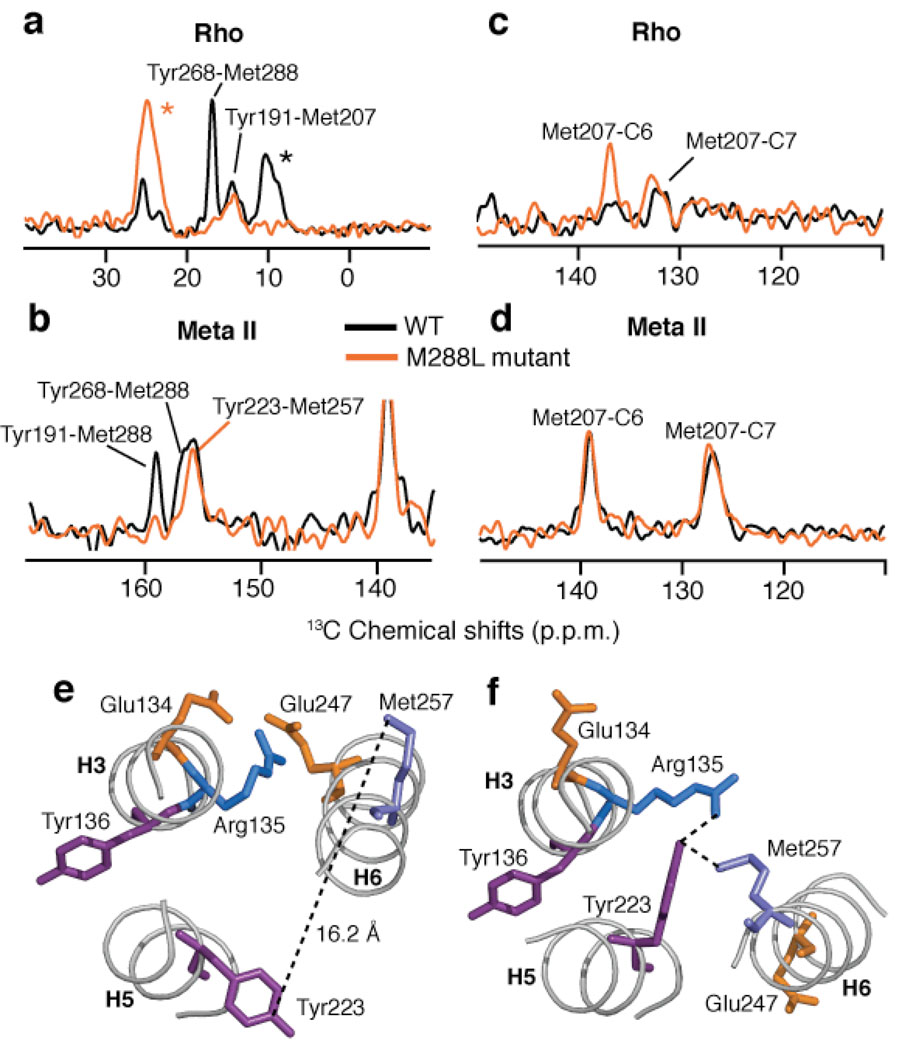
The loss of the 159.3 ppm resonance in the E181Q mutant and its sensitivity to mutation of Tyr268 and Tyr191 strongly suggest an assignment to one of the tyrosines associated with EL2. This assignment is supported by 2D DARR data obtained on rhodopsin labeled with 13Cζ-tyrosine and 13Cε-methionine. In the rhodopsin crystal structure (1U19), there are 5 Met(13Cε) - Tyr(13Cζ) pairs (Met288-Tyr268, 3.9 Å; Met207-Tyr191, 4.8 Å; Met288-Tyr191, 5.2 Å; Met253-Tyr306, 5.5 Å; Met288-Tyr192, 5.7 Å). In Fig. 5a, we observe two crosspeaks between tyrosine and methionine that we assign to the closest Met-Tyr pairs (i.e. Met288-Tyr268 and Met207-Tyr191). Conversion to meta II generates a crosspeak between the tyrosine resonance at 159.3 ppm and a methionine resonance at 12.8 ppm. We can assign this methionine to Met288 on H7 based on the loss of this crosspeak in the M288L mutant (orange, Fig. 5b).
Figure 5.
2D DARR NMR of Tyr(Cζ) - Met(Cε) contacts in rhodopsin and the M288L rhodopsin mutant. (a) Rows through the 13Cζ-Tyr diagonal resonance from 2D DARR NMR spectra of rhodopsin (black) and the M288L rhodopsin mutant (orange) labeled with 13Cζ-tyrosine and 13Cε-methionine. (b) Rows through the 13Cε-Met diagonal resonance from 2D DARR NMR spectra of meta II (black) and the M288L rhodopsin mutant (orange) following conversion to meta II. (c) Rows through the 13Cε-methionine diagonal resonance of rhodopsin (black) and the M288L rhodopsin mutant (orange) showing the crosspeaks to the retinal 13C6 and 13C7 resonances. (d) Same as in (c) following conversion to meta II. In the M288L mutant of rhodopsin, we observe a contact between Met207 and C6 that is not present in wild type rhodopsin. This change in the Met207-retinal contact in the M288L mutant of rhodopsin can be interpreted as either a change in the position of the retinal or in the position of Met207 on H5 upon mutation of Met288 (H7) to a leucine. Upon activation, the Met207 – retinal interactions in the M288L mutant are identical to those in wild-type meta II. (e) A view of the ionic lock between Arg135 and Glu247 from the crystal structure of rhodopsin20. The Tyr223-Met257 distance is well beyond the range of the DARR NMR experiment. (f) Structure of the ionic lock from the recent crystal structure of opsin21,22 showing the close proximity between Tyr223 and Met257. Asterisks correspond to MAS sidebands.
The M288L data along with the tyrosine difference spectra above indicate that the 159.3 ppm resonance belongs to either Tyr191 or Tyr268 in meta II. We assume that the strong H-bonding interaction for a tyrosine at 159.3 ppm is due to its interaction with Glu181 and that the appearance of a resonance at 159.3 ppm in the Y268F rhodopsin spectrum and in the Y191F meta II spectrum is because these mutations lead to the rearrangement of the EL2 H-bonding network. We assign the 159.3 ppm resonance in meta II to Tyr191 since we observe a crosspeak at 156.5 ppm between a tyrosine and the retinal C20 methyl group19 that we assign to Tyr268. The C20 methyl group is closer to Tyr268 (4.2 Å) than to Tyr191 (8.0 Å) in rhodopsin, and we expect that motion of EL2 away from the retinal would only increase the 13C20-Tyr191(13Cζ) distance. Together these data argue that Tyr191 becomes more strongly H-bonded in meta II, and that the H-bonding network involving the tyrosines and Glu181 on EL2 remains intact.
Coupling of EL2 displacement to rotation of helix H5
The data presented above on the E181Q and M288L mutants and in Supplementary Fig. 3 on the Y206F mutant suggest that light-induced structural changes in EL2 are strongly coupled to the H-bonding network centered on H5. First, in the E181Q mutant (Fig. 4d) the resonance at 153.6 ppm assigned to Tyr206 (H5) is lost. Second, in the M288L (H7) mutant a contact is gained between the ε-CH3 group of Met207 (H5) and the retinal C6 carbon (Fig. 5c). Third, in the Y206F (H5) mutant a Tyr-Gly contact is lost that likely involves Tyr10 or Tyr29 on the extracellular loops of rhodopsin since there are no glycines in the vicinity of Tyr206.
We propose that the functional unit is the EL2-H5 sequence. The crystal structure of rhodopsin shows that the β-strands in EL2 are extensively knit together by H-bonding interactions, which extend to Tyr268 on H6 and Glu113 on H366,20. If the motion of EL2 is coupled to motion of H5, then the Pro170-Pro171 sequence at the H4-β3 boundary may serve as a hinge leading to observable changes in the H-bonding interactions linking β3-to-H4 and H4-to-H5. We observe that many of the H-bonding contacts involving the extracellular ends of H4 and H5 change in meta II (Supplementary Fig. 1a).
We have previously shown that H5 undergoes a change in orientation in the region of His211 (Supplementary Fig. 6). Figs. 5e and 5f illustrate how rotation of H5 leads to disruption of the ionic lock between H3 and H6. In the dark-state of rhodopsin (Fig. 5e), Arg135 of the conserved ERY sequence on H3 interacts with Glu247 (H6). In the recent structure of opsin (Fig. 5f) with21 and without22 the Gα peptide bound, H5 has rotated and Tyr223 (a residue that is highly conserved across the GPCR family) now interacts directly with Arg135 and Met257 on H6. The Tyr223-Arg135 interaction is thought to be one element in breaking the ionic lock and stabilizing the active conformation of rhodopsin. In Fig. 5b, we observe a new Tyr-Met contact in meta II that we can assign to the Tyr223-Met257 interaction, consistent with the proposal that this active state geometry is maintained in the opsin structure22. Importantly, mutation of the Tyr223 to phenylalanine results in an appreciable increase in the decay rate of meta II to opsin (Supplementary Fig. 7) in agreement with the Tyr223-Arg135 interaction stabilizing the active conformation of the receptor. These results indicate that there are two critical H3–H5 interactions that hold helix H5 in an active geometry: Glu122-His21123,24 and Arg135-Tyr22321,22. The model of activation that emerges from these studies is one where steric contacts between the retinal β-ionone ring with H5 and the retinal C19 methyl group with EL2 shift the EL2-H5 sequence into an active geometry stabilized by H3–H5 contacts; retinals lacking either the ring25 or the C19 methyl group26 fail to activate rhodopsin, and mutation of Tyr223 to phenylalanine leads to rapid meta II decay.
DISCUSSION
EL2 controls access to the retinal binding site
The major conclusion from our studies is that EL2 changes position upon activation and that this change is coupled to motion of TM helix H5. We have recently defined the location of the retinal chromophore in meta II (S.A., Evan Crocker, M.E., P.J.R., M.S., S.O.S, unpublished data) and our current measurements between the β4 strand and the retinal indicate that there must be an increase in separation between the retinal and EL2 upon activation. The H-bonding network involving Glu181 appears to remain intact in meta II, and consequently the displacement of EL2 does not appear to be large.
Our observations can be compared with the crystal structures of opsin22 and a ‘photoactivated’ (deprotonated) intermediate of rhodopsin27. In these structures, EL2 has not moved to any appreciable extent. The differences between meta II and opsin suggest that the all-trans retinal Schiff base is holding EL2 in an active conformation in meta II. Release of retinal to form opsin allows the binding site residues to rearrange and EL2 to shift back to roughly its position in rhodopsin.
Displacement of EL2 away from the retinal is consistent with studies showing that the retinal binding site becomes more accessible to water and hydroxylamine in meta II28,29. Mutation of many of the residues in the H-bonding network involving EL2, such as Glu18130 and Tyr19231, results in increased accessibility of the retinal PSB to hydroxylamine in the dark. Also, Furutani et al.32 observed the appearance of an N-D amide A vibration at 2366 cm−1 in meta II that they attributed to access of the EL2 β-hairpin to hydrogen-deuterium exchange in the meta I-to-meta II transition. Interestingly, neither disruption of the Cys110-Cys187 disulfide bond by mutation to alanine nor disruption of the salt bridge between Arg177 and Asp190 on EL2 increases hydroxylamine accessibility33,34 suggesting that the H-bonding network involving Glu181 is alone sufficient to keep EL2 tightly capped over the retinal binding site.
In a parallel fashion, EL2 may serve to control the access of small molecule ligands to interior binding sites within the ligand-activated GPCRs. For example, alanine scanning mutagenesis of the M1 muscarinic acetylcholine receptor revealed that the access of ligands to the binding site was increased by mutation of EL2 residues35. Furthermore, substituted cysteine accessibility studies of the dopamine D2 receptor showed that the extracellular part of H5 is accessible to hydrophilic reagents36. Finally, the recent crystal structure of the β2AR with a bound partial inverse agonist14 shows that EL2 does not cap the amine-binding site, as in rhodopsin. Taken together, the studies on GPCRs activated by small molecule ligands suggest that there is a dynamic role of EL2 in allowing water and ligands to enter the interior binding sites.
EL2 as a negative regulator in GPCR activation
Several studies have suggested that EL2 serves a role as a negative regulator in the class A GPCRs. The simple idea is that EL2 has multiple interactions with the extracellular ends of the TM helices in the inactive state and that displacement of EL2 upon ligand binding allows H5, H6 and H7 to adopt active conformations. For example, Baranski and colleagues3 showed that a high degree of constitutive activity is associated with the mutation of residues in EL2 of the C5a receptor. They proposed that mutation of EL2 increases the flexibility of the loop and releases inhibitory constraints. The high degree of basal activity in the melanocortin receptor, which has a short EL2 and lacks the conserved disulfide bond, was explained by a related mechanism13. Finally, crosslinking in the putative ligand binding site37,38 and metal binding sites39 in the vicinity of EL2 modulate receptor activity. These modifications were designed to mimic the movement of the TM helices, and for this to occur, EL2 was envisioned to change conformation or position.
In rhodopsin, EL2 has also been implicated as a negative regulator of receptor activity. It is known that mutation of Tyr191 and Tyr192 to leucine decreases the stability of the binding pocket leading to faster meta II decay rates40, and that mutation of Ser186 to alanine and Glu181 to phenylalanine strongly perturbs the kinetics of rhodopsin activation41. However, none of the EL2 mutants tested in rhodopsin have been shown to display constitutive activity. This may be due to the presence of additional regulatory elements, such as the interaction between the retinal PSB and its Glu113 counterion and the tight packing between the 11-cis retinal and conserved Trp265 (H6), which all contribute to low dark noise in rhodopsin.
EL2-H5 as a structural unit in GPCRs
One of the challenges in understanding the mechanism of GPCR activation is to establish how retinal isomerization42,43 or ligand binding39,44 produces rigid body motion of the TM helices. Our results suggest that motion of EL2 is coupled to motion of H5 and breaking of the ionic lock.
Tight coupling between EL2 and H5 is supported in studies on ligand-activated GPCRs45–48. Wurch et al.46 addressed the coupling of EL2 and H5 by replacing the EL2-H5 sequence from the 5HT1D serotonin receptor with the corresponding sequence from the 5HT1B serotonin receptor. They found that it was necessary to replace the entire EL2-H5 sequence in order to recover antagonist binding; replacing either the EL2 or H5 sequence alone markedly decreased binding. Also, the idea that EL2 is a structured unit is reflected in GnRH receptor studies where Pfeger et al.48 showed that exchange of the entire EL2 from another species had less effect on ligand binding affinity than point mutations of EL2 within a species.
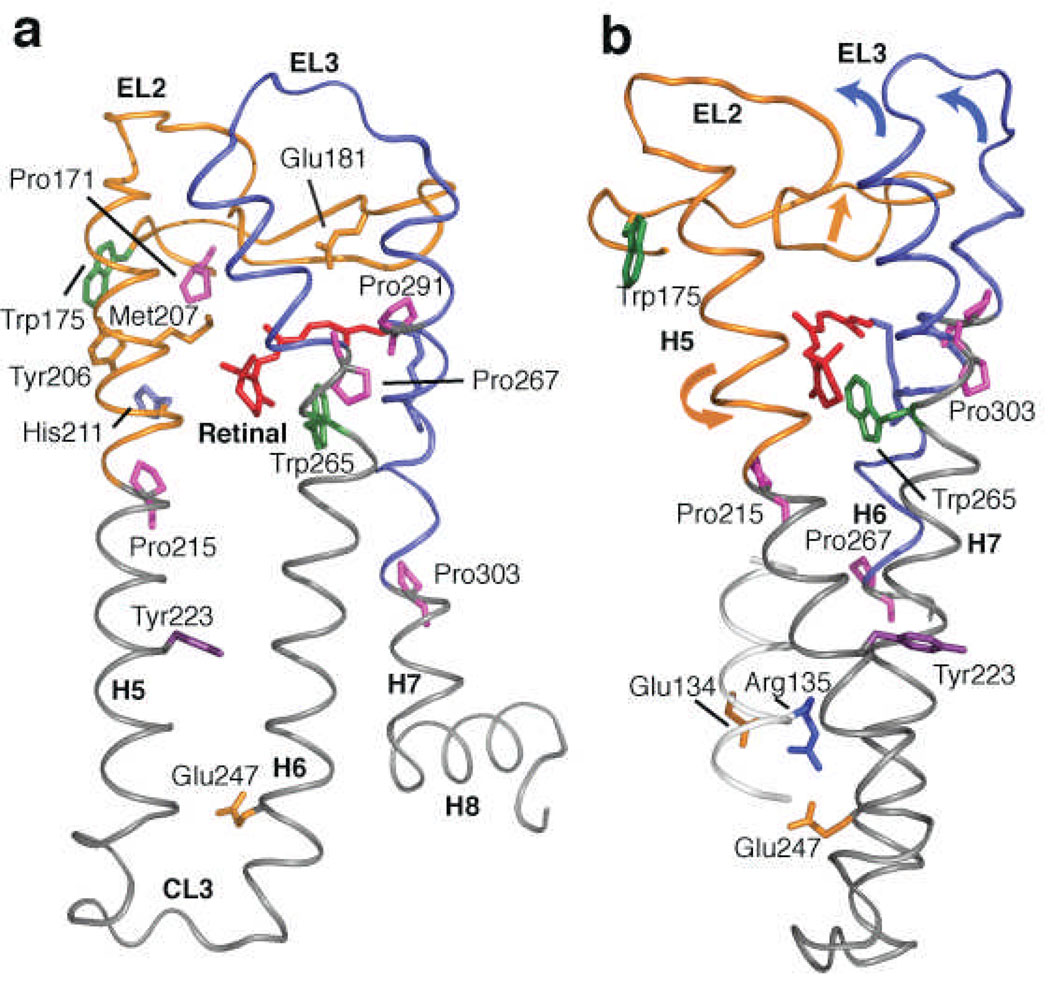
Fig. 6 highlights the helix-loop-helix (HLH) segments involving EL2 and EL3. Motion of EL2 away from the retinal-binding site is coupled to the outward displacement of the extracellular end of H5 and the inward displacement and rotation of the intracellular end of H521,22. The outward displacement of H5 is driven by steric interaction with the retinal β-ionone ring and is stabilized by a direct Glu122-His211 interaction. Motion of EL2 may allow the extracellular end of the H6-EL3-H7 segment to pivot toward the center of the protein and conversely allow the intracellular end of H6 to rotate outward42,49. Inward motions of the extracellular ends of H6 and H7 are captured in the global toggle switch model of GPCR activation39.
Figure 6.
Crystal structure of rhodopsin20 highlighting EL2 and H5. (a) Retinal isomerization within the tightly packed binding site results in steric contacts between the β-ionone ring and H5, and between the retinal C19 and C20 methyl groups and EL2. These interactions trigger the simultaneous displacement of EL2 and H5. Motion of the β-ionone ring is also coupled to the motion of Trp265. Trp265 is packed against the β-ionone ring and C20 of the retinal, as well as Gly121 on H3 and Ala295 on H7. Movement of the Trp265 sidechain away from these critical contacts allows helices H6 and H7 to shift into active conformations. The coupled motions of helices H5–H7, in turn, are coupled to the rearrangement of electrostatic interactions involving the conserved ERY sequence at the cytoplasmic end of H3, exposing the G protein binding site on the cytoplasmic surface of the protein. (b) View of the rhodopsin crystal structure highlighting the interaction between EL2 and EL3 on the extracellular side of the receptor, and the positions of Tyr223 and the conserved Glu135-Arg135-Tyr136 sequence on the intracellular side of the receptor.
Additionally, Fig. 6 shows the positions of key tryptophan residues in rhodopsin, Trp265 (H6) and Trp175 (H4). Trp265 is conserved throughout the class A GPCRs and is an important element of the activation mechanism of rhodopsin50. Trp175 is at the junction of EL2 with H4 and H5. In rhodopsin, the W175F mutation is one of the only mutations in the H4-EL2-H5 segment that leads to constitutive activity51. The fact that this tryptophan is highly conserved in the visual receptors, but not in other class A GPCRs, suggests that the H4-EL2-H5 sequence up to Pro215 is specific to different subfamilies of class A GPCRs.
In conclusion, the structural constraints described above provide insights into how EL2 and its extensive H-bonding interactions play a role in coupling retinal isomerization to the activation of rhodopsin. The subfamily specific H4-EL2-H5 unit in rhodopsin holds H5 and the extracellular ends of H6 and H7 in inactive conformations. Retinal isomerization and displacement of EL2 from the retinal-binding site is coupled to motion of H5 and to the inward motion of the H6-EL3-H7 unit. Similar motions are likely to occur in other GPCRs39,52 suggesting that EL2 may act as a plug or cork that must be released or rearranged for receptor activation.
METHODS
Expression and Purification of 13C-Labeled Rhodopsin
A stable tetracycline-inducible HEK293S cell line53 containing the bovine opsin gene and its mutants54 was used to express rhodopsin. The cells were grown in Dulbecco’s modified Eagle’s medium55 prepared from cell culture-tested components (Sigma, St. Louis, MO). Suspension cultures were grown using a bioreactor in medium with specific 13C-labeled amino acids (Cambridge Isotope Laboratories, Andover, MA), dialyzed (3 times against 20 L phosphate buffered saline (PBS) per liter serum56), heat-inactivated fetal bovine serum (10% v/v), Pluronic F-68 (0.1% w/v), dextran sulfate (300 mg L−1), penicillin (100 units mL−1) and streptomycin (100 µg mL−1). On day 4 after incubation, cells were fed with 2.4 g L−1 glucose. Opsin gene expression was induced 5 days after inoculation by addition of both tetracycline (2 mg L−1) and 5 mM (final concentration) sodium butyrate to the growth medium53 and cells were harvested on day 7.
The HEK293S cell pellets were resuspended in 40 mL PBS per liter cell culture + protease inhibitors54 and unlabeled 11-cis retinal was added in two steps to a final concentration of 15 µM. The rhodopsin containing cells were solubilized in 40 mL L−1 of PBS + DDM (1% w/v) for 4 hours at room temperature. Subsequent purification by immunoaffinity chromatography using the rho-1D4 antibody (National Cell Culture Center, Minneapolis, MN) was carried out as described previously54. The eluted rhodopsin fractions were pooled and concentrated to a final volume of ~400 µL using 10 kDa MWCO Centricon devices (Amicon, Bedford, MA).
Synthesis of 13C-Labeled Retinals and Regeneration into Rhodopsin
Specific 13C-labeled retinals were synthesized by previously described methods57,58 and purified using High Pressure Liquid Chromatography (HPLC) as previously described50.
Regeneration of the rhodopsin pigments with 13C-labeled retinal in DDM micelles was performed by illumination of the concentrated samples containing a 2:1 molar ratio of labeled retinal-to-protein, as described previously19. Typically, greater than 85% of the sample was regenerated with labeled retinal. Different regeneration rates were observed for wild type and mutant opsins. A stream of argon gas was used to evaporate the regenerated sample down to a volume of 60 µL.
Solid-State NMR Spectroscopy
Concentrated samples (7–10 mgs) were loaded into 4 mm MAS zirconia rotors. All NMR spectra were acquired at a static magnetic field strength of 14.1 T (600 MHz) on a Bruker AVANCE spectrometer using double-channel 4 mm MAS probes as described previously50. Typically, MAS spinning rates of 8–12 KHz were used. 1D 13C spectra were acquired using ramped amplitude cross polarization with contact times of 2 ms and acquisition times of the order of 16 ms for all experiments. Intermolecular 13C…13C distance constraints on rhodopsin in the inactive and the active state were obtained using Dipolar Assisted Rotational Resonance (DARR) recoupling technique with a mixing time of 600 ms to maximize homonuclear recoupling between different 13C labels. The 1H radiofrequency field strength during mixing was matched to the MAS speed for each sample, satisfying the n=1 matching condition. Two-pulse phase modulated or SPINAL64 proton decoupling was typically used during the evolution and acquisition periods with a radiofrequency field strength of 80–90 kHz. In each 2D data set, 1024 time domain points in the f2 (direct) dimension and 64 points in the f1 (indirect) dimension were acquired. All experiments were conducted at −80°C. 13C spectra were referenced externally to the carbonyl resonance of powdered glycine at 176.46 ppm relative to neat TMS at 0.0 ppm.
Trapping of the Metarhodopsin II Intermediate
Samples were illuminated for 45–60 sec at room temperature in the NMR rotor using a 400 W lamp with a >495 nm cutoff filter and immediately placed in the NMR probe with the probe stator warmed to 5°C. Under slow spinning (~2 kHz) the sample was frozen within 3 min of illumination using N2 gas cooled to −80°C. To confirm that meta II conversion was complete and stably trapped, chemical shift changes of the 13C labeled carbons of the polyene chain of the retinal were monitored as they are sensitive to both protonation and isomerization. The linewidths of the resolved protein and retinal NMR resonances are generally between 1 and 2 ppm in both rhodopsin and meta II. The absence of line broadening or resonance splitting indicates we have trapped a spectroscopically well-defined meta II state. The time between illumination and freezing of the sample is approximately 3 min indicating that the proton uptake in our sample is complete; the intermediate we have trapped is functionally equivalent to meta II in ROS membranes as it can activate transducin59. Also, it has been shown that the vibrational frequencies observed in FTIR difference spectrum of meta II minus rhodopsin are identical for rhodopsin in DDM or ROS membranes 60.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH-NSF instrumentation grants (S10 RR13889 and DBI-9977553), a grant from the NIH to S.O.S (GM-41412), and from US-Israel Binational Science Foundation to M.S. We thank Chikwado A Opefi for help with the M288A and M288L mutants, and gratefully acknowledges the W.M. Keck Foundation for support of the NMR facilities in the Center of Structural Biology at Stony Brook. MS acknowledges support from Kimmelman center for Biomolecular Structure and Assembly.
References
- 1.Samson M, et al. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J. Biol. Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 2.Shi L, Javitch JA. The second extracellular loop of the dopamine D-2 receptor lines the binding-site crevice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:440–445. doi: 10.1073/pnas.2237265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klco JM, Wiegand CB, Narzinski K, Baranski TJ. Essential role for the second extracellular loop in C5a receptor activation. Nat. Struct. Mol. Biol. 2005;12:320–326. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 4.Scarselli M, Li B, Kim SK, Wess J. Multiple residues in the second extracellular loop are critical for M-3 muscarinic acetylcholine receptor activation. J. Biol. Chem. 2007;282:7385–7396. doi: 10.1074/jbc.M610394200. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 7.Karnik SS, Khorana HG. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J. Biol. Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- 8.Hwa J, Klein-Seetharaman J, Khorana HG. Structure and function in rhodopsin: Mass spectrometric identification of the abnormal intradiscal disulfide bond in misfolded retinitis pigmentosa mutants. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4872–4876. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg G, Ottolenghi M, Sheves M. pKa of the protonated Schiff base of bovine rhodopsin: A study with artificial pigments. Biophys. J. 1993;64:1499–1502. doi: 10.1016/S0006-3495(93)81518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakmar TP, Franke RR, Khorana HG. The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength absorbance and Schiff base pKa. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3079–3083. doi: 10.1073/pnas.88.8.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen GB, Oprian DD, Robinson PR. Mechanism of activation and inactivation of opsin: Role of Glu113 and Lys296. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 12.Rader AJ, et al. Identification of core amino acids stabilizing rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holst B, Schwartz TW. Molecular mechanism of agonism and inverse agonism in the melanocortin receptors - Zn2+ as a structural and functional probe. Ann. N. Y. Acad. Sci. 2003;994:1–11. doi: 10.1111/j.1749-6632.2003.tb03156.x. [DOI] [PubMed] [Google Scholar]
- 14.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto H, Yoshizawa T. Recognition of opsin to longitudinal length of retinal isomers in formation of rhodopsin. Vision Res. 1978;18:607–609. doi: 10.1016/0042-6989(78)90212-2. [DOI] [PubMed] [Google Scholar]
- 16.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J. Biomol. NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 17.Herzfeld J, et al. Solid-state 13C NMR study of tyrosine protonation in dark-adapted bacteriorhodopsin. Biochemistry. 1990;29:5567–5574. doi: 10.1021/bi00475a022. [DOI] [PubMed] [Google Scholar]
- 18.DeLange F, et al. Tyrosine structural changes detected during the photoactivation of rhodopsin. J. Biol. Chem. 1998;273:23735–23739. doi: 10.1074/jbc.273.37.23735. [DOI] [PubMed] [Google Scholar]
- 19.Patel AB, et al. Coupling of retinal isomerization to the activation of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 21.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 22.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 23.Patel AB, et al. Changes in interhelical hydrogen bonding upon rhodopsin activation. J. Mol. Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 24.Imai H, et al. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2322–2326. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jäger F, et al. Interactions of the β-ionone ring with the protein in the visual pigment rhodopsin control the activation mechanism. An FTIR and fluorescence study on artificial vertebrate rhodopsins. Biochemistry. 1994;33:7389–7397. doi: 10.1021/bi00189a045. [DOI] [PubMed] [Google Scholar]
- 26.Ganter UM, Schmid ED, Perez-Sala D, Rando RR, Siebert F. Removal of the 9-methyl group of retinal inhibits signal transduction in the visual process. A Fourier transform infrared and biochemical investigation. Biochemistry. 1989;28:5954–5962. doi: 10.1021/bi00440a036. [DOI] [PubMed] [Google Scholar]
- 27.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 30.Yan ECY, et al. Function of extracellular loop 2 in rhodopsin: Glutamic acid 181modulates stability and absorption wavelength of metarhodopsin II. Biochemistry. 2002;41:3620–3627. doi: 10.1021/bi0160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janz JM, Farrens DL. Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J. Biol. Chem. 2004;279:55886–55894. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- 32.Furutani Y, Shichida Y, Kandori H. Structural changes of water molecules during the photoactivation processes in bovine rhodopsin. Biochemistry. 2003;42:9619–9625. doi: 10.1021/bi034592k. [DOI] [PubMed] [Google Scholar]
- 33.Davidson FF, Loewen PC, Khorana HG. Structure and function in rhodopsin: Replacement by alanine of cysteine residues 110 and 187, components of a conserved disulfide bond in rhodopsin, affects the light-activated metarhodopsin II state. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janz JM, Fay JF, Farrens DL. Stability of dark state rhodopsin is mediated by a conserved ion pair in intradiscal loop E-2. J. Biol. Chem. 2003;278:16982–16991. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin JA, Hulme EC, Langmead CJ, Tehan BG. Roof and floor of the muscarinic binding pocket: Variations in the binding modes of orthosteric ligands. Mol. Pharmacol. 2007;72:1484–1496. doi: 10.1124/mol.107.038265. [DOI] [PubMed] [Google Scholar]
- 36.Javitch JA, Fu D, Chen J. Residues in the fifth membrane-spanning segment of the dopamine D2 receptor exposed in the binding-site crevice. Biochemistry. 1995;34:16433–16439. doi: 10.1021/bi00050a026. [DOI] [PubMed] [Google Scholar]
- 37.Struthers M, Yu HB, Oprian DD. G protein-coupled receptor activation: Analysis of a highly constrained, "straitjacketed" rhodopsin. Biochemistry. 2000;39:7938–7942. doi: 10.1021/bi000771f. [DOI] [PubMed] [Google Scholar]
- 38.Han SJ, et al. Identification of an agonist-induced conformational change occurring adjacent to the ligand-binding pocket of the M-3 muscarinic acetylcholine receptor. J. Biol. Chem. 2005;280:34849–34858. doi: 10.1074/jbc.M506711200. [DOI] [PubMed] [Google Scholar]
- 39.Elling CE, et al. Metal ion site engineering indicates a global toggle switch model for seven-transmembrane receptor activation. J. Biol. Chem. 2006;281:17337–17346. doi: 10.1074/jbc.M512510200. [DOI] [PubMed] [Google Scholar]
- 40.Doi T, Molday RS, Khorana HG. Role of the intradiscal domain in rhodopsin assembly and function. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4991–4995. doi: 10.1073/pnas.87.13.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan ECY, et al. Photointermediates of the rhodopsin S186A mutant as a probe of the hydrogen-bond network in the chromophore pocket and the mechanism of counterion switch. J. Phys. Chem. C. 2007;111:8843–8848. [Google Scholar]
- 42.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 43.Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 44.Sheikh SP, et al. Similar structures and shared switch mechanisms of the β2-adrenoceptor and the parathyroid hormone receptor - Zn(II) bridges between helices III and VI block activation. J. Biol. Chem. 1999;274:17033–17041. doi: 10.1074/jbc.274.24.17033. [DOI] [PubMed] [Google Scholar]
- 45.Olah ME, Jacobson KA, Stiles GL. Role of the 2nd extracellular loop of denosine receptors in agonist and antagonist binding - Analysis of Chimeric A1/A3-adenosine receptors. J. Biol. Chem. 1994;269:24692–24698. [PMC free article] [PubMed] [Google Scholar]
- 46.Wurch T, Colpaert FC, Pauwels PJ. Chimeric receptor analysis of the ketanserin binding site in the human 5-hydroxytryptamine1D receptor: Importance of the second extracellular loop and fifth transmembrane domain in antagonist binding. Mol. Pharmacol. 1998;54:1088–1096. doi: 10.1124/mol.54.6.1088. [DOI] [PubMed] [Google Scholar]
- 47.Conner M, et al. Systematic analysis of the entire second extracellular loop of the V-1a vasopressin receptor - Key residues, conserved throughout a G-protein-coupled receptor family, identified. J. Biol. Chem. 2007;282:17405–17412. doi: 10.1074/jbc.M702151200. [DOI] [PubMed] [Google Scholar]
- 48.Pfleger KDG, Pawson AJ, Millar RP. Changes to gonadotropin-releasing hormone (GnRH) receptor extracellular loops differentially affect GnRH analog binding and activation: Evidence for distinct ligand-stabilized receptor conformations. Endocrinology. 2008;149:3118–3129. doi: 10.1210/en.2008-0002. [DOI] [PubMed] [Google Scholar]
- 49.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crocker E, et al. Location of Trp265 in metarhodopsin II: Implications for the activation mechanism of the visual receptor rhodopsin. J. Mol. Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 51.Madabushi S, et al. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J. Biol. Chem. 2004;279:8126–8132. doi: 10.1074/jbc.M312671200. [DOI] [PubMed] [Google Scholar]
- 52.Holst B, Elling CE, Schwartz TW. Partial agonism through a zinc-ion switch constructed between transmembrane domains III and VII in the tachykinin NK1 receptor. Mol. Pharmacol. 2000;58:263–270. doi: 10.1124/mol.58.2.263. [DOI] [PubMed] [Google Scholar]
- 53.Reeves PJ, Kim JM, Khorana HG. Structure and function in rhodopsin: A tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeves PJ, Thurmond RL, Khorana HG. Structure and function in rhodopsin: High level expression of a synthetic bovine opsin gene and its mutants in stable mammalian cell lines. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dulbecco R, Freeman G. Plaque production by the polyoma virus. Virology. 1959;8:396–397. doi: 10.1016/0042-6822(59)90043-1. [DOI] [PubMed] [Google Scholar]
- 56.Eilers M, Reeves PJ, Ying WW, Khorana HG, Smith SO. Magic angle spinning NMR of the protonated retinylidene schiff base nitrogen in rhodopsin: Expression of 15N-lysine and 13C-glycine labeled opsin in a stable cell line. Proc. Natl. Acad. Sci. U.S.A. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lugtenburg J. The synthesis of 13C-labeled retinals. Pure Appl. Chem. 1985;57:753–762. [Google Scholar]
- 58.Crocker E, et al. Dipolar assisted rotational resonance NMR of tryptophan and tyrosine in rhodopsin. J. Biomol. NMR. 2004;29:11–20. doi: 10.1023/B:JNMR.0000019521.79321.3c. [DOI] [PubMed] [Google Scholar]
- 59.Han M, Groesbeek M, Smith SO, Sakmar TP. Role of the C9 methyl group in rhodopsin activation: Characterization of mutant opsins with the artificial chromophore 11-cis-9-demethylretinal. Biochemistry. 1998;37:538–545. doi: 10.1021/bi972060w. [DOI] [PubMed] [Google Scholar]
- 60.Fahmy K, et al. Protonation states of membrane-embedded carboxylic acid groups in rhodopsin and metarhodopsin II: A Fourier-transform infrared spectroscopy study of site-directed mutants. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.