SUMMARY
The power of genetic association studies to identify disease susceptibility alleles fundamentally relies on the variants studied. The standard approach is to determine a set of tagging-SNPs (tSNPs) that capture the majority of genomic variation in regions of interest by exploiting local correlation structures. Typically, tSNPs are selected from neutral discovery panels, collections of individuals comprehensively genotyped across a region. We investigated the implications of discovery panel design on tSNP performance in association studies using realistically-simulated sequence data. We found that discovery panels of 24 sequenced ‘neutral’ individuals (similar to NIEHS or HapMap ENCODE data) were sufficient to select well-powered tSNPs to identify common susceptibility alleles. For less common alleles (0.01–0.05 frequency) we found neutral panels of this size inadequate, particularly if low-frequency variants were removed prior to tSNP selection; superior tSNPs were found using panels of diseased individuals. Only large neutral panels (200 individuals) matched diseased panel performance in selecting well-powered tSNPs to detect both common and rarer alleles. The 1000 Genomes Project initiative may provide larger neutral panels necessary to identify rarer susceptibility alleles in association studies. In the interim, our results suggest investigators can boost power to detect such alleles by sequencing diseased individuals for tSNP selection.
Keywords: genetic association study, discovery panel, tagging-SNP
INTRODUCTION
Extensive coverage of human genetic variation is now readily available (Frazer et al., 2007). The existence of publicly available sequencing data from the NIEHS SNPs Program (Livingston et al., 2004), SeattleSNPs (Carlson et al., 2004), HapMap ENCODE (HapMap Consortium, 2005) and dense HapMap data (HapMap Consortium, 2003) have provided investigators with the means to more rigorously select variants for subsequent association study. The utility of currently available genetic data resources is immense, and many successful findings have resulted (Eeles et al., 2008, Salonen et al., 2007, Klein et al., 2005). However, it is also important to recognize the limitations, especially with respect to identifying the less common underlying variants that are sure to exist (Eberle et al., 2007, Khoury et al., 2007). The common-disease, common-variant hypothesis is undergoing scrutiny (Campbell & Manolio, 2007); some researchers suggest many rare functional SNPs are likely major contributors to common disease susceptibility (Gorlov et al., 2008), and genomewide association studies have uncovered only a very small proportion of the total number of variants hypothesized to be involved (Iles, 2008). Until such time as full genomic sequencing of all individuals is technically and financially viable in association studies, approaches to selecting tSNPs that increase power to detect rarer variants are needed.
Current resources were designed to target common population variants and sequencing and genotyping map data are only publicly available for a limited number of individuals. For a single ethnic/racial group, the maximum number with sequence data is 24 individuals (Livingston et al., 2004, HapMap Consortium, 2005), and the maximum with map data is 60 unrelated individuals (Carlson et al., 2004, HapMap Consortium, 2003). The individuals in these ‘discovery panels’ are ‘neutral’, that is, chosen without regard to disease status. The distinct advantage of neutral discovery panels is that they are universally applicable. However, it has been shown that small, neutral panels are inadequate to detect and characterize the genomic variation surrounding less common alleles (Zeggini et al., 2005, Iles, 2008), leading to sub-optimal tSNPs. This problem is exacerbated using tSNP selection procedures that pre-screen variants by restricting consideration to only more common variants (Zeggini et al., 2005). Larger neutral panels are better able to detect and characterize genomic structure around less common variants, and we hypothesize that so too will discovery panels consisting of affected individuals (diseased panels) as they are enriched for susceptibility alleles and structure through more recently shared common ancestry.
In our unique investigation, we simulated 1,000 replicates of a 250 kb genomic region using a realistic coalescent model to study the limitations of current discovery panels. For a spectrum of scenarios including a variety of underlying disease models, with varying tagging-SNP protocols and data densities (sequencing or map data), we compared the efficacy of tSNPs selected from diseased and neutral discovery panels of different sizes.
METHODS
In each simulation, a population of 100,000 haplotypes were generated using ms software (Hudson, 2002) and standard parameters: mutation rate of 10−8 per base per meiosis, uniform recombination rate of 1 cM per Mb, and an effective population size of 104 (HapMap Consortium, 2005, Phillips et al., 2003). For a spectrum of scenarios including a variety of underlying disease models, with varying tagging-SNP protocols and data densities (sequencing or map data), we compared the efficacy of tSNPs selected from diseased and neutral discovery panels of different sizes. Nine single-locus disease models were explored based on dSNP minor allele frequencies (MAFs) of approximately 0.20, 0.05, and 0.01 and multiplicative genotypic relative risks (GRRs) of 1.2, 2.0 and 4.0 to represent small to moderate effect sizes (Khoury et al., 2007, Ioannidis). A constant sporadic affected rate of 0.05 was assumed (Gloeckler Ries et al., 2003, Jemal et al., 2007). From the coalescent-generated haplotypes in each simulation, a variant of appropriate MAF was randomly selected as the dSNP. A range around each targeted MAF was determined to yield approximately 20–30 variants within the specified range from which the dSNP was randomly selected: for MAF of 0.20, 0.17–0.23; for MAF of 0.05, 0.47 to 0.53; and for MAF of 0.01, 0.009 to 0.011. Diploid individuals were created by sampling with replacement from the haplotype population. Disease status was assigned based on the dSNP genotype and disease model considered. Discovery panels of neutral individuals ignored disease status; individuals in diseased panels were diseased. For each model, neutral and diseased discovery panels of size 200, 100, 60 and 24 individuals and an independent sample of 1,000 cases and 1,000 controls were generated. One thousand replicates were generated for each.
SNP tagging software based on the pairwise linkage disequilibrium statistic r2, ldSelect (Carlson et al., 2004) was used to select tSNPs from marker genotypes (with phase removed) for each panel in each replicate. Either all variants were considered in the tagging procedure (unrestricted), or only those above a MAF threshold of 0.01 or 0.05 (restricted). An r2 threshold of 0.8 for binning SNPs was used. Hence, tSNPs selected should represent all variants identified in the discovery panel with a minimum correlation of 0.8. For each scenario, the correlation between tSNPs selected in each panel and the known disease SNP (dSNP) were calculated in the 4,000 phase-known haplotypes from an independent sample of 1,000 cases and 1,000 controls, and the best tSNP noted (highest r2). We chose r2 as our measure for correlation because it has been widely used in assessing tSNP performance (HapMap Consortium, 2005, Frazer et al., 2007, Eberle et al., 2007), and is inversely proportional to the sample size multiplier necessary to approximately maintain equivalent power to the dSNP itself in an association analysis (Amos, 2007, Klein, 2007, Pritchard & Przeworski, 2001). Across all 1,000 data replicates, the r2 achieved or exceeded by 80% of best tSNPs (20th percentile r2 value) was determined (r280). The r280 measure was used to compare the different scenarios and can be interpreted as the correlation that will be achieved by the best tSNP with 80% probability. Due to non-normality, differences in r280 between panels were assessed using a two-sample Kolmogorov-Smirnov test (SAS v.9.1). A Monte Carlo estimate (10,000 simulations) of the p-value was used as asymptotic results may be unreliable when the distribution of the data is skewed, or heavily tied (Agresti, 1992). To assess the relative impact on power, 1/r280 was used as the ‘sample size’ multiplier necessary to achieve a specified level of power (Thompson et al., 2003, Pritchard & Przeworski, 2001).
RESULTS
In our realistically simulated regions, the frequency distribution and total number of variants simulated closely matched that observed in the HapMap ENCODE regions. For the neutral discovery panel of size 24, we observed an average of 441 SNPs in 1,000 replicates of 250 kb, or 1 per 567 bp (95%CI, 1 per 563 to 571 bp), compared to 11,974 SNPs across 6,777,685 bp, or 1 per 566 bp averaged over all ENCODE regions in 16 CEU samples (Thorisson et al., 2005).
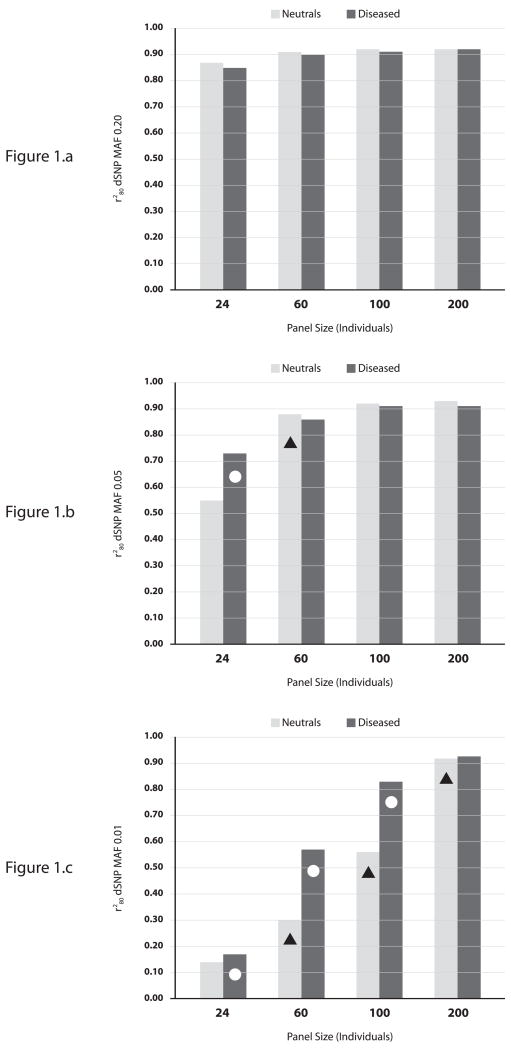
Figure 1 illustrates findings for models with GRR of 2.0. If the dSNP is common (0.20), neutral panels of all sizes performed well (all r280≈0.90); diseased panels did not improve tagging performance. For dSNP MAF=0.05 and a panel size of 24, the diseased panel had a significantly higher r280 than the neutral panel (p<0.0001). Increasing the neutral panel size to 60 significantly improved the performance (p<0.0001) and matched that of the disease panel (both r280≈0.90). Further increasing panel size provided no significant advantage. For dSNP MAF=0.01, neutral panels of successive larger size significantly outperformed smaller neutral panels (all p<0.0001). Diseased panels significantly outperformed same-sized neutral panels until the largest panel (200 individuals), when their performance was comparable (both r280≈0.90). For models with GRR=1.2, diseased panels only outperformed neutral for dSNP MAF=0.05 at a panel size of 24. Successively larger neutral panels increased performance; only a panel of 200 was able to achieve an r280≈0.90 for both dSNP MAF 0.05 and 0.01 (Supplemental Figure 1 online). For GRR=4.0 results were qualitatively the same as for GRR=2.0, but with comparably larger r280 (Supplemental Figure 2 online).
Figure 1.
Comparison of discovery panels using r280 (multiplicative GRR 2.0). (a) dSNP MAF of 0.20. (b) dSNP MAF of 0.05. (c) dSNP MAF of 0.01. Circles indicate a two-sample Kolmogorov-Smirnov (KS) D-statistic p-value<0.0001, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of the next smallest size.
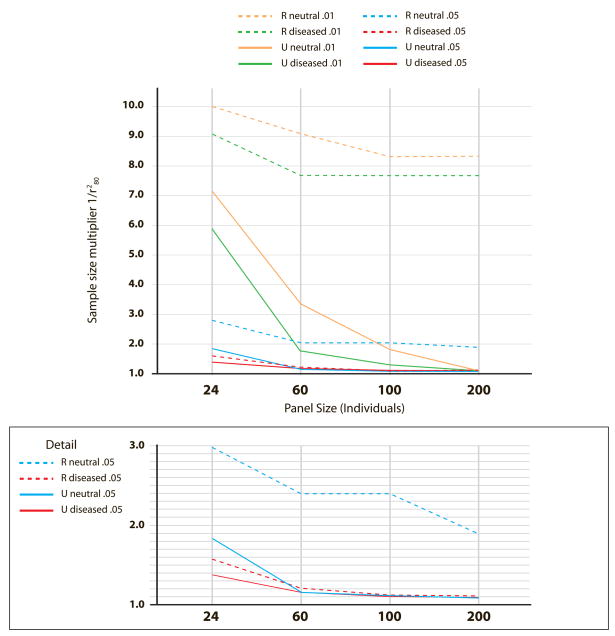
We have thus far assumed that all variants would be considered in the tSNP selection procedure (unrestricted selection). However, it is common practice to pre-screen and exclude variants from the selection procedure based on MAF; for restricted selection, we find the discovery panel type becomes increasingly important. For less common dSNPs, restricted selection based on a pre-screen MAF threshold of 0.05 had severe consequences. To illustrate the potential impact on sample size to maintain power, Figure 2 shows the sample-size multiplier 1/r280 for neutral and diseased panels of varying sizes, and dSNP MAFs of 0.05 and 0.01 (GRR=2.0). Note that a binning criteria of r2>0.8 should lead to a sample-size multiplier no higher than 1.25 if the dSNP is appropriately tagged. The multiplier is <1.25 for unrestricted situations with panel size 200. For dSNP MAF=0.05, only the diseased panel of size 200 performs well with restricted selection. For dSNP MAF=0.01 (MAF threshold of 0.05) the tSNPs available are so poor that the multiplier can increase to 8–10 in both neutral and diseased panels. Using restricted selection with a pre-screen MAF threshold of 0.01 mitigates the loss of power, particularly in larger panels; diseased panels of size 100–200 perform well, whereas the sample size multiplier was as high as ~3 for same-sized neutral panels (Supplemental Figure 3 online).
Figure 2.
Comparison of sample size multiplier 1/r280 for restricted (R) and unrestricted (U) tSNP selection procedures. Restriction is based on a pre-screen MAF threshold of 0.05. Genetic models with dSNP MAFs of 0.01 and 0.05 (multiplicative GRR 2.0) are illustrated.
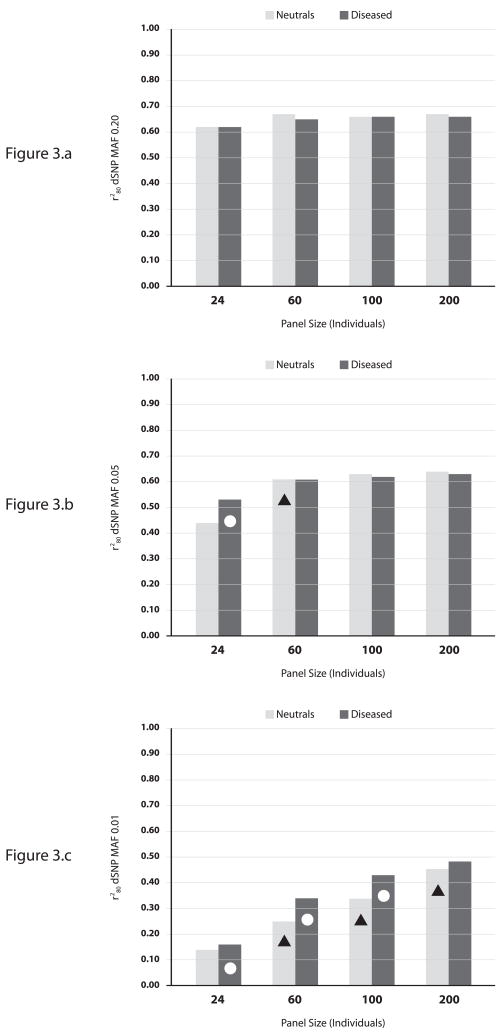
Our simulations assume full sequencing with the dSNP position contained in the sequence data. This fairly represents the HapMap ENCODE data and the 1000 Genomes Project (National Institutes of Health, 2008) sequencing initiative; however, our results will be optimistic for map data and or incomplete sequence data (unsequenced intronic or regulatory regions). To address this, we repeated analyses removing the dSNP from the data (Figure 3). The same relative patterns between panels were evident, but with substantially lower, ~30% decreased r280 values.
Figure 3.
Comparison of discovery panels using r280 and dSNP removed from the data (multiplicative GRR 2.0). (a) dSNP MAF of 0.20. (b) dSNP MAF of 0.05. (c) dSNP MAF of 0.01. Circles indicate two-sample KS D-statistic p-value<0.0001, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of the next smallest size.
DISCUSSION
There is much empirical and theoretical evidence that rarer variants are involved in complex diseases. Indeed, in studies with large sample sizes, targeting rare SNPs may be a better strategy for identifying causal alleles than targeting common variation (Gorlov et al., 2008). Based on ENCODE data, Gorlov, et al. (Gorlov et al., 2008) estimated that ~60% of SNPs have MAF<0.05, and suggested that numerous rare functional SNPs are likely major contributors to common disease susceptibility. Also, genomewide association analyses performed thus far have uncovered only a very small proportion of the total number of variants hypothesized to be involved in disease susceptibility (Iles, 2008). The ENCODE data are based on a maximum sequencing panel size of 16 individuals per racial/ethnic group(HapMap Consortium, 2005) and the NIEHS/SeattleSNPs on sequencing panels of maximum size 24 (Carlson et al., 2004, Livingston et al., 2004). Our results, and those of others (Zeggini et al., 2005, Iles, 2008), indicate that currently available neutral discovery panels are adequate to tag common variants (e.g. MAF 0.20), but not less common variants (MAF 0.01–0.05). Of course, full genomic sequencing for all individuals will eventually obviate the need for the use of tSNPs in analyses. However, until this is technically and financially viable, approaches to selecting tSNPs that increase power to detect rarer variants are needed. Our approach, similar to others, was to use r2 to assess power differences. In contrast to single-locus, pairwise tagging strategies, it has been demonstrated that multi-locus tagging approaches may tag rarer variants with greater success (Zeggini et al., 2005). We explored the efficacy of diseased vs. neutral discovery panels in a multi-locus framework using Haploview (Barrett, et al. 2005) with a subset of simulated data. Similar to our observations based on a pairwise approach, diseased panels outperformed neutral panels when panel size was small (24 individuals) and the underlying susceptibility allele was less common (MAF 0.01–0.05).
Our novel investigation suggests that although not a panacea, using diseased discovery panels can produce superior tSNPs for association studies when compared to same-sized neutral panels when the underlying susceptibility allele is rare. In the majority of scenarios, diseased panels of 24–60 individuals provide significantly better tSNPs for alleles in the 0.01–0.05 range compared to equivalent neutral panels. A large neutral discovery panel of ~200 individuals is required to provide ≥0.8 probability that a tSNP will be highly correlated with a rare dSNP (0.01–0.05), compared to a panel of 100 diseased individuals. Similarly to others (Zeggini et al., 2005, Klein, 2007) we also find that the common practice of pre-screening based on MAF to exclude variants from the selection procedure below a specified threshold leads to tSNPs with much lower correlation with underlying dSNPs and can therefore have severe consequences for the ability to detect less common susceptibility alleles. We show that this problem persists even if diseased discovery panels are used, although the negative effect is not as exaggerated. This questions the benefit of genotyping fewer tSNPs with a restricted selection since to regain equivalent power it necessitates genotyping on much larger resources –increasing to sample sizes only available via large collaborative efforts. Finally, it is clear that full sequence data is essential to adequately tag a region of interest (Tantoso et al., 2006); our results also show that tSNP performance declines substantially if the underlying susceptibility allele is unavailable as might occur with map or incomplete sequence data, in both neutral and diseased panels. The 1000 Genomes Project aims to resequence the genomes of at least 1000 unrelated individuals across several racial/ethnic populations (National Institutes of Health, 2008). This effort will provide a valuable resource to develop the next generation tSNPs sets, especially for populations represented by at least 200 individuals. However, before this new resource is realized, our results indicate that investigators can increase their power to detect rarer disease susceptibility variants by sequencing disease discovery panels to supplement tSNP selection. This approach may be especially helpful for following up specific regions of interest.
As we embrace a paradigm that includes rare alleles as some researchers suggest (Campbell & Manolio, 2007, Iles, 2008, Gorlov et al., 2008), tSNP selection will need to be revisited. Our novel investigation indicates that sequencing 24–60 diseased individuals and implementing unrestricted tSNP selection may be a useful endeavor to significantly increase the efficacy of the tSNPs for detecting rarer susceptibility variants for many genetic models. Furthermore, our study clearly illustrates the superiority of full sequencing in the discovery panel and unrestricted tSNP selection procedures in this regard, which are necessary for adequate tSNP performance even if diseased panels are used. The completion of the production phase of the 1000 Genomes Project anticipated in 2011 will certainly provide a new generation of universally useful tagging sets, and provided that the population-specific panels sequenced are at least of size 200, these sets will adequately tag not only common susceptibility alleles, but also rare variants that may have low to moderate effect sizes. In the interim, however, we must be aware of the limitations, or use alternate approaches to combat them. Including diseased individuals in discovery panels is a unique approach that investigators can use to boost their power to detect rarer alleles.
WEB RESOURCES
The National Institute of Environmental Health Sciences Environmental Genome Project homepage is available at http://egp.gs.washington.edu/. The HapMap homepage is available at http://www.hapmap.org/. The SeattleSNPs homepage is available at http://pga.gs.washington.edu/. The 1000 Genomes Project homepage is available at http://www.1000genomes.org/.
Supplementary Material
Comparison of discovery panels using r280 (multiplicative GRR 1.2). (a) dSNP MAF of 0.20. (b) dSNP MAF of 0.05. (c) dSNP MAF of 0.01. Circle indicates a two-sample KS D-statistic p-value=0.004, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of the next smallest size.
Comparison of discovery panels using r280 (multiplicative GRR 4.0). (a) dSNP MAF of 0.05. (b) dSNP MAF of 0.01. Circles indicate a two-sample KS D-statistic p-value<0.0001, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of next smallest size.
Comparison of sample size multiplier 1/r280 for restricted (R) and unrestricted (U) tSNP selection procedures. Restriction is based on a pre-screen MAF threshold of 0.01. Genetic models with dSNP MAFs of 0.01 and 0.05 (multiplicative GRR 2.0) are illustrated.
Acknowledgments
This study was funded by National Institute of Health Grants CA123550-01 and CA098364-01 (to N.J.C.). M.M.I. is supported by a Cancer Research UK senior research fellowship. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute.
Footnotes
AUTHOR CONTRIBUTIONS
N.J.C., M.M.I., and K.C. conceived the study. The simulations and data analyses were performed by K.C. The paper was written primarily by K.C. and N.J.C. with contributions from M.M.I.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- Agresti A. A survey of exact inference for contingency tables. Statistical Science. 1992;7:131–177. [Google Scholar]
- Amos CI. Successful design and conduct of genome-wide association studies. Hum Mol Genet. 2007;16 Spec No 2:R220–5. doi: 10.1093/hmg/ddm161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Campbell H, Manolio T. Commentary: rare alleles, modest genetic effects and the need for collaboration. International Journal of Epidemiology. 2007;36:445–8. doi: 10.1093/ije/dym055. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle MA, Ng PC, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS genetics. 2007;3:1827–37. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckler Ries LA, Reichman ME, et al. Cancer survival and incidence from the Surveillance, Epidemiology, and End Results (SEER). program. Oncologist. 2003;8:541–52. doi: 10.1634/theoncologist.8-6-541. [DOI] [PubMed] [Google Scholar]
- Gorlov IP, Gorlova OY, et al. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–12. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapmap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Hapmap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–8. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Iles MM. What Can Genome-Wide Association Studies Tell Us about the Genetics of Common Disease? PLoS genetics. 2008;4:e33. doi: 10.1371/journal.pgen.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Commentary: grading the credibility of molecular evidence for complex diseases. International journal of epidemiology. 2006;35:572–8. doi: 10.1093/ije/dyl003. discussion 593–6. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Little J, et al. On the synthesis and interpretation of consistent but weak gene-disease associations in the era of genome-wide association studies. International journal of epidemiology. 2007;36:439–45. doi: 10.1093/ije/dyl253. [DOI] [PubMed] [Google Scholar]
- Klein RJ. Power analysis for genome-wide association studies. BMC Genet. 2007;8:58. doi: 10.1186/1471-2156-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, et al. Science. Vol. 308. New York, N.Y: 2005. Complement factor H polymorphism in age-related macular degeneration; pp. 385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston RJ, Von Niederhausern A, et al. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–31. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health, National Human Genome Research Institute. International Consortium Announces the 1000 Genomes Project 2008 [Google Scholar]
- Phillips MS, Lawrence R, et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet. 2003;33:382–7. doi: 10.1038/ng1100. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen JT, Uimari P, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81:338–45. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantoso E, Yang Y, Li KB. How well do HapMap SNPs capture the untyped SNPs? BMC genomics. 2006;7:238. doi: 10.1186/1471-2164-7-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Stram D, et al. Haplotype tagging single nucleotide polymorphisms and association studies. Hum Hered. 2003;56:48–55. doi: 10.1159/000073732. [DOI] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, et al. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Rayner W, et al. An evaluation of HapMap sample size and tagging SNP performance in large-scale empirical and simulated data sets. Nat Genet. 2005;37:1320–2. doi: 10.1038/ng1670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of discovery panels using r280 (multiplicative GRR 1.2). (a) dSNP MAF of 0.20. (b) dSNP MAF of 0.05. (c) dSNP MAF of 0.01. Circle indicates a two-sample KS D-statistic p-value=0.004, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of the next smallest size.
Comparison of discovery panels using r280 (multiplicative GRR 4.0). (a) dSNP MAF of 0.05. (b) dSNP MAF of 0.01. Circles indicate a two-sample KS D-statistic p-value<0.0001, diseased vs. neutral panel of the same size. Triangles indicate two-sample KS D-statistic p-value<0.0001, neutral panel vs. neutral panel of next smallest size.
Comparison of sample size multiplier 1/r280 for restricted (R) and unrestricted (U) tSNP selection procedures. Restriction is based on a pre-screen MAF threshold of 0.01. Genetic models with dSNP MAFs of 0.01 and 0.05 (multiplicative GRR 2.0) are illustrated.