Abstract
Human cancers often carry many somatically acquired genomic rearrangements, some of which may be implicated in cancer development. However, conventional strategies for characterizing rearrangements are laborious and low-throughput and have low sensitivity or poor resolution. We used massively parallel sequencing to generate sequence reads from both ends of short DNA fragments derived from the genomes of two individuals with lung cancer. By investigating read pairs that did not align correctly with respect to each other on the reference human genome, we characterized 306 germline structural variants and 103 somatic rearrangements to the base-pair level of resolution. The patterns of germline and somatic rearrangement were markedly different. Many somatic rearrangements were from amplicons, although rearrangements outside these regions, notably including tandem duplications, were also observed. Some somatic rearrangements led to abnormal transcripts, including two from internal tandem duplications and two fusion transcripts created by interchromosomal rearrangements. Germline variants were predominantly mediated by retrotransposition, often involving AluY and LINE elements. The results demonstrate the feasibility of systematic, genome-wide characterization of rearrangements in complex human cancer genomes, raising the prospect of a new harvest of genes associated with cancer using this strategy.
Somatic genetic changes involved in cancer causation include point mutations, genomic rearrangements and changes in copy number1. Most of the currently identified genes associated with cancer contribute to oncogenesis as a result of somatic rearrangements that result either in fusion transcripts or in transcriptional deregulation by apposing enhancer or promoter elements to intact protein coding sequences1. The large majority of the known somatically rearranged genes associated with cancer are found in the small minority of human cancers comprising leukemias, lymphomas and soft tissue tumors (see URLs section in Methods). Fusion genes may be more frequent than previously thought in epithelial cancers as well2, a prediction substantiated by recent discoveries of an EML4-ALK fusion in non-small-cell lung cancer3 and ETS fusion genes in prostate cancer4,5.
The relatively late discovery of commonly rearranged genes associated with cancer in solid tumors reflects the difficulty of systematically characterizing rearrangements in highly rearranged cancer genomes. G-banded cytogenetics, spectral karyotyping and FISH lack sensitivity to detect anything less than gross genomic rearrangements and provide limited resolution of breakpoint positions. Copy number arrays can detect breakpoints associated with copy number imbalances; however, they will not report on balanced rearrangements or the fusion events that have occurred following chromosome breakage. Both end-sequencing of BAC libraries derived from cancer genomes6,7 and hybridization of flow-sorted chromosomes to arrays8 are labor-intensive and therefore not applicable to large numbers of cancer genomes.
Massively parallel sequencing strategies offer the potential to carry out genuinely genome-wide screening for point mutations, copy number changes and rearrangements on a single platform. Here, we use a massively parallel technology—incorporating a procedure in which short reads are generated from both ends of millions of DNA fragments up to 500 bp in size—to systematically detect and characterize rearrangements. Using this strategy, we explore and compare patterns of somatic and germline rearrangement in human cancer genomes.
RESULTS
Massively parallel paired-end sequencing
In order to identify acquired genomic rearrangements that may be involved in cancer development, we undertook massively parallel sequencing on the GenomeAnalyzer platform of both ends of genomic DNA fragments that had been randomly sheared by nebulization and then size-selected by gel electrophoresis (Fig. 1a). As proof of principle, we used two lung cancer cell lines (NCI-H2171, a small-cell lung cancer cell line, and NCI-H1770, a neurendocrine cell lung cancer line9). We have previously characterized 61 and 9 acquired genomic rearrangements, respectively, in these two cell lines to the base-pair level by shotgun sequencing of BAC libraries7. For NCI-H2171, 400-bp fragments were used for library production and sequencing, and for NCI-H1770, 200-bp fragments were used.
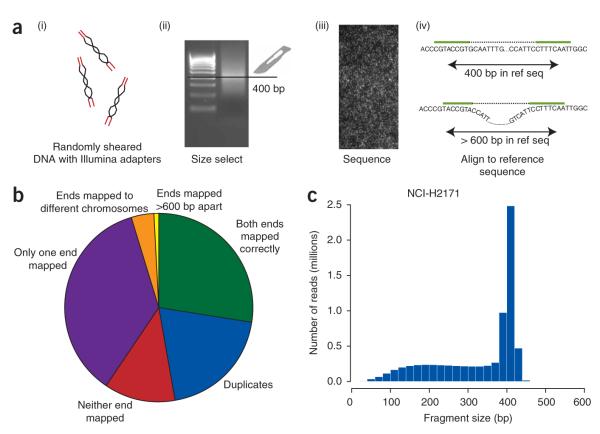
Figure 1.
Experimental protocol and outcome of sequencing. (a) Genomic DNA was sheared by nebulization and primers ligated to either end of the DNA fragments (i). These were size selected on an agarose gel (ii) and sequenced from either end on a GenomeAnalyzer instrument (iii). We mapped the short sequence reads to the genome and determined spacing and whether or not the two ends aligned in the correct orientation (iv). (b) The final disposition of reads for NCI-H2171. (c) Distribution of fragment size for correctly mapping reads from NCI-H2171.
From the NCI-H2171 library, we generated 36.2 million paired reads of 29–36 bases at each end (Fig. 1b and Supplementary Table 1 online). Of these, ~18.8 million (52%) reads were of sufficient quality that both ends mapped back to the current reference genome assembly (NCBI build 36), whereas there were ~13.0 million reads (36%) for which only one end mapped back and ~4.4 million (12%) where neither end mapped back. Of the reads for which both ends mapped back to the genome, ~7.2 million were excluded from further analysis because they precisely duplicated other sequences from the library, and therefore probably arose from preferential PCR amplification during library production. We also excluded several other classes of sequence reads from further analysis, including reads with mispriming of the sequencing (~50,000 reads), reads from mitochondrial DNA (~9,000 reads) and reads from contaminating nonhuman DNA (191 reads). Sequences from gaps in the reference genome (estimated to encompass ~160 Mb in total) generated many mapping errors, as the repetitive DNA in these regions is often similar to heterochromatic sequence in the reference genome build. These sequences manifested as substantial increases in copy number near centromeres and telomeres. We therefore excluded regions within 1 Mb of a telomeric or centromeric sequence gap from copy number analysis or rearrangement screens (~1.2 million reads). The outcomes of sequencing and mapping for NCI-H2171 were very similar to those for NCI-H1770, which had a library of smaller inserts (Supplementary Table 1), indicating that using larger DNA fragments in the paired-end protocol does not compromise sequence quality, mapping or evenness of genome coverage.
We calculated insert size for paired ends that mapped to the genome in the correct orientation and spacing (Fig. 1c) and found that the size selection of fragments was accurate, with a peak insert size of ~400 bp for NCI-H2171 and 190 bp for NCI-H1770 (data not shown) but a shoulder of smaller fragments for both. Notably, the upper limit of fragment size for both cell lines was clearly demarcated. As a consequence, deletions as small as 300 bp could readily be detected. In total, for NCI-H2171, >700 Mb of sequence was generated, which, when combined with the insert size, generates an overall effective coverage of 2.4 Gb (7.3 million high-quality reads × 328-bp mean insert). Similar calculations for NCI-H1770 gave a lower overall coverage of 1.8 Gb as a result of the smaller insert size.
Rearrangements
There were ~1.2 million reads from NCI-H2171 and 770,000 from NCI-H1770 in which mapping of the two ends to the genome suggested a possible rearrangement. Most of these were presumably due to incorrect mapping induced by sequencing errors, as the quality scores of the base-calling and alignment algorithms were, on average, substantially lower than those for correctly mapping reads. We therefore focused on aberrantly mapping reads for which both ends mapped with high uniqueness scores (7,042 read pairs for NCI-H2171 and 19,071 for NCI-H1770). Reads were then prioritized for confirmatory screening on the basis of criteria listed in Methods. A total of 1,152 aberrantly mapping reads from the NCI-H2171 library and 495 from NCI-H1770 were evaluated on cancer and constitutional DNA by genomic PCR using primers on either side of potential breakpoints. We then conventionally sequenced the PCR products for the annotation of rearrangements to the base-pair level.
Using this strategy, we identified a total of 325 rearrangements in NCI-H2171 (Table 1, Fig. 2a, Supplementary Note and Supplementary Table 2 online) and 84 in NCI-H1770 (Table 1, Fig. 2b, Supplementary Note and Supplementary Table 3 online). Of these, 244 and 62 were germline, respectively. Most of these were deletions, including 156 300- to 350-bp deletions of AluY elements and 38 deletions of LINE repeats, a much higher proportion than that found using paired-end sequencing with larger insert sizes10. These probably reflect insertions in the reference genome (by retrotransposition) rather than genuine rearrangements in the test samples, as they were generally present in the homozygous state. Moreover, there is no known mechanism for the specific removal of an Alu element from the germline11.
Table 1. Summary of confirmed rearrangements in NCI-H2171 and NCI-H1770.
| NCI-H2171 | NCI-H1770 | |
|---|---|---|
| Somatic rearrangements | 81 | 22 |
| Intrachromosomal | 59 | 22 |
| Deletions | 2 | 0 |
| Tandem duplications | 9 | 2 |
| Inversions | 2 | 1 |
| Inverted duplication | 2a | 0 |
| Within an amplicon | 44 | 19 |
| Interchromosomal | 22 | 0 |
| Between amplicons | 15 | 0 |
| Amplicon to nonamplified region | 7 | 0 |
| Outside amplicons | 0 | 0 |
| Germline rearrangements | 244 | 62 |
| Intrachromosomal | 243 | 62 |
| Deletions | 233 | 58b |
| AluY elements | 136 | 20 |
| Other Alu elements | 8 | 1 |
| LINE elements | 29 | 9 |
| Other repeats | 8 | 3 |
| Not repeat mediated | 52 | 25 |
| Tandem duplications | 1 | 4 |
| Inversions | 9 | 2 |
| Interchromosomal | 1 | 0 |
The two breakpoints identified represent both ends of the same inverted duplication (see Supplementary Table 2).
Not all apparent deletions <1kb were screened in NCI-H1770 because of the high frequency of germline AluY deletions in NCI-H2171.
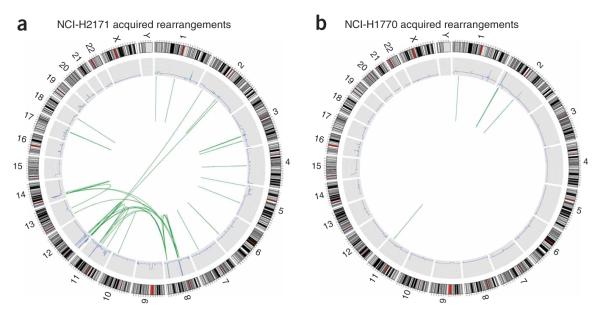
Figure 2.
Genome-wide acquired rearrangements. (a) NCI-H2171. (b) NCI-H1770. The outer ring shows a representation of the normal karyotype (red indicates centromeres). The blue line in the middle ring indicates copy number as determined by short read data. The inner circle shows the two endpoints of each somatic rearrangement identified, joined by green lines. Very small rearrangements appear as single lines.
We identified 103 somatically acquired rearrangements in the two cell lines. Among these were 27 (44%) of the 61 acquired rearrangements we previously characterized in NCI-H2171 and 4 (44%) of the 9 found in NCI-H1770 by BAC shotgun sequencing7. Rearrangements involving heavily amplified regions predominated for both tumors (69 of 81 for NCI-H2171 and 19 of 22 for NCI-H1770) and included both inter- and intrachromosomal rearrangements. However, somatically acquired intrachromosomal rearrangements outside amplicons were also identified in the two cancers. These included deletions, inversions, an inverted duplication and several tandem duplications (see below).
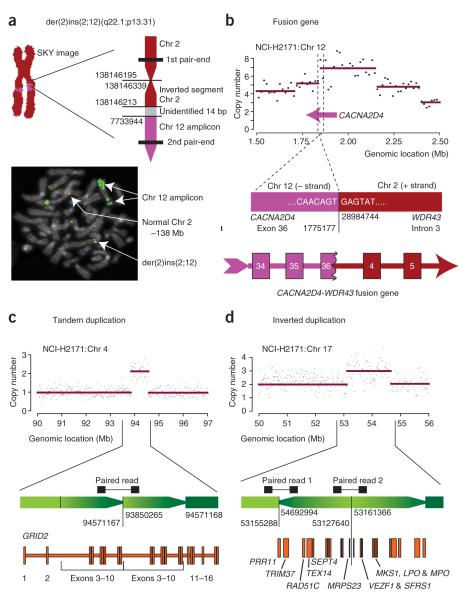
The spectral karyotypes of the cancer cell lines show several gross interchromosomal aberrations, a pattern often observed in solid tumors (see URLs section in Methods)12. Among these is an insertion of sequence from a homogeneously staining region of chromosome 12 into chromosome 2q in NCI-H2171, which we were able to map to the base-pair level using a paired-end read that spanned the breakpoint (Fig. 3a). Sequencing showed that the breakage and repair at this insertion had resulted in inversion of a small fragment or ‘genomic shard’ from chromosome 2 adjacent to the breakpoint. Indeed, such shards are a feature of many of the rearrangements in NCI-H2171 identified both in this screen (Supplementary Table 2) and in our previous BAC sequencing7. The region of chromosome 12 that is inserted into 2q corresponds to one of the most heavily amplified regions of the NCI-H2171 genome (Supplementary Fig. 1 online). FISH using BAC probes selected on the basis of the sequences in the rearrangement showed a fusion signal at the 2;12 junction (Fig. 3a), supporting the hypothesis that the sequenced breakpoint corresponds to that seen on the spectral karyotype. Thus, systematic sequencing using the paired-end strategy can resolve somatic rearrangements observed at low resolution by conventional karyotyping or FISH-based techniques.
Figure 3.
Rearrangements in NCI-H2171. (a) Mapping to the base-pair level of an acquired insertion of sequence from a homogeneously staining region on chromosome 12p13 into chromosome 2q22. The insertion was evident on spectral karyotype, and a paired-end read spanned the breakpoint. Confirmatory PCR and sequencing showed a small 127-bp fragment (shard) from chromosome 2 inverted at the breakpoint. FISH using a probe (RP11-444J21) from the chromosome 12p13 amplicon adjacent to the breakpoint (green) and a probe (RP11-58C7) from the chromosome 2q22 region (red) generated a fusion signal (yellow), confirming that the breakpoint identified corresponded to that seen on the spectral karyotype. (b) A CACNA2D4-WDR43 fusion gene. The 5′ portion of the CACNA2D4 gene is amplified, and the paired-end reads showed a rearrangement that breaks the gene in exon 36, fusing it into intron 3 of WDR43. The sequence at the breakpoint creates an almost perfect splice donor site, resulting in the production of a fusion transcript with a shortened exon 36 from CACNA2D4. (c) An acquired tandem duplication resulting in an aberrant mRNA transcript. A 700-Mb region of increased copy number on chromosome 4 was identified on the copy number analysis. A single paired-end read mapped with one end at the 5′ border of the amplification and the other end at the 3′ border. The breakpoints fell within introns 2–10 of GRID2 and would be expected to give rise to a partial tandem duplication of exons 3–10 of the gene, confirmed by RT-PCR. (d) An inverted duplication of a gene-rich region of chromosome 17 was identified by a localized increase in copy number together with two paired-end reads spanning both inverted breakpoints.
One of the primary aims of our strategy is the identification of rearrangements that result in fusion transcripts that may be implicated in oncogenesis. We were able to identify a fusion transcript in NCI-H2171 resulting from another t(2;12) rearrangement (Fig. 3b). The 5′ partner gene, CACNA2D4, lies in an amplified region near the telomere of chromosome 12p. The rearrangement breaks this gene within exon 36 and juxtaposes it to intron 3 of a gene on chromosome 2p, WDR43. Of note, the sequence lying across the breakpoint junction creates an almost perfect splice donor site (…GTGAGT…). By RT-PCR from NCI-H2171 mRNA and sequencing of the fusion transcript, we confirmed that exon 36 of CACNA2D4-WDR43 is indeed shorter than the wild-type exon and splices at the created donor site into exon 4 of WDR43. The fusion transcript is predicted to be out-of-frame on the basis of the canonical transcripts in the RefSeq database. Nevertheless, this result demonstrates that systematic pairedend sequencing can identify previously unknown fusion transcripts arising from rearranged cancer genomes.
In addition to noting the numerous somatic rearrangements associated with regions of amplification, we scrutinized our data for previously unreported patterns of somatic rearrangement. Of note, we found nine acquired tandem duplications in NCI-H2171 and two in NCI-H1770 (Fig. 3c), ranging in size from <1 kb to 2.7 Mb. Apart from the smallest, all showed a discernible increase in copy number. The sequencing data predicted that two adjacent copies of the region were aligned in the same orientation. Four of these rearrangements duplicated a subset of exons within a single gene, of which two were duplications of internal exons (Fig. 3c). For the other two, the duplicated segment encompassed the promoter and first few exons of a single gene, with the tandem orientation arranging these exons immediately upstream of the intact, full-length gene. A further five tandem duplications amplified multiple genes, one duplicated part of an intron and one contained no annotated genes. By RT-PCR and sequencing, we found that the two rearrangements that duplicated internal exons within a single gene (GRID2 and CNTNAP5) resulted in aberrant transcripts in the tumor RNA that were not found in the matched constitutional B-cell line (Fig. 3c and data not shown). Both of the newly identified transcripts are predicted to be out-of-frame. We were unable to find aberrant transcripts from the two rearrangements duplicating the first few exons of a gene (WDR34 and CCBE1).
Tandem duplications are not the only cause of small, localized increases in copy number. We found an example of an acquired inverted duplication, 1.5 Mb in size, on chromosome 17q23 of NCI-H2171 (Fig. 3d). In contrast to tandem duplications, where only one breakpoint can be detected, inversions and inverted duplications are associated with two breakpoints. In this example, both breakpoints were covered by the massively parallel sequencing, and their locations exactly demarcate the increase in copy number. The orientation of the paired-end reads, confirmed by sequencing of the breakpoints, suggests that the duplicated and inverted segment lies 5′ to the noninverted segment. The duplicated segment is rich in genes and overlaps with a frequently amplified region in breast cancer, including the DNA-repair gene RAD51C13.
Copy number
We assessed whether high-resolution information on copy number changes across the genome could be obtained from reads that mapped uniquely to the genome with correct orientation and spacing of the two ends. As the density of repetitive elements shows substantial regional variation across the genome, we used in silico simulations to define unequal width intervals of the genome that contained a standardized amount of unique or ‘mappable’ DNA, and counted the number of sequences mapping to each window. Windows containing 15 kb of mappable sequence gave robust estimates of copy number from the data generated, as exemplified by chromosome 11 of NCI-H2171 compared to Affymetrix SNP6 (1.85 million loci) array data (Fig. 4). Point-to-point variability was less with the paired-end data than for SNP arrays, suggesting that the sensitivity to detect small regional changes in copy number should be at least equivalent. For heavily amplified regions of the genome, such as the area around 85 Mb on chromosome 11, the paired-end data gave substantially higher estimates of copy number than SNP arrays, probably because of signal saturation on the array platform.
Figure 4.
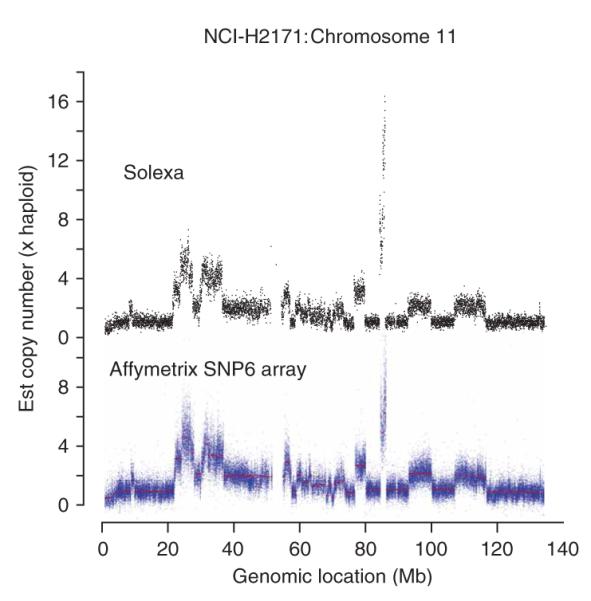
Copy number. Comparison of copy number plots for chromosome 11 of NCI-H2171 between massively parallel paired-end sequencing (upper panel) and Affymetrix SNP6 genomic array data (lower panel).
We adapted a circular binary segmentation algorithm developed for SNP array data14 to generate statistical predictions of copy number changes. This method estimates both the copy number in each region and the location of predicted change-points in copy number (Supplementary Figs. 1 and 2 and Supplementary Tables 4 and 5 online). Unlike conventional technologies for measuring copy number, however, the rearrangement screen allowed us to prove the accuracy of the copy number algorithm through the identification of the breakpoints. For example, of the nine tandem duplications identified in NCI-H2171, all were predicted by the algorithm, with all breakpoints correctly estimated to within 30 kb (Supplementary Tables 2 and 4). However, the 289-kb chromosome 1 duplication in NCI-H1770 was not identified by the copy number algorithm, possibly because this chromosome is tetraploid. The smallest correctly identified duplication in NCI-H2171 was only 30 kb in size, suggesting that the resolution obtained is at least comparable to that of the currently available SNP arrays.
Amplicons
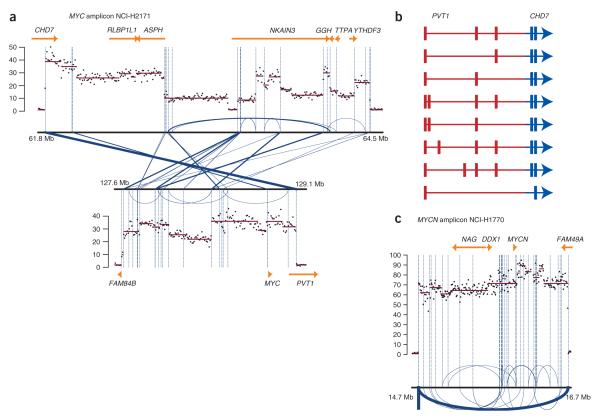
We explored whether combining data on copy number and rearrangement breakpoints could provide insights into the genomic architecture of complex amplicons (Fig. 5a). NCI-H2171 has liberal amplification of a 1.4-Mb region of chromosome 8q24, which includes the MYC proto-oncogene, together with regions on chromosomes 8q12, 11q14, 12p12–12p13 and 14q12 where the target genes (if any) have not been identified. For the two regions on chromosome 8q, the paired-end read data show marked variation in copy number at high resolution (Fig. 5a). MYC is located in the region of greatest amplification on chromosome 8q24 (~35-fold amplification). We identified a substantial number of breakpoints (29 in total) linking the 8q12 and the 8q24 amplicons, the locations of which often corresponded precisely to change-points in copy number. Notably, the 29 rearrangements showed considerable variability in the number of paired-end reads that spanned the breakpoint (ranging from 1 to 14; Supplementary Table 2 and Fig. 5a), suggesting that some of the individual breakpoints have themselves been amplified.
Figure 5.
Amplicons in NCI-H2171 and NCI-H1770. (a) The MYC amplicon of NCI-H2171 involves two regions of chromosome 8q, both of which show extensive variation in copy number. Breakpoints between and within the two regions are shown in blue lines, with the thickness of the line proportional to the number of paired-end reads spanning the same breakpoint. The locations of the breakpoints frequently correspond to changes in copy number. (b) A PVT1-CHD7 fusion gene is created by the most commonly seen breakpoint in the MYC amplicon. Extensive splicing of the PVT1 moiety was seen on RT-PCR, but the three most common transcripts accounted for 23 of 30 of colonies sequenced. (c) The NMYC amplicon of NCI-H1770 showed up to 85-fold amplification of a 2-Mb region. The most common breakpoint observed demarcated the 5′ and 3′ borders of the amplicon and suggested tandem insertion of the 2-Mb region, but several other rearrangements were seen within the amplicon, both inverted (arcs above the line) and noninverted (arcs below the line).
The high-resolution structural detail that results from combining copy-number data with quantitative estimates of breakpoint frequency allows us to hypothesize which rearrangements occurred early in the genesis of the amplicon. The most heavily amplified portion of chromosome 8q12 includes the 3′ portion of the gene CHD7. In the third intron of this gene, there is a breakpoint (row 21, Supplementary Table 2) that results in juxtaposition of the 3′ portion of CHD7 with the 5′ portion of the noncoding RNA gene PVT1. This breakpoint demarcates both the boundary between diploid DNA and the amplicon on 8q12 and the boundary between the 8q24 amplicon and haploid DNA. There were 14 different sequencing reads that spanned this breakpoint, compared to seven reads for the next most common breakpoint. These observations suggest that an early—and possibly the first—event in the genesis of these amplicons was a t(8;8)(q12;q24) translocation resulting in fusion of the PVT1 and CHD7 genes.
RT-PCR using a forward primer in exon 1 of PVT1 and a reverse primer in exon 4 or exon 5 of CHD7 showed multiple bands (data not shown). Cloning and sequencing of the RT-PCR reaction products showed eight variants, arising from differential splicing of the PVT1 moiety (Fig. 5b). The three most common variants (accounting for 23 of 30 clones sequenced) would all be predicted to give rise to a small upstream open reading frame from the PVT1 gene, followed by a CHD7 open reading frame which, if translated, would result in a CHD7 protein truncated at the N-terminal (amino acids 806–2,997).
In contrast, the MYCN amplicon on chromosome 2 of NCI-H1770 has a somewhat different pattern from that seen in NCI-H2171 (Fig. 5c), with a relatively homogeneous but massive increase in copy number (60–85 copies per cell) across the 2-Mb region. One rearrangement delineates this amplicon exactly (row 20, Supplementary Table 3) and was seen 17 times in the sequencing reads. The orientation of the rearrangement, together with its frequency compared to the others observed, suggest that this amplicon arose through creation of a double-minute chromosome by excision and circularization of a 2-Mb loop of DNA, which then amplified extrachromosomally before reinserting into the genome as a homogeneously staining region7. We found 18 other acquired breakpoints within this amplicon, occurring in both inverted and noninverted orientations, suggesting either that the amplicon has undergone further rearrangement after reinsertion or that the double-minute chromosomes were subject to error-prone replication.
Breakpoints to the base-pair resolution
We have characterized 103 acquired and 306 germline structural variants to the base-pair level. This has shown diverse sequence contexts, with differences between somatic and germline rearrangements evident. In particular, the occurrence of nontemplated sequence 1–57 bp in length at the breakpoints was much more frequent in acquired than in germline rearrangements (30% versus 9.4%; P < 0.0001). Similarly, small fragments of genomic DNA 20–150 bp in length, known as ‘genomic shards’, captured within the rearrangements (shaded boxes, Supplementary Tables 2 and 3) were much more common in somatic than in germline rearrangements. We found that 53% of the acquired rearrangements showed short stretches of microhomology between the two ends fused together, ranging from 1 to 10 bp in length, with similar patterns in the two cell lines. These short stretches of microhomology are generally believed to reflect nonhomologous end-joining mechanisms of DNA break repair15. In contrast, the germline AluY variants generally showed longer stretches of 10–20 bp of AT-rich microhomologous sequences, thought to form when the transposon inserts into the genome11. These were also seen in the LINE and SVA repeat–mediated variants, but less so in the non–repeat-mediated germline polymorphisms.
DISCUSSION
This study demonstrates the potential of massively parallel sequencing technologies for the investigation of cancer genomes. By using a paired-end strategy, we were able to identify and characterize to the base-pair level acquired deletions, tandem duplications, inverted duplications, inversions and interchromosomal rearrangements, as well as obtain high-resolution copy-number information. The data have shown that the patterns of somatic structural variation encountered in the two cancers studied differ markedly from those found in the germline, allowed resolution of rearrangements previously detected by cytogenetic approaches, yielded previously unknown fusion transcripts, shown that many rearrangements occur within or between amplicons and uncovered a distinctive pattern of somatic tandem duplication operative outside amplified regions. The results therefore illustrate the substantial amount of information pertaining to somatic structural change that will emerge by application of such approaches to the genomes of many cancer classes.
We used relatively small insert sizes in this study (200–500 bp), a strategy that has the advantages of tight size selection of DNA fragments (and therefore greater sensitivity for small intrachromosomal rearrangements) and straightforward PCR confirmation of variants. In contrast, using larger insert sizes, as in a recently published study of germline structural variants10, has the advantage of greater genomic coverage per sequenced fragment, though with the drawback of increased difficulty with sequence annotation of breakpoints. It is likely that complete characterization of all rearrangements in a cancer cell line will require paired-end sequencing of several libraries of different insert sizes, allowing reads to fall outside any repetitive elements at the breakpoints, while maintaining the capacity to identify small insertions, deletions and genomic shards. Approaches using cDNA, such as paired-end diTags16, show promise for identifying fusion genes and could readily be adapted to new sequencing technologies and used in combination with our protocol to annotate genomic and transcriptional consequences of rearrangements.
The digital read-out of copy number predicted aberrations as small as 30 kb in size that were proven to be genuine through mapping of the actual breakpoints from paired sequence reads. This provides comparable sensitivity to the current generation of array-CGH platforms, and the paired-end strategy has the additional potential to identify the actual breakpoints underlying a given copy number change. This additional information can be important for determining transcriptional effects associated with copy number changes. From the copy number data alone, it would be impossible to distinguish the genomic arrangement of the 11 acquired tandem duplications (Fig. 3c) from the inverted duplication (Fig. 3d). Moreover, copy number analyses are blind to rearrangements such as balanced translocations and inversions. The capacity of massively parallel sequencing to reconstruct such rearrangements is important for the identification of oncogenic fusion genes.
Moreover, in contrast to other strategies for studying copy number such as array CGH, the paired-end strategy allows resolution to be improved simply by increasing the amount of sequence generated. This is effectively the situation in amplicons, permitting detailed annotation of copy number changes across the amplified regions that can be correlated with breakpoint locations. The complexity that emerges from the analysis of the NCI-H2171 amplicons implies that amplification involved an iterative process during which aberrant sister chromatid exchange to repair double-stranded DNA breaks led to progressive reorganization and expansion of the amplicons under selection pressure. It can be argued that the earliest rearrangements in the genesis of the amplicon will be those breakpoints that are themselves most amplified and that demarcate the greatest changes in copy number, as exemplified by both the PVT1-CHD7 fusion in the MYC amplicon of NCI-H2171 and the tandem insertion evident in the MYCN amplicon of NCI-H1770. The ability to extract quantitative read-out of breakpoint frequency and correlate this with copy number changes will be a powerful application of new sequencing technologies to explore the evolution of cancer amplicons.
Our screen has identified four previously unknown rearrangements leading to structural alterations in mRNA transcripts, including two fusion genes and two tandem exon duplications. None of the genes involved has previously been implicated in cancer development, with the exception of PVT1 (refs. 17,18), and it is difficult to ascertain the role these aberrations play here. There is precedent, for example, for partial tandem duplications of exons to produce oncogenic proteins, most convincingly with MLL in acute leukemia19,20. However, it is notable that one of the tandem duplications observed here affected a gene found in a chromosomal fragile site, GRID2 (refs. 21,22), raising the possibility that its occurrence is more a reflection of genome instability and abnormal DNA repair pathways than oncogenicity. It may well be that a proportion of acquired genomic rearrangements, including those that generate abnormal or fusion transcripts, are ‘passenger’ events not associated with cancer development, as has been observed for point mutations23.
This study demonstrates the potential of massively parallel sequencing technologies to annotate large numbers of somatically acquired genome-wide rearrangements in cancer to the base-pair level. In addition to the insights this provides into the diversity of aberrant processes sculpting the genome that underlie the evolution and development of cancer, it is anticipated that these technologies will lead to the identification of previously unknown fusion genes and other rearrangements that may be future therapeutic targets.
METHODS
Paired-end sequencing
We prepared and sequenced DNA using the Solexa sequencing technology platform (GenomeAnalyzer, Illumina) following the manufacturer’s instructions. We randomly sheared 5 μg of NCI-H2171 and NCI-H1770 genomic DNA using the nebulizer supplied with the GenomeAnalyzer instrument according to the manufacturer’s instructions. The fragmented DNA was end-repaired using T4 DNA polymerase and Klenow polymerase with T4 polynucleotide kinase to phosphorylate the 5′ ends. A 3′ overhang was created using a 3′–5′ exonuclease-deficient Klenow fragment, and Illumina paired-end adaptor oligonucleotides were ligated to the sticky ends thus created. We electrophoresed the ligation mixture on an agarose gel and size-selected fragments by stabbing a scalpel blade into the gel at 470 bp for NCI-H2171 and 270 bp for NCI-H1770. DNA was washed from the blade and enriched for fragments with Solexa primers on either end by an 18-cycle PCR reaction. We used different fragment sizes in the two libraries because, although the manufacturer recommends 200-bp inserts for the paired-end protocol (hence the size used for NCI-H1770), in order to increase the genomic coverage per paired-end sequenced, we wanted to assess whether the insert size could be increased to 400 bp without affecting the evenness of genome representation (especially any bias against GC-rich regions), the quality of sequencing, or the ability to perform confirmatory PCR and sequencing.
We prepared the GenomeAnalyzer paired-end flowcell on the supplied cluster station according to the manufacturer’s protocol. Clusters of PCR colonies were then sequenced on the GenomeAnalyzer platform using recommended protocols from the manufacturer. Images from the instrument were processed using the manufacturer’s software to generate FASTQ sequence files. These were aligned to the human genome (NCBI build 36.2) using the MAQ algorithm v0.4.3 (see URLs section below). Reads in which the two ends failed to align to the genome in the correct orientation and distance apart were further screened with the SSAHA algorithm24.
Artifact removal
Reads where the two ends mapped back to within 500 bp of one another, but with one of the two ends in the incorrect orientation were excluded from analysis, as these were likely to be artifacts resulting from either mispriming of the Solexa sequencing oligonucleotide within the PCR colony or intramolecular rearrangements generated during library amplification. Reads that were exact duplicates of one another (created during the PCR enrichment step) were identified by the fact that the two ends of the sequences mapped to identical genomic locations: only the fragment with the higher mapping quality was retained. Spurious mapping of DNA from sequence gaps in NCBI build 36 was reduced by excluding regions within 1 Mb of a centromeric or telomeric sequence gap from copy number and rearrangement analyses (for a list of current sequence gaps, see URLs section below).
Copy number algorithm
To correct for varying levels of uniqueness across the genome, we carried out an in silico simulation of paired-end short reads by creating paired sequences of 35 bases at each end, 500 bp apart, with different simulated pairs located every 35 bp along the genome. These simulated reads were mapped to the genome using the MAQ algorithm. On the basis of this, we divided the genome into nonoverlapping, unequal width windows that contained a constant number of in silico reads mapped with high uniqueness (alternative mapping quality ≥35). The choice of the constant number of mappable reads per window is somewhat arbitrary, but we found that values of about 425 reads per window (equivalent to ~15 kb mappable sequence per window) yielded reliable modeling for our data. With greater sequence yields or for heavily amplified regions, the window size can be reduced to allow greater sensitivity for small copy number variations.
With the window boundaries set, the number of paired-end reads mapping (with alternative mapping quality ≥35) within each window is counted. This forms the raw input to a binary circular segmentation algorithm originally developed for genomic hybridization microarray data14. This algorithm, implemented in R as the DNAcopy library of the Bioconductor project, identifies change-points in copy number by iterative binary segmentation. We used α = 0.01, together with a smoothing parameter of 2 and 2 s.d., for pruning of probable false positives after segmentation, although the modeling gave generally similar results for different parameter choices.
Confirmatory screening
The following criteria were used to prioritize incorrectly mapping reads for confirmatory screening: (i) ≥2 reads spanning the same rearrangement; (ii) generation of a potential fusion gene, defined below (NCI-H2171 only); (iii) very high mapping quality (uniqueness) scores for both ends (NCI-H2171 only); (iv) ends mapping within an amplicon; (v) reads mapping <100 kb apart on the same chromosome (excluding putative deletions <750 bp in size in NCI-H1770); and (vi) both ends mapping to within 100 kb of a change-point in copy number identified by the segmentation algorithm.
When considering these criteria, we screened only reads that mapped with high uniqueness (alternative mapping quality ≥35), except for reads mapping near change-points in copy number (when a threshold for alternative mapping quality of ≥10 was used). The yield of confirmed rearrangements from criteria 2 and 3 above was very low for NCI-H2171, and therefore these criteria were not used for NCI-H1770.
To identify rearrangements representing potential fusion genes, we compared read pair mapping locations in the genome (NCBI build 36) to annotated Ensembl genes (version 47) and Vega genes (release 28). We identified instances in which both ends of the rearrangement fell within the coding footprint of two different genes in the correct orientation to produce a fusion gene; these were then analyzed to determine if an in-frame fusion gene could be formed. Only breakpoints falling within introns were considered, as in these cases, the exact exon phase could be determined. Each combination of transcripts from the two genes was examined to determine if the exon phases were compatible. Only rearrangements bringing together different genes in the correct orientation and with compatible transcript combinations to produce coding regions in frame were considered potential fusion genes.
Primers were designed to span the possible breakpoint by locating them in the 1 kb outside the paired-end reads, for a maximum product size of 1 kb. PCR reactions were done on tumor and normal genomic DNA for each set of primers at least twice, once using a touch-down protocol for GC-rich templates, and once using a low-GC protocol. Products giving a band were sequenced by conventional Sanger capillary methods and compared to the reference sequence to identify breakpoints. Acquired rearrangements were defined as those PCR reactions giving a convincing band in the tumor DNA with no matching band in the normal DNA, seen in at least two separate reactions, together with unambiguously mapping sequence data suggesting a rearrangement. It should be noted that genomic changes acquired during the EBV transformation of the constitutional B cells could cause problems with interpretation. For example, loss of heterozygosity in the B-cell line could theoretically cause a heterozygous, intrachromosomal germline variant to mimic an acquired rearrangement.
FISH
The FISH experiments were done as described in detail elsewhere8. Briefly, BACs were from the 1-Mb clone set (Sanger Institute). Purified BAC DNA was labeled by nick translation with digoxigenin-11-dUTP or biotin-16-dUTP. We then hybridized mixtures of labeled BACs to metaphase spreads and used the appropriate antibody (FITC sheep anti-digoxigenin) or streptavidin (streptavidin-cy5 and biotinylated goat anti-streptavidin) conjugate for detection.
RT-PCR and cloning
Total RNA from the tumor and matched constitutional B-cell lines was reverse transcribed using random priming and SuperScript III (Invitrogen) according to the manufacturer’s instructions. To detect aberrant transcripts created by tandem duplications, we used a forward primer in the most 3′ of the duplicated exons and a reverse primer in the most 5′ of the duplicated exons in a PCR reaction. Resulting bands were sequenced to confirm the specificity of the reaction and the presence of the aberrant transcript. To detect fusion transcripts, we used forward primers in the putative 5′ partner gene and reverse primers from the 3′ partner. When multiple bands suggestive of splice variants were detected, the RT-PCR products were cloned and sequenced using a TOPO TA cloning kit (Invitrogen).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by the Wellcome Trust. P.J.C. is a Kay Kendall Leukaemia Fund fellow, and T.S. has a fellowship from the Michael and Betty Kadoorie Cancer Genetics Research Programme. GlaxoSmithKline provided financial support for the SNP v6.0 microarray analysis for copy number.
Footnotes
URLs Cancer gene census, http://www.sanger.ac.uk/genetics/CGP/Census/; Lung carcinoma cell lines, http://www.path.cam.ac.uk/~pawefish/cell%20line%20catalogues/lung-cell-lines.htm; MAQ algorithm v0.4.3, http://maq.sourceforge.net/maq-man.shtml; UCSC table browser, http://genome.ucsc.edu/cgi-bin/hgTables.
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 3.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Volik S, et al. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc. Natl. Acad. Sci. USA. 2003;100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howarth KD, et al. Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene. 2007 Dec 17; doi: 10.1038/sj.onc.1210993. advance online publication, doi: 10.1038/sj.onc.1210993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J. Cell. Biochem. 1996;24(Suppl.):1–11. doi: 10.1002/jcb.240630502. [DOI] [PubMed] [Google Scholar]
- 10.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 12.Grigorova M, Lyman RC, Caldas C, Edwards PA. Chromosome abnormalities in 10 lung cancer cell lines of the NCI-H series analyzed with spectral karyotyping. Cancer Genet. Cytogenet. 2005;162:1–9. doi: 10.1016/j.cancergencyto.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Wu GJ, et al. 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer Res. 2000;60:5371–5375. [PubMed] [Google Scholar]
- 14.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 15.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front. Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 16.Ruan Y, et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using paired-end diTags (PETs) Genome Res. 2007;17:828–838. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huppi K, Siwarski D. Chimeric transcripts with an open reading frame are generated as a result of translocation to the Pvt-1 region in mouse B-cell tumors. Int. J. Cancer. 1994;59:848–851. doi: 10.1002/ijc.2910590623. [DOI] [PubMed] [Google Scholar]
- 18.Cory S, Graham M, Webb E, Corcoran L, Adams JM. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985;4:675–681. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basecke J, Whelan JT, Griesinger F, Bertrand FE. The MLL partial tandem duplication in acute myeloid leukaemia. Br. J. Haematol. 2006;135:438–449. doi: 10.1111/j.1365-2141.2006.06301.x. [DOI] [PubMed] [Google Scholar]
- 20.Dorrance AM, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J. Clin. Invest. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson KO, Petersen AM, Morrison SN, Elso CM, Stubbs L. Two reciprocal translocations provide new clues to the high mutability of the Grid2 locus. Mamm. Genome. 2005;16:32–40. doi: 10.1007/s00335-004-2423-z. [DOI] [PubMed] [Google Scholar]
- 22.Rozier L, El-Achkar E, Apiou F, Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: possible association with an inherited disease and cancer. Oncogene. 2004;23:6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- 23.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.