Abstract
During apoptotic execution, chromatin undergoes a phase change from a heterogeneous, genetically active network to an inert highly condensed form that is fragmented and packaged into apoptotic bodies. We have previously used a cell-free system to examine the roles of caspases or other proteases in apoptotic chromatin condensation and nuclear disassembly. But so far, the role of DNase activity or ATP hydrolysis in this system has not yet been elucidated. Here, in order to better define the stages of nuclear disassembly in apoptosis, we have characterized the apoptotic condensation using a cell-free system and time-lapse imaging. We demonstrated that the population of nuclei undergoing apoptosis in vitro appears to follow a reproducible program of nuclear condensation, suggesting the existence of an ordered biochemical pathway. This enabled us to define three stages of apoptotic chromatin condensation: Stage 1 ring condensation; Stage 2 necklace condensation; and Stage 3 nuclear collapse/disassembly. Electron microscopy revealed that neither chromatin nor detectible subnuclear structures were present inside the stage 1 ring-condensed structures. DNase activity was not essential for stage 1 ring condensation, which could occur in apoptotic extracts depleted of all detectible DNase activity. However, DNase(s) were required for stage 2 necklace condensation. Finally, we demonstrated that hydrolysable ATP is required for stage 3 nuclear collapse/disassembly. This requirement for ATP hydrolysis further distinguished stage 2 from stage 3. Together, these experiments provide the first steps towards a systematic biochemical characterization of chromatin condensation during apoptosis.
Keywords: chromatin condensation, cell-free system, time-lapse imaging, Dnase, ATP
Introduction
Dynamic changes in the compaction of nuclear chromatin are one of the characteristic phenomena of apoptotic execution [1]. In healthy cells, the genomic DNA, ~2m in size, is packaged into nuclei several micrometers in diameter. At mitosis, the chromatin is further condensed and reorganized into discrete chromosomes, in a process that remains poorly understood. During apoptosis, the DNA, which often attains a level of condensation even greater than that observed in mitosis, is packaged together with nuclear proteins into so-called “apoptotic bodies”. The remarkable degree of compaction observed, together with the fact that apoptotic chromatin condensation does not involve the formation of discrete chromosomes, suggests that cells have an apoptosis-specific system for the induction of chromatin condensation.
Remarkably little is known about the factors responsible for apoptotic chromatin condensation. AIF (apoptosis inducing factor), an essential flavoprotein released from mitochondria during apoptotic execution, can cause nuclear chromatin to condense into a ring at the nuclear periphery [2]. Although AIF has been reported to activate nuclease activity capable of generating high molecular weight (HMW) DNA fragments [3,4], its requirement for the generation of those fragments has been disputed [5,6], and the underlying mechanism by which it promotes peripheral chromatin condensation is unknown. If AIF is depleted by RNAi, nuclear disassembly occurs normally, or is even enhanced in cells with a functional CAD/ICAD system [6]. In cells lacking CAD activity, AIF knockdown causes a hypercondensation of the chromatin, apparently without an intervening peripheral-condensation stage.
A second factor termed acinus was also shown to be able to induce apoptotic chromatin condensation in vitro [7]. However, in a recent study, apoptotic chromatin condensation was shown to occur normally even when acinus had been depleted by RNAi knockdown [8]. Thus the role of acinus in apoptotic chromatin condensation is uncertain. More recently, phospholipase A2 (PLA2) was reported to induce nuclear shrinkage without fragmentation during caspase-independent cell death induced by hypoxia [9].
We [10] and many others [11–18] have developed cell-free systems with which to study the molecular mechanisms of apoptotic execution. Our cell-free system has previously allowed us to reveal critical roles of caspases and downstream activities in apoptotic execution [19–22].
In the present study, we characterize in detail the dynamic changes in the chromatin that occur during cell-free apoptosis. These observations lead us to propose the following nomenclature for the characteristic stages of nuclear disassembly during apoptotic execution: stage 0 - uncondensed; stage 1 - ring condensation; stage 2 - necklace condensation; stage 3–nuclear collapse/disassembly. We go on to show that DNase activity is not required for stage 1, but is essential for stage 2 condensation. We further show that hydrolysable ATP is required for stage 3 nuclear disassembly. This in vitro study provides a framework, which in the future may lead to the identification of the physiological factors that drive chromatin condensation during apoptosis in vivo.
Materials and Methods
Cell Treatment and Preparation of Extracts
Chicken DU249 cells were presynchronized in S phase with aphidicolin for 12 h, released from the block for 6 h, and synchronized in mitosis with nocodazole for 3 h as described previously [10,23]. S/M extracts were prepared from floating cells obtained by selective detachment after the nocodazole treatment [22].
Fractionation of S/M extracts with Heparin-Agarose Resin
Roller S/M extracts (typically 7 mg of protein) prepared in KPM buffer containing 60 mM KCl was mixed with 360 μl of heparin-agarose resin (HiTrap Heparin HP; Piscataway, NJ, USA) [22]. The mixture was rotated at 4°C for 1h, centrifuged in a microcentrifuge at 5,000 rpm for 5 min, and the supernatant was recovered (fraction-1). Three cycles of absorption with heparin-agarose were needed to completely remove all detectable DNase activities from S/M extracts. Pretreatment of fraction-1 (Fr-1) with caspase inhibitor, Ac-DEVD-CHO (Peptide Institutes, Minoh, Osaka, Japan) was performed at 100 μM for 20 min. at room temperature.
In Vitro Apoptosis Reaction
MDA-AF8 (derived from MDA-435) or HeLa S3 nuclei prepared as previously described [10,23] were added to a 10 μl reaction (S/M extracts or Fr-1) supplemented with or without ATP or ATP analogues (final 2 mM) plus a regeneration system and incubated at 37°C for the indicated time. One μl of DNase-1 ( 171~213 U/μl, BRL, Invitrogen Corp., Carlsbad, CA, USA) was added when necessary to a 10 μl reaction containing either MDB buffer [10] or Fr-1 plus nuclei. After incubation, nuclei were either stained with DAPI to observe chromatin condensation or solubilized for analysis of DNA ladder formation.
DNA Fragmentation Assay
Conventional Agarose Gel Electrophoresis
HeLa nuclei (1~2 x106 per loading) were incubated for the indicated period in S/M extract or with other partially purified fractions, solubilized, processed and the resulting genomic DNA was loaded onto 1% agarose gels as previously described[22].
Pulse-Field Gel Electrophoresis
HeLa nuclei (1~2 x106 per loading) were incubated in the cell-free system, solubilized, processed and analyzed by Contour-clamped homogenous (CHEF) electrophoresis, as previously described [22].
TUNEL Assay
TUNEL assays [24,25] were performed using the In Situ Cell Death Detection Kit (Roche Diagnostics GmbH; Penzberg, Germany) according to the manufacturer’s protocol.
Electron Microscopy
After the cell-free apoptotic reaction, nuclei were centrifuged in microfuge tubes and subsequently fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), containing 0.1% CaCl2. After washing in the same buffer and post-fixation in 1% OsO4 for 1 hr, the specimens were washed again, dehydrated in an ethanol series, and embedded in Epon 812. Thin sections were cut with a Leica Ultracut UCT microtome, post-stained with 2% uranyl acetate and Reynolds lead citrate and examined using a JEOL JEM-2000EX operated at 80 kV.
Real Time Imaging and Analysis
Human MDA-435 cells were transfected with EGFP-CENP-A expression plasmid and a stable cell line expressing EGFP-CENP A protein was isolated (MDA-AF8 ) [26]. Nuclei derived from MDA-AF8 cells were stained with SYTO 59 (Molecular Probes, Invitrogen Corp., Carlsbad, CA, USA) for 20 min at 0.5 μM at RT, centrifuged, mixed with S/M extracts and placed on the bottom of a Iwaki glass base dish (Asahi Techno Glass, Chiba, Japan) on the stage of a fluorescent microscope (Eclipse TE300, Nikon) with a PlanApo 60x objective, in a temperature-controlled humid chamber. Real time imaging was performed as described previously [27]. Briefly, images were obtained with Lumina Vision software (Mitani Corp., Tokyo, Japan), controlling a cooled CCD digital camera (C4742-95, Hamamatsu Photonics, Shizuoka, Japan) and a BioPoint MAC3000 controller system for the filter wheel and Z-axis motor (Ludl Electronic Products Ltd., Hawthone, NY, USA), for collecting Z-series optical sections (0.2 μm intervals). The distances between any two green dots (EGFP-CENP-A) representing centromeres were measured using the MacScope software (Mitani Corp.).
Results
Nuclei exhibit consistent structural changes during cell-free apoptosis
The morphological changes that occur in HeLa S3 nuclei undergoing cell-free apoptosis in S/M extracts in the presence of ATP are shown in Figure 1A. These observations allow us to identify three distinct stages in apoptotic chromatin condensation (defining the starting condition with uncondensed chromatin as stage 0– see Fig. 7). Stage 1 ring condensation (Fig. 1Ab), is characterized by a continuous ring of condensed chromatin at the interior surface of the nuclear envelope. Stage 1 was completed within ~15 minutes in our extract. In stage 2 necklace condensation (Fig. 1Ac), discontinuities are seen in the ring (arrows in Fig. 1Ac), which begins to adopt a beaded appearance over the course of 15–30 minutes. During this time, the nucleus begins to shrink. In stage 3 nuclear collapse/disassembly (Fig. 1Ad), final apoptotic bodies form as the nucleus rapidly completes its shrinkage and separates into individual fragments.
Figure 1. Time-course of cell-free apoptosis.
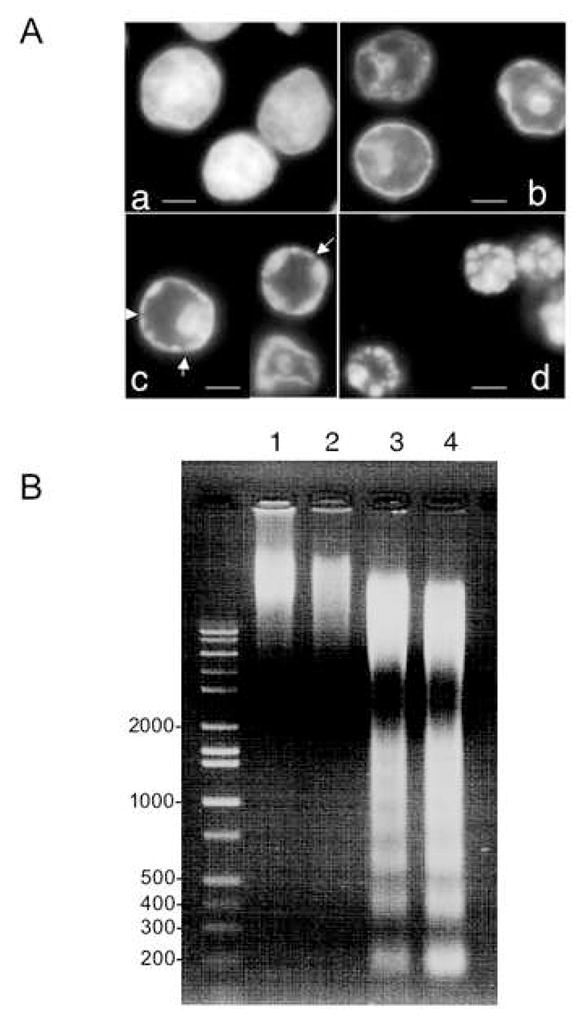
A Morphological changes of HeLa S3 nuclei incubated in S/M extract for 0 min (a), 15 min (b), 30 min (c) and 60 min (d) and then stained with DAPI. Bars in (a-d) represent 5 μm. Arrows in (c) represent slits on the ring.
B Conventional agarose gel analysis of DNA prepared from nuclei incubated in S/M extract for 0 min (lane 1), 15 min (lane 2), 30 min (lane 3) and 60 min (lane 4).
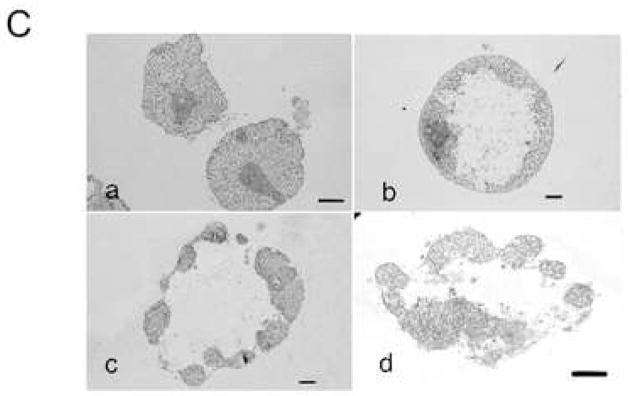
C Ultrastructure of nuclei incubated with S/M extracts in the presence of exogenous ATP. Transmission electron microscopy (TEM) analysis of HeLa S3 nuclei incubated with S/M extracts in the presence of ATP for 0 min (a), 15 min (b), 30 min (c) or 60 min (d). Bars: 2 μm (a, d) or 1 μm (b, c ).
Figure 7. Proposed stages of nuclear condensation during cell-free apoptosis and model showing factors acting each steps.
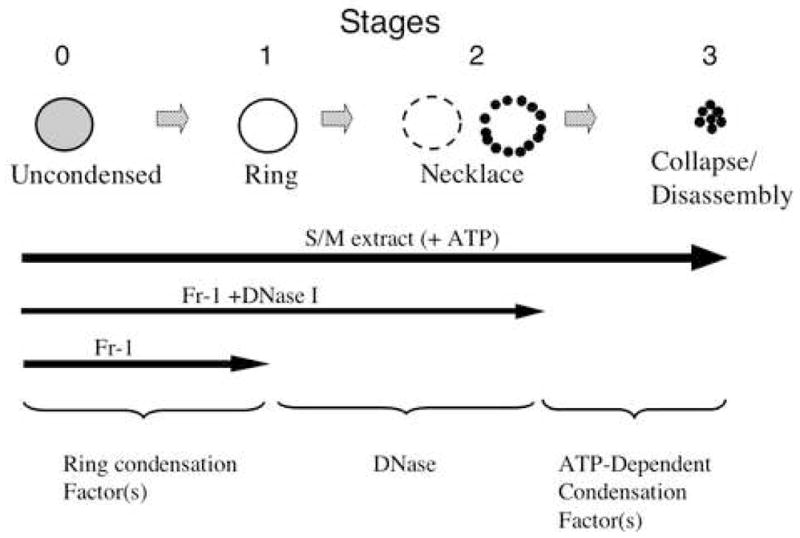
We have described three stages in nuclear condensation in cell-free apoptosis, Stage 1: ring condensation; Stage 2: necklace condensation; Stage 3: nuclear collapse/disassembly. Transit from stage 0 to 1 requires putative condensation factors that may include AIF. Transit from stage 1 to 2 requires DNase activity, while transit from stage 2 to 3 additionally requires ATP hydrolysis.
These morphological transitions were accompanied by degradation of the DNA. Although some slight fragmentation of genomic DNA could be detected at 15 minutes, extensive digestion of the DNA to yield a nucleosomal ladder was much more evident at 30 minutes (Figure 1B).
Ultrastructural changes of nuclei during cell-free apoptosis
To gain further information about the ultrastructural changes to the nucleus that occur during apoptotic chromatin condensation, we examined nuclei before and after exposure to S/M extract in the presence of ATP (Fig. 1C). Electron microscopy revealed that the peripheral condensed ring formed at stage 1 contains not only the chromatin, but the bulk of the nuclear components. Therefore, the stage 1 structure is a hollow-ball containing a large central cavity. The central cavity lacks recognizable substructures, with only some fibrous materials to be seen (Fig. 1C b). Similar electron microscope analyses of stage 2 necklace condensation and stage 3 nuclear collapse/disassembly were both consistent with the DAPI stained images (Fig. 1C c, d). At stage 2, nuclear envelope (NE) apparently remains intact, while in stage 3 the NE appears to be broken/disassembled. Therefore, we call stage 3 “collapse/disassembly”.
The structural changes occur with consistent timing and sequence during cell-free apoptosis
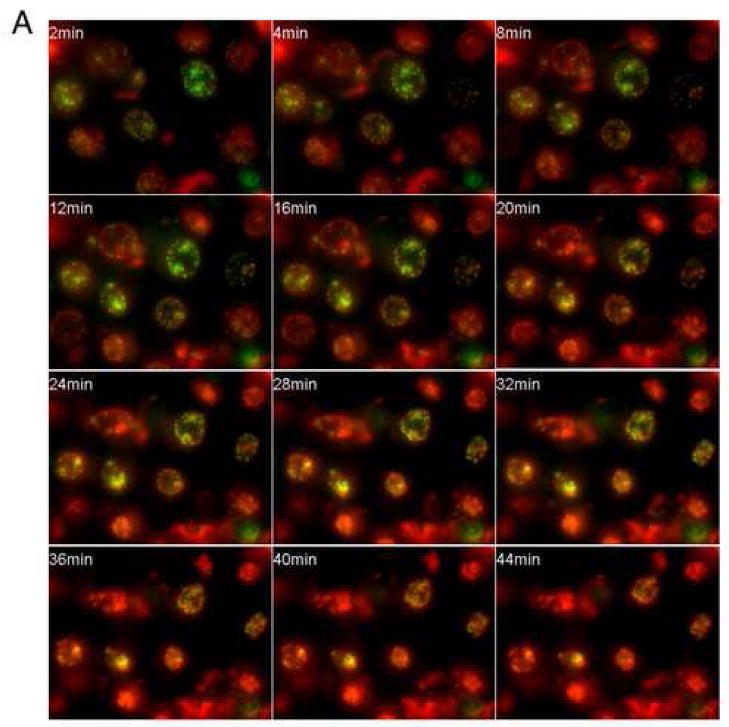
We used simultaneous two color visualization of bulk DNA (stained with SYTO 59) and centromeres (revealed by EGFP-CENP-A [26]) to conduct a time-lapse analysis of apoptotic DNA condensation in MDA-AF8 cells (Fig. 2A). As the time required for the transition from uncondensed to stage 1 ring condensation of the chromatin was very short (within 2 min of addition of nuclei to the extract), it was not possible to follow this transition, which was already completed by the time specimens had been prepared and mounted on the microscope. Therefore, in Fig. 2A, nuclei already exhibited stage 1 ring condensation at the earliest time points. Consistent with data obtained using HeLa S3 nuclei (ref. [10] and our unpublished data), the total time required for cell-free apoptosis of MDA-AF8 nuclei in the presence of ATP was ~30–40 min. Using this time-lapse approach, we could confirm that the discrete changes in appearance previously observed in individual fixed nuclei did indeed represent a temporal sequence. The duration of stages 1 and 2 was highly reproducible from experiment to experiment, and all of the nuclei condensed faithfully from stage 1 to 3.
Figure 2. Time-lapse imaging of cell-free apoptosis and quantification of nuclear condensation.
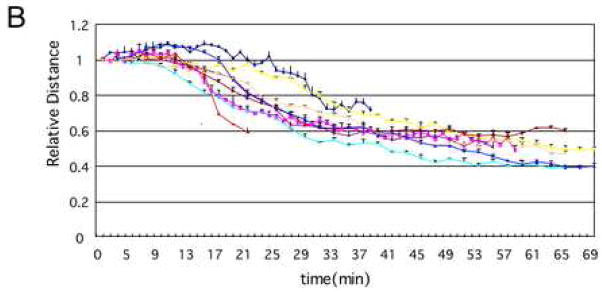
A MDA-AF8 nuclei were incubated with S/M extract and imaged by time-lapse fluorescence microscopy. DNA was visualized using SYTO 59 (red), and centromeres with EGFP-CENP-A (green). Movie data are available at Experimental Cell Research online. Representative images are shown. B Quantification of nuclear condensation in MDA-AF8 nuclei incubated with S/M extract in the presence of ATP. The distances between pairs of centromeres (green dots - EGFP-CENP-A) were measured using MacScope software. Relative values show the average of three measurements of the distance between arbitrarily chosen pairs of green dots at each time point divided by the distance between the same two dots at the initial time point.
During stage 1 condensation, nuclei retained their size. During stage 2, as slits and breaks began to appear in the peripheral ring of condensed chromatin, centromeres (visualized by EGFP-CENP-A fusion protein) slowly moved toward the nuclear interior. This reflected a movement of the underlying bulk chromatin.
In order to quantify the rate and extent of chromatin movements during apoptotic nuclear condensation, we measured the distances between pairs of randomly selected centromeres (Fig. 2B). The distance between selected centromeres within most nuclei began to decrease after 10 to 14 minutes in apoptotic extract. This movement reflected an overall compaction (shrinkage) of the chromatin. The shrinkage was accelerated later in stage 2 and ultimately reached a plateau after approximately 30 minutes as nuclei underwent stage 3 nuclear disassembly. In a small percent of nuclei, the shrinkage began later (~20 min) and reached the plateau after 40 min.
DNase activity is not required for stage 1 ring condensation of chromatin
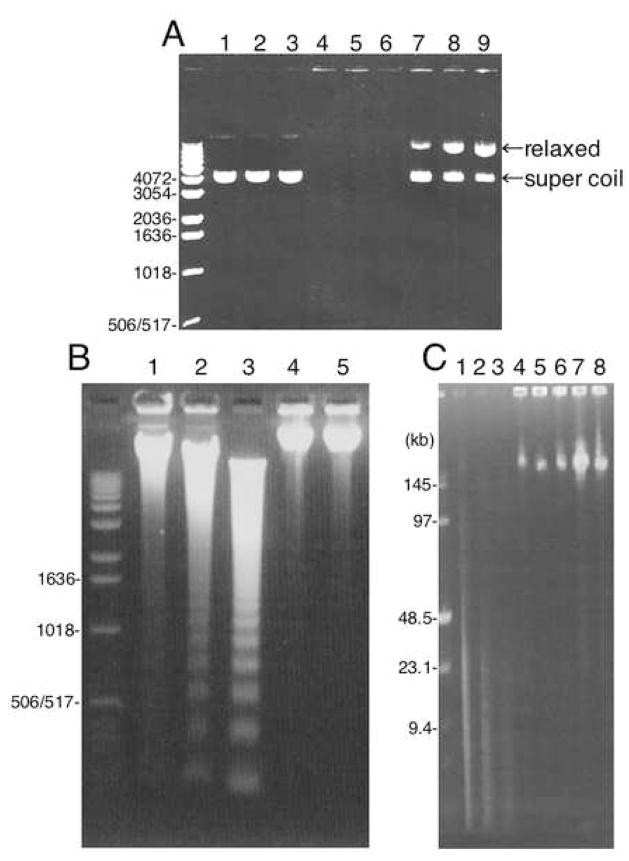
In order to gain insight into the biochemical properties of factors responsible for each stage in nuclear condensation, we performed biochemical fractionation of large-scale roller S/M extracts prepared from cells growing in roller bottles as previously reported [22]. When S/M extracts were fractionated on heparin agarose, DNases bound to the column in the presence of 50 mM KCl. After three cycles of binding and elution, all detectable DNase activity had been removed from the extract. The eluate from the heparin agarose column designated Fraction 1 (Fr-1) was unable to cleave plasmid DNA, although it did appear to have topoisomerase activity capable of relaxing the supercoiled plasmid DNA (Fig. 3A lanes 7–9). Fr-1 was also unable to trigger either internucleosomal DNA fragmentation (Fig. 3B lane 5) or large DNA fragmentation revealed by pulse-field gel electrophoresis (Fig. 3C lanes 4–6) in added nuclei.
Figure 3. DNase activity of Fraction-1 and S/M extracts.
A Plasmid pUC18 DNA was incubated with MDB buffer (lanes 1–3), S/M extract (lanes 4–6) or Fraction-1 (lanes 7–9) for 10 min (lanes 1, 4 or 7), 20 min (lanes 2, 5 or 8) or 30 min (lanes 3, 6 or 9) and analyzed by agarose electrophoresis. B HeLa S3 nuclei were incubated with S/M extract (lanes 1–3), MDB buffer (lane 4) or Fraction-1 (lane 5) for 15 min (lane 1), 30 min (lane 2) or 60 min (lanes 3–5) and analyzed by agarose gel electrophoresis. C HeLa S3 nuclei were incubated with S/M extract (lanes 1–3), Fraction-1 (lanes 4–6) or MDB buffer (lanes 7–8) for 0 min (lane 7), 15 min (lanes 1, 4), 30 min (lanes 2, 5) or 60 min (lanes 3, 6, 8) and analyzed by pulse-field gel electrophoresis.
Remarkably, Fr-1 could rapidly and efficiently induce stage 1 ring condensation in added nuclei (Fig. 4b). That this dramatic structural reorganization of the nucleus occurred without DNA fragmentation was supported not only by the electrophoresis studies of Fig. 3, but also by the results of TUNEL staining. While nuclei treated with S/M extract were strongly TUNEL-positive (Fig. 4g, h), nuclei treated with Fr-1 or buffer alone were negative for TUNEL staining (Fig. 4e, f and i, j, respectively).
Figure 4. Morphology of nuclei incubated with Fraction-1 and/or DNase I.
HeLa S3 nuclei were incubated with MDB buffer (a), Fraction-1 (b), Fraction-1 plus DNase I (c) or DNase I only (d) for 60 min and stained with DAPI. TUNEL analysis was also performed using nuclei incubated with Fraction-1 (e, f), S/M extract (g, h) or MDB buffer (i, j) for 60 min and stained with DAPI (e, g, i) or the TUNEL reaction (f, h, j). After Fraction-1 (Fr-1) was pretreated with DMSO ( k ) or Ac-DEVD-CHO ( l ), HeLa S3 nuclei were incubated with preteated Fr-1 and stained with DAPI. Bars represent 5 μm.
When exogenous Fr-1 was supplemented with DNase I, this caused added nuclei to progress from stage 1 to stage 2 necklace condensation (Fig. 4c). Interestingly, when DNase I alone was added to nuclei, stage 2 necklace–like structures were not formed. Instead, most nuclei were characterised by irregular patchy internal chromatin clumping (Fig. 4d). We conclude that both nucleases and non-nuclease activities are essential for full apoptotic chromatin condensation. In addition, pretreatment of Fr-1 with the caspase inhibitor Ac-DEVD-CHO did not suppress ring condensation by Fr-1 (Fig. 4k and l), suggesting that caspase activities are dispensable for stage 1 condensation.
The final stages of chromatin condensation and nuclear collapse/disassembly in vitro require ATP hydrolysis
The completion of nuclear collapse/disassembly in cell-free apoptotic extracts requires hydrolyzable ATP. If nuclei were incubated in S/M extracts in the absence of exogenous ATP, the pattern of morphological changes was normal only through the beginning of stage 2 necklace condensation. However, in the absence of ATP, the process was arrested at this stage and the final stages of chromatin condensation and shrinkage towards the nuclear interior did not occur (Fig. 5A a, b). The failure of further chromatin condensation and overall nuclear shrinkage was confirmed by electron microscopy, where in the presence of ATP, nuclei showed shrunken stage 3 structures (Fig. 5B,a), while in the absence of exogenous ATP nuclei showed stage 2 necklace condensation but failed to shrink further (Fig. 5B,b).
Figure 5. Exogenous ATP is essential for transition from Stage 2 to 3.
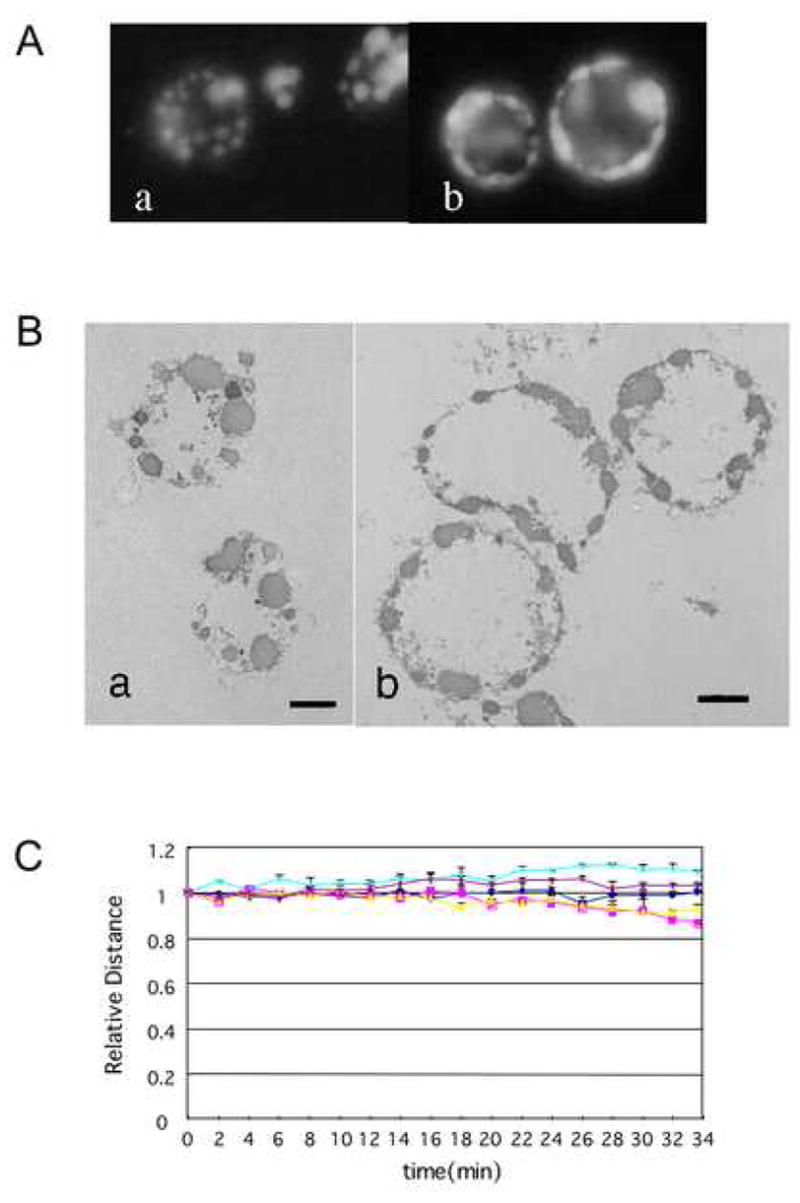
A Morphology of apoptotic nuclei incubated with or without exogenous ATP. HeLa S3 nuclei were incubated with S/M extracts in the presence (a) or absence (b) of ATP and stained with DAPI. B Ultrastructure of nuclei incubated with S/M extracts in the presence or absence of ATP. TEM analysis of HeLa S3 nuclei incubated with S/M extracts for 1 hour in the presence (a) or absence (b) of ATP. Bars: 2 μm C Quantification of nuclear condensation (centromere movement) without exogenous ATP. MDA-AF8 nuclei were incubated with S/M extract in the absence of exogenous ATP and analyzed as in Figure 2B.
To confirm the requirement for ATP in chromatin condensation during apoptosis, we measured the spacing between centromeres (identified by EGFP-CENP-A) in nuclei undergoing apoptotic chromatin condensation in vitro in the presence or absence of exogenous ATP. As described earlier, when this analysis was performed in the presence of ATP, the centromeres gradually approached one another as the chromatin condensed overall (Fig. 2B). In the absence of ATP, the distances between EGFP-CENP-A foci did not change significantly throughout the incubation period (Fig. 5C). Therefore, ATP is required for chromatin compaction and nuclear collapse/disassembly during apoptotic execution.
The requirement for ATP in apoptotic chromatin condensation appears to reflect a requirement for ATP hydrolysis. Amongst the ATP analogues examined, only ATP-γ-S, which is hydrolyzed, albeit poorly, could partially replace ATP (Fig. 6a). If ATP was replaced by dATP (Fig. 6b), GTP (Fig. 6c) or AMP-CPP (Fig. 6d), nuclear condensation was arrested at stage 2 (necklace condensation) with no final shrinkage and disassembly of the nuclei.
Figure 6. Completion of nuclear condensation in S/M extracts requires hydrolysable exogenous ATP.
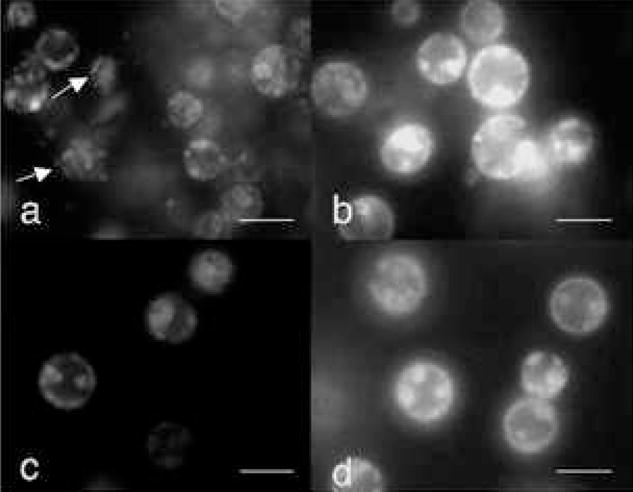
HeLa S3 nuclei were incubated with S/M extract in the presence of ATP-γ-S (a), dATP (b), GTP (c) or AMP-CPP (d) at a concentration of 2 mM. Arrows in (a) indicate nuclei in stage3. Bars represent 10 μm.
Discussion
Here, we demonstrate that there are three distinct stages in nuclear/chromatin condensation in a well studied cell-free apoptotic system. Starting with stage 0 (uncondensed chromatin), these stages can be classified as stage 1 - ring condensation; stage 2 - necklace condensation; and stage 3–nuclear collapse/disassembly (summarized in Fig. 7). Time lapse observations of populations of isolated nuclei at fifteen minute intervals showed that most nuclei seemed to follow this scenario of condensation with a remarkable degree of reproducibility, suggesting that nuclear changes in apoptosis represent a defined biochemical pathway.
Stage 1 ring condensation is remarkable because within about 2 minutes, virtually the entire contents of the nucleus collapses outward against the nuclear periphery. This includes not only the chromatin, but also the bulk of the nuclear components. This global reorganization of the nucleus is all the more remarkable as it does not require either DNA cleavage, ATP hydrolysis or caspase activity (see below). Clearly, if the nuclear interior is organized by some sort of nuclear matrix or scaffold, either this must undergo a rapid dissolution or contents are released from it within minutes of exposure of the nucleus to apoptotic extract.
Biochemical fractionation experiments demonstrated that depletion of DNase activities from S/M extracts did not block stage 1 ring condensation of the chromatin. Although other activities in Fraction-1 might be responsible for ring condensation, DNase activities are evidently dispensable for it. This is consistent with a previous report that chromatin condensation during apoptosis occurred without functional DNase in a Jurkat cell line expressing caspase-resistant ICAD but that the condensed chromatin remained near the nuclear periphery [28]. The results are also consistent with our previous observation of stage 1 ring condensation in DT40 cells lacking CAD/DFF40 [29].
One activity proposed to be important for stage 1 ring condensation is the flavoprotein AIF [2]. Although apparently not itself a nuclease, AIF has been proposed to recruit another nuclease(s) that is responsible for HMW DNA cleavage [2]. Since we have shown here that stage 1 ring condensation can occur in the absence of detectible DNase activity, if AIF is involved in our system, it must act by some mechanism other than nuclease activation/recruitment.
The mechanism underlying the very rapid stage 1 ring condensation is presently unknown. The unidentified factor(s) involved could potentially include enzymes that modify chromatin, or caspases, which might cleave underlying structural elements in the isolated nuclei. Indeed, although Fraction-1 does have high levels of caspase activity, it is unlikely that caspase(s) drive stage 1 ring condensation, since the condensation was not inhibited by prior treatment of Fr-1 with the potent caspase inhibitor Ac-DEVD-CHO (Fig. 4l). Enzymes that modify the chromatin could include kinases such as MST1, which modifies histone H2B [30], and may help to induce the rapid remodeling of nuclear structures. The transition from stage 1 to stage 2 chromatin condensation requires DNase activity, which in this system appears to be principally provided by CAD/DFF [29], although other nucleases have been implicated in other systems (for review, see ref [31] ). Caspase-6 is also essential for the transition from stage 1 to 2 nuclear condensation in nuclei of cells expressing lamin A [32]. Cells lacking lamin A (which is cleaved solely by caspase-6) completed nuclear apoptosis normally in the absence of caspase 6, suggesting that the role of the caspase in this instance was to release the chromatin from sites of tethering at the nuclear lamina.
Published evidence has disagreed over whether ATP is [17] or is not [18] required for nuclear/chromatin condensation during apoptosis. This controversy arose in part due to the lack of a clear nomenclature describing nuclear condensation during apoptosis. Some researchers have regarded the ring-condensation as final apoptotic condensation, while others did not discriminate between the different morphologies at different stages of the process. Therefore, it was useful for us to define the stages clearly and propose the nomenclature in Figure 7. Given this conceptual framework, our results clearly demonstrated that exogenous ATP is not essential through stage 2 (necklace condensation), but that it is essential for stage 3 (nuclear collapse/disassembly).
At present, the reason for the ATP dependency of stage 3 nuclear disassembly is completely unknown. Protein kinases such as MST1 are obvious candidates for factors that would utilize ATP, but it cannot be ruled out that other activities, including chromokinesin motor proteins could be involved. It is tempting to speculate that nuclear disassembly during apoptosis requires two independent activities, one of which remodels the chromatin and is responsible for stage 1 and 2 condensation. The second may drive the shrinkage of the necklace into compact aggregates (responsible for stage 3 condensation). In the absence of exogenous ATP, the shrinkage activity is not fully functional. In this sense, whether PLA2 [9] is also involved in the shrinkage of apoptotic nuclei or not is very interesting.
Chromatin condensation in both mitosis and apoptosis was observed well over 100 years ago by Flemming and others, and after decades of intense study, neither process is understood in molecular detail. Studies of mitotic chromosome condensation implicated the condensin complex[33], however a variety of studies including a genetic knockout of condensin in chicken DT40 cells [34] have revealed that condensin is not essential for mitotic condensation, and that the critical factors remain to be identified. Condensin also has no observable role in apoptotic chromatin condensation (Damien Hudson & WCE, unpublished). Apoptotic condensation, which is efficiently reproduced in vitro, may prove to be an ideal system for isolation of the factors that drive these phase transitions in chromatin. Identification of the activity underlying stage 1 ring condensation and the ATPase required for stage 3 nuclear disassembly could potentially yield insights into the mechanism of mitotic chromatin condensation as well.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the Wellcome Trust and from the NIH to WCE, and in part supported by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan and a Project Research Grant from Kawasaki Medical School to ST and YM. WCE is a Principal Research Fellow of the Wellcome Trust.
Abbreviations
- CAD
caspase-activated DNase
- DFF
DNA fragmentation factor
- ICAD
inhibitor of CAD
- AIF
apoptosis inducing factor
- acinus
apoptotic chromatin condensation inducer in the nucleus
- EGFP
enhanced green fluorescent protein
- TUNEL
TdT-mediated dUTP-biotin nick end labeling
- AMP-CPP
alpha, beta-methylene adenosine 5′-triphosphate
Footnotes
Supplemental movies showing cell-free apoptosis are available at Experimental Cell Research’s website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wyllie AH, Morris RG, Smith AL, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J Pathol. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 2.Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16:264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Susin SA, Daugas E, Ravagnan L, Samejima K, Zamzami N, Loeffler M, Costantini P, Ferri KF, Irinopoulou T, Prevost MC, Brothers G, Mak TW, Penninger J, Earnshaw WC, Kroemer G. Two distinct pathways leading to nuclear apoptosis. J Exp Med. 2000;192:571–580. doi: 10.1084/jem.192.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CY, Sasaki T, Elia AJ, Cheng HY, Ravagnan L, Ferri KF, Zamzami N, Wakeham A, Hakem R, Yoshida H, Kong YY, Mak TW, Zuniga-Pflucker JC, Kroemer G, Penninger JM. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 5.Slagsvold HH, Rosseland CM, Jacobs C, Khuong E, Kristoffersen N, Gaarder M, Fallgren AB, Huitfeldt HS, Paulsen RE. High molecular weight DNA fragments are processed by caspase sensitive or caspase independent pathways in cultures of cerebellar granule neurons. Brain Res. 2003;984:111–121. doi: 10.1016/s0006-8993(03)03119-6. [DOI] [PubMed] [Google Scholar]
- 6.Yuste VJ, Sanchez-Lopez I, Sole C, Moubarak RS, Bayascas JR, Dolcet X, Encinas M, Susin SA, Comella JX. The contribution of apoptosis-inducing factor, caspase-activated DNase, and inhibitor of caspase-activated DNase to the nuclear phenotype and DNA degradation during apoptosis. J Biol Chem. 2005;280:35670–35683. doi: 10.1074/jbc.M504015200. [DOI] [PubMed] [Google Scholar]
- 7.Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- 8.Joselin AP, Schulze-Osthoff K, Schwerk C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J Biol Chem. 2006;281:12475–12484. doi: 10.1074/jbc.M509859200. [DOI] [PubMed] [Google Scholar]
- 9.Shinzawa K, Tsujimoto Y. PLA2 activity is required for nuclear shrinkage in caspase-independent cell death. J Cell Biol. 2003;163:1219–1230. doi: 10.1083/jcb.200306159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazebnik YA, Cole S, Cooke CA, Nelson WG, Earnshaw WC. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solary E, Bertrand R, Kohn KW, Pommier Y. Differential induction of apoptosis in undifferentiated and differentiated HL-60 cells by DNA topoisomerase I and II inhibitors. Blood. 1993;81:1359–1368. [PubMed] [Google Scholar]
- 12.Newmeyer DD, Farschon DM, Reed JC. Cell-free apoptosis in Xenopus egg extracts: inhibition by Bcl-2 and requirement for an organelle fraction enriched in mitochondria. Cell. 1994;79:353–364. doi: 10.1016/0092-8674(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 13.Enari M, Hase A, Nagata S. Apoptosis by a cytosolic extract from Fas-activated cells. EMBO J. 1995;14:5201–5208. doi: 10.1002/j.1460-2075.1995.tb00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin SJ, Newmeyer DD, Mathias S, Farschon DM, Wang H-G, Reed JC, Kolesnick RN, Green DR. Cell-free reconstitution of Fas-, UV radiation- and ceramide-induced apoptosis. EMBO J. 1995;14:5191–5200. doi: 10.1002/j.1460-2075.1995.tb00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlegel J, Peters I, Orrenius S. Isolation and partial characterization of a protease involved in Fas-induced apoptosis. FEBS Lett. 1995;364:139–142. doi: 10.1016/0014-5793(95)00374-i. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 17.Kass GE, Eriksson JE, Weis M, Orrenius S, Chow SC. Chromatin condensation during apoptosis requires ATP. Biochem J. 1996;318:749–752. doi: 10.1042/bj3180749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widlak P, Palyvoda O, Kumala S, Garrard WT. Modeling apoptotic chromatin condensation in normal cell nuclei. Requirement for intranuclear mobility and actin involvement. J Biol Chem. 2002;277:21683–21690. doi: 10.1074/jbc.M201027200. [DOI] [PubMed] [Google Scholar]
- 19.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly (ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazebnik YA, Takahashi A, Moir R, Goldman R, Poirier GG, Kaufmann SH, Earnshaw WC. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc Nat Acad Sci (USA) 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi A, Alnemri E, Lazebnik YA, Fernandes-Alnemri T, Litwack G, Moir RD, Goldman RD, Poirier GG, Kaufmann SH, Earnshaw WC. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Nat Acad Sci (USA) 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samejima K, Toné S, Kottke TJ, Enari M, Sakahira H, Cooke CA, Durrieu F, Martins LM, Nagata S, Kaufmann SH, Earnshaw WC. Transition from caspase-dependent to caspase-independent mechanisms at the onset of apoptotic execution. J Cell Biol. 1998;143:225–239. doi: 10.1083/jcb.143.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood ER, Earnshaw WC. Mitotic chromatin condensation in vitro using somatic cell extracts and nuclei with variable levels of endogenous topoisomerase II. J Cell Biol. 1990;111:2839–2850. doi: 10.1083/jcb.111.6.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toné S, Tanaka S, Minatogawa Y, Kido R. DNA fragmentation during the programmed cell death in the chick limb buds. Exp Cell Res. 1994;215:234–236. doi: 10.1006/excr.1994.1337. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto K, Fukuda R, Himeno M. Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct Funct. 2000;25:253–261. doi: 10.1247/csf.25.253. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto K, Tasaka H, Dotsu M. Molecular behavior in living mitotic cells of human centromere heterochromatin protein HPLalpha ectopically expressed as a fusion to red fluorescent protein. Cell Struct Funct. 2001;26:705–718. doi: 10.1247/csf.26.705. [DOI] [PubMed] [Google Scholar]
- 28.Sakahira H, Enari M, Ohsawa Y, Uchiyama Y, Nagata S. Apoptotic nuclear morphological change without DNA fragmentation. Curr Biol. 1999;9:543–546. doi: 10.1016/s0960-9822(99)80240-1. [DOI] [PubMed] [Google Scholar]
- 29.Samejima K, Toné S, Earnshaw WC. CAD/DFF40 nuclease is dispensable for high molecular weight DNA cleavage and stage I chromatin condensation in apoptosis. J Biol Chem. 2001;276:45427–45432. doi: 10.1074/jbc.M108844200. [DOI] [PubMed] [Google Scholar]
- 30.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 31.Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 32.Ruchaud S, Korfali N, Villa P, Kottke TJ, Dingwall C, Kaufmann SH, Earnshaw WC. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002;21:1967–1977. doi: 10.1093/emboj/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 34.Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.