Abstract
The RAG1 recombinase, which participates in DNA manipulation during rearrangement of antigen receptor genes in developing immune cells, possesses ubiquitin ligase activity. The nuclear transport protein karyopherin alpha 1 (KPNA1) binds to RAG1 upstream of its ubiquitin ligase domain, but this interaction is not required for nuclear localization of RAG1. We found that the isolated ubiquitin ligase domain of RAG1 (amino acids 218-389) promoted ubiquitylation of purified KPNA1. While RAG1 auto-ubiquitylation is dependent on the ubiquitin conjugating enzyme CDC34, ubiquitylation of KPNA1 was best supported by UbcH2/Rad6 and UbcH5a. Ubiquitylation of KPNA1 required the lysine/arginine-rich region spanning RAG1 amino acids 218-263 upstream of the RAG1 ubiquitin ligase domain, but RAG1 was still able to undergo auto-ubiquitylation in this region even in the presence of KPNA1. This is the first putative substrate identified for the RAG1 ubiquitin ligase, and to our knowledge it is the first reported case of ubiquitylation of KPNA1.
1. Introduction
The production of intact antigen receptor genes in B and T cells requires rearrangement of the immunoglobulin and T cell receptor loci on the chromosomes by the process of V(D)J recombination (Tonegawa, 1983). Recombination is initiated by a lymphocyte-specific apparatus comprising the RAG1 and RAG2 proteins (Oettinger et al., 1990; Schatz et al., 1989), which together bind DNA sequences flanking the antigen receptor gene segments and introduce double stranded breaks in the DNA (van Gent et al., 1996). These breaks are subsequently joined by the general non-homologous end joining DNA repair machinery (Taccioli et al., 1993) to form diverse coding joints (CJ), which encode the antigen receptors, and signal joints (SJ), which are frequently lost from the cell. Such a process is inherently dangerous, and many chromosomal translocations associated with lymphoid malignancy arise from inappropriate activity of the RAG1/2 recombinase (Showe and Croce, 1987; Tsujimoto et al., 1985). V(D)J recombination is subject to regulation on many levels, including cell-type and developmentally stage specific expression of the recombinase genes at the transcriptional level (Geier and Schlissel, 2006; Llorian et al., 2007; Verkoczy et al., 2007). In addition, the RAG2 protein is phosphorylated, ubiquitylated and degraded at the G1/S transition (Lee and Desiderio, 1999; Lin and Desiderio, 1993; Mizuta et al., 2002), restricting recombinase activity to the G1 phase of the cell cycle. The zinc-binding RING domain of RAG1 displays ubiquitin ligase (E3) activity (Jones and Gellert, 2003; Yurchenko et al., 2003), but RAG1 is not required for ubiquitylation of RAG2 (Jiang et al., 2005; Lee and Desiderio, 1999; Mizuta et al., 2002). Instead it is hypothesized that RAG1 E3 activity participates in some other aspect of regulation of V(D)J recombination or lymphocyte development.
Ubiquitin conjugation is a multi-step process that participates in regulation of virtually every biochemical system in eukaryotic cells (Ciechanover et al., 1980; Hershko et al., 1980; Pickart, 2004). The small ubiquitin protein is first captured by the ubiquitin activating enzyme (E1) then passed to one of several ubiquitin conjugating (E2) enzymes (Hershko et al., 1983). The E3 assists in the final step of the process, transfer of the ubiquitin moiety to the epsilon amino group of lysine residues on the protein substrate (Hershko et al., 1983), thus it is the E3 that is primarily responsible for specificity. Repeated iterations of the cascade can lead to the formation of poly-ubiquitin chains that most often target the substrate for degradation by the 26S proteasome (Baumeister et al., 1998). In other cases, mono- or poly-ubiquitylation serves as a regulatory signal in a manner similar to phosphorylation or methylation (Pickart, 2000). A purified RAG1 fragment (amino acids 218-389) including the E3 domain when combined with ubiquitin, E1 and the E2 CDC34 promotes its own ubiquitylation primarily at two conserved lysine residues in the basic region just upstream of the RING ((Jones and Gellert, 2003) Simkus C. and Jones J. M. manuscript in preparation). Full length RAG1 undergoes ubiquitylation in intact cells (Jones and Gellert, 2003), but it is not known whether this is auto-catalyzed. To date, no other substrates of RAG1 E3 activity have been identified, but known RAG1-interacting proteins are likely candidates.
RAG1 is a 1040 amino acid protein with multiple domains and the potential to interact with a variety of partners. The above-mentioned basic region and RING domain as well as several cysteine-rich segments comprise much of the amino terminal third of the protein (Bellon et al., 1997; Rodgers et al., 1996), while the multi-domain “core”, amino acids 383-1008, covers much of the remainder (Sadofsky et al., 1993; Silver et al., 1993). The core includes both DNA binding and cleavage activities in complex with RAG2 (van Gent et al., 1996). RAG1 has been shown to bind to two cellular proteins in addition to RAG2, karyopherin alpha 1 (KPNA1, a.k.a. importin alpha 5, SRP1 and RAG cohort protein 2; (Cortes et al., 1994)) and karyopherin alpha 2 (KPNA2, a.k.a. RAG cohort protein 1; (Cuomo et al., 1994)), both of which are belived to be involved in nuclear trafficking (Fried and Kutay, 2003). KPNA2 binds to a nuclear localization sequence within the core, and is necessary and sufficient for nuclear import of RAG1 (Spanopoulou et al., 1995). KPNA1 binds to the basic region upstream of the RING, and neither this sequence nor interaction with KPNA1 is required for import (Spanopoulou et al., 1995). The significance of this interaction is unknown.
Import into the nucleus begins with recognition of the target protein by one of the karyopherin alpha family members (Goldfarb et al., 2004). A ternary complex is formed including the target, karyopherin alpha and a karyopherin/importin beta family member. The complex passes through the nuclear pore (Goldfarb et al., 2004), at which point it disassociates in a manner dependent on RAN-GTP (Gilchrist et al., 2002; Gilchrist and Rexach, 2003), releasing the cargo into the nucleus. There is increasing evidence that both karyopherin alpha and beta proteins are involved regulation of nuclear processes outside of their roles in import (Gorjanacz et al., 2002; Kussel and Frasch, 1995; Loeb et al., 1995; Tabb et al., 2000). Since import of RAG1 into the nucleus requires only interaction with KPNA2 (Spanopoulou et al., 1995), it is possible that interaction with KPNA1 may play another regulatory function.
We found that a fragment of RAG1 (amino acids 218-389) comprising the RING and basic region promoted ubiquitylation of KPNA1 in a purified system including ubiquitin, E1 and E2. We have found previously that RAG1 E3 activity is required for efficient recombination by the full length protein when RAG1 is present at relatively low levels (Simkus et al., 2007), and suggested that ubiquitylation of negative regulator may be required to enable recombination. The previously established interaction between RAG1 and KPNA1 in vivo along with RAG1’s ability to ubiquitylate KPNA1 suggests that it may be that regulator.
2. Experimental
2.1 Materials
2.1.1 Cloning and expression of KPNA1
A clone including the cDNA of KPNA1 (GenBank ID AW245333, AW245690, BC002374) was obtained from the American type Culture Collection (Manassas, VA, USA), and the entire coding region was sub-cloned into the bacterial expression vector pMAL-2cx (New England Biolabs, Boston, MA, USA) downstream and in frame with the maltose binding protein (MBP) protein coding region. Correct reading frame was confirmed by sequencing (Retrogen, SanDiego, CA, USA). This plasmid, pMAL-Kα, was transformed into Rosetta DE3 bacterial cells (Novagen, USA), which were grown in LB medium (0.5 l) supplemented with 100 μg/ml ampicillin to mid-log phase (OD = 0.5). Expression was induced by the addition of IPTG to 1 mM, and allowed to proceed for 4 hrs. Cells were collected by centrifugation, and resuspended in 10 ml Buffer A (20 mM Tris [pH 7.4], 0.2 M NaCl, 1 mM EDTA, 1 mM DTT) supplemented with 50 μg/ml lysozyme and 1 mM PMSF. After 10 min incubation on ice, the suspension was subjected to a single round of freeze-thaw, and subjected to 5 bursts of sonication (15 sec each). Cell debris was collected by high speed centrifugation, and the supernatant (FrI) was loaded onto a 4.6 ml amylose resin column (New England Biolabs, Boston, MA, USA) equilibrated in Buffer A. After extensive washing with Buffer A, bound proteins were eluted with Buffer A plus 10 mM maltose. Fractions containing maltose binding protein-KPNA1 (MBP-KPNA1) were pooled, and a portion (0.5 ml) was loaded onto a Sephadex 200 10/300 GL column (GE Healthcare, USA) equilibrated in Buffer B (20 mM Tris [pH7.4], 0.2M NaCl, 1 mM DTT). Fractions containing MBP-KPNA1 were pooled and flash frozen in small aliquots.
2.1.2 Additional proteins
RAG1[218-389], RAG1[264-389], and protein-kinase tagged ubiquitin (PK-Ubi) were expressed and purified as previously described (Jones and Gellert, 2003; Simkus et al., 2007). Recombinant human UbcH5a was sub-cloned, expressed and purified as previously described for CDC34 (Jones and Gellert, 2003; Simkus et al., 2007). Leporine E1 and all remaining E2 enzymes were purchased from Boston Biochem (Boston, MA, USA).
2.1.3 Reagents
Chemical reagents were purchased from Fisher Scientific unless otherwise indicated. Anti-MBP antibody was purchased from New England Biolabs. Anti-Xpress epitope antibody was purchased from Invitrogen (USA). Anti-mouse horse radish peroxidase conjugate and SuperSignal West Femto detection reagent were purchased from Thermo Scientific (Rockford, IL, USA).
2.2 Methods
2.2.1 Ubiquitylation
For ubiquitylation of KPNA1, purified MBP-KPNA1 (1 μM as monomer), E1 (45 nM), the E2 indicated (0.3 μM), PK-Ubi (500 μM), and RAG1 (4 μM as dimer) were incubated in reaction buffer (50 mM Tris [pH 7.4], 0.001% Brij, 2 mM Mg-ATP, 50 nM NaCl, 0.4 mM DTT, and 0.02 mM ZnCl2) for up to 16 hrs at room temperature. Products were separated on 4–12% NuPAGE gels (Invitrogen, USA), transfered to nitrocellulose and subjected to Western blot with anti-MBP antibody (1:10,000), followed by anti-mouse-HRP conjugate (1:10,000), and SuperSignal West Femto detection reagent. ECL was detected and quantified with a Kodak ImageStation 3000 (Kodak, Cambridge, MA, USA).
Ubiquitylation of RAG1[218-389] was performed essentially as described above, except that KPNA1 concentration was increased (8 μM). Products were detected with anti-Xpress epitope antibody as previously described.
2.2.2 Gel Filtration
Equimolar concentrations (80 μM) of KPNA1, RAG1[218-389], or RAG1[264-389] as indicated were combined in 0.2 ml and loaded onto a Sephadex 200 10/300 GL column equilibrated in buffer C (20 mM tris [pH 7.4], 0.5 M NaCl, 1 mM DTT, 10% glycerol). Protein elution was monitored by absorbance at 280 nm and Western blot. The column was calibrated with standards from Bio-Rad (Hercules, CA, USA).
3. Results
3.1 Ubiquitylation of KPNA1 in a reconstituted system
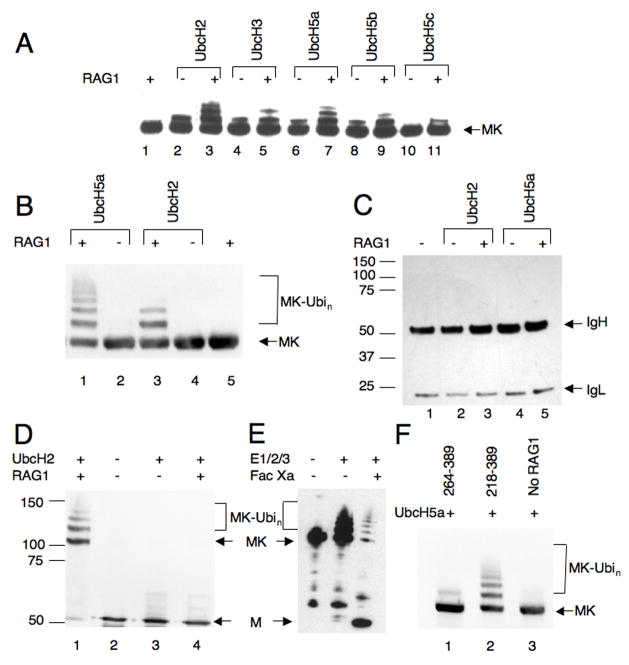
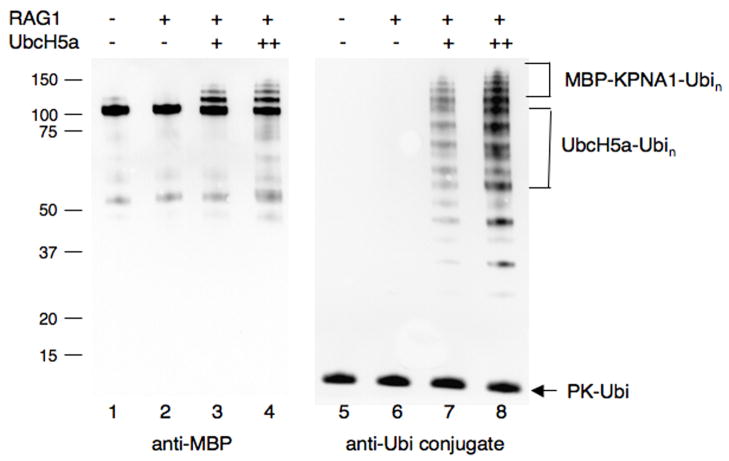
RAG1[218-389] promoted ubiquitylation of purified maltose binding protein (MBP) tagged KPNA1 (Fig 1). MBP-KPNA1 was combined with RAG1[218-389], the E2 and E1 enzymes, and protein-kinase tagged ubiquitin (PK-Ubi). After incubation, reaction products were separated on denaturing polyacrylamide gels, blotted and probed with anti-MBP antibody. In a screen of various E2 enzymes, both UbcH2 and UbcH5a were found to support robust modification of MBP-KPNA1 in a RAG1-dependent manner (Fig 1A, lanes 3 and 7). In these reactions we observed anti-MBP reactive species of a molecular weight consistent with ubiquitylation of MBP-KPNA1. Presumptive RAG1-promoted ubiquitylation of MBP-KPNA1 could be supported to a small extent by CDC34 and UbcH5b, but not by UbcH5c, UbcH6, UbcH7 and UbcH10 (Fig 1A and data not shown). In confirmatory experiments, we found little difference between results obtained with UbcH2 and UbcH5a (Fig 1B, lanes 1 and 3), and subsequently both were used as indicated. To confirm that these higher molecular weight products were indeed the result of ubiquitylation, MBP-KPNA1 was treated with RAG1[219-389] and two different levels of UbcH5a. A Western blot of products was probed with anti-MBP (Fig 2 lanes 1–4), then stripped and re-probed with anti-ubiquitin conjugate (Fig 2, lanes 5–8). Only the bands corresponding in molecular weight to ubiquitylated MBP-KPNA1 were recognized by both antibodies, although the anti-ubiquitin conjugate antibody also recognized several other components of the reaction such as PK-ubiquitin and auto-ubiquitylated UBCH5a (Fig 2, lanes 5–8). KPNA1 sometimes underwent mono-ubiquitylation to a small extent in the absence of RAG1[218-389] (data not shown), but RAG1 strongly stimulated this reaction and promoted formation of di-, tri- and poly-ubiquitylated species.
Figure 1.
Figure 2.
RAG1-dependent ubiquitylation is specific for the KPNA1 substrate and does not occur promiscuously with other proteins that bind near the ubiquitin ligase domain. RAG1 did not promote ubiquitylation of MBP alone (Fig 1D, cf. lanes 1 and 4), and when MBP-KPNA1 was allowed to undergo ubiquitylation and the MBP moiety was subsequently cleaved from KPNA1, there was no evidence that MBP had undergone extensive modification (Fig 1E, lane 3; approximately 90% of the MBP-KPNA1 underwent cleavage). RAG1 did not promote ubiquitylation of an anti-Xpress antibody that bound to the Xpress epitope fused in frame upstream of the RING finger (Fig 1C).
3.2 Requirements for RAG1-supported ubiquitylation of KPNA1
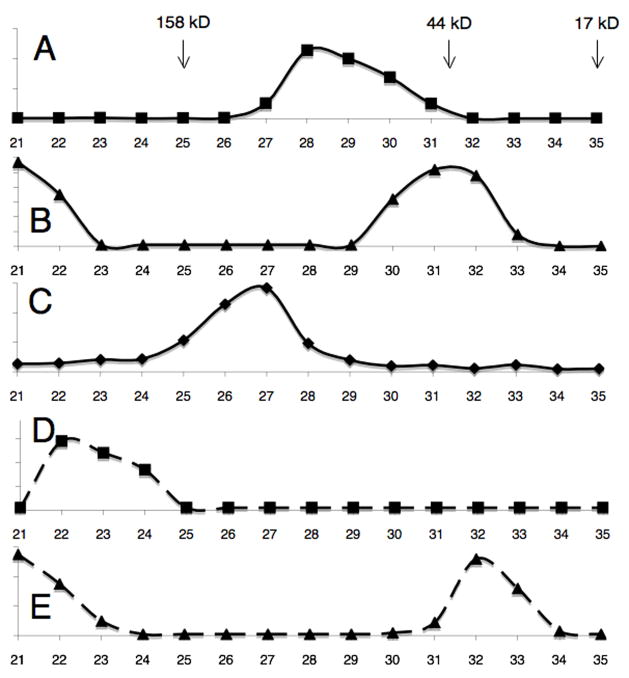
The lysine-arginine rich segment (amino acids 218-263) upstream of the RAG1 RING domain was required for interaction with KPNA1 and for its ubiquitylation. RAG1[264-389] which spans the RING domain and closely associated zinc finger promoted only minimal ubiquitylation of MBP-KPNA1 when compared to RAG1[218-389] (Fig 1F, cf. lanes 1 and 2). Physical interaction between the RAG1 fragment and KPNA1 was examined by gel filtration. When analyzed by themselves, RAG1[218-389] and RAG1[264-389] eluted at 14 and 15.5 ml, respectively (Fig 3A and 3B). Based on comparison to molecular weight standards, this suggested an apparent molecular weight of 39 kD for RAG1[264-389]. This is consistent with previously published results indicating that this domain forms a dimer (Rodgers et al., 1996). A portion of RAG1[264-389] also occurred as a high molecular weight species that nevertheless eluted within the included volume; the amount of high molecular weight complex could be decreased by decreasing the concentration of RAG1[264-389] (data not shown). It has been shown previously that isolated RING domains from larger proteins will self-assemble into large, soluble, dodecameric complexes (Kentsis et al., 2002a; Kentsis et al., 2002b); our data were consistent with a mixed population of dimer and dodecamer. It is possible that the presence of the basic extension from 218-263 blocked self-assembly, as we did not see these very large complexes with RAG1[218-389] alone. RAG1[218-389] eluted as a single peak with an apparent molecular weight of 81 kD. This is somewhat larger than expected for a dimer of 24 kD protomers, indicating either that a) RAG1[218-389] exists as trimer or tetramer complex or b) the highly basic, largely unstructured region upstream of the RING leads to anomalous behavior on the gel filtration column. The trailing edge of the peak profile lends support to the second explanation. MBP-KPNA1 eluted at 13.5 ml (Fig 3C), consistent with it being a 103 kD monomer.
Figure 3.
When RAG1[218-389] was combined with a molar excess of MBP-KPNA1, they formed a high molecular weight complex that eluted at 11 ml, approximately 350 kD (Fig 3D). The shape of the peak was similar to RAG1[218-389] alone. If a dimer of RAG1[218-389] provides two binding sites for KPNA1 monomers (RAG1[218-389]2-2KPNA1), we would expect a 253 kD complex. Based on the anomalous behavior of RAG[218-389] by itself, the somewhat larger apparent molecular weight of the eluted complex is consistent with the RAG1[218-389]2-2KPNA1 stoichiometry. Unlike RAG1[218-389], RAG1[264-389] eluted at 15.5 ml regardless of the presence of MBP-KPNA1 (Fig 3E), indicating that the two proteins do not form a stable complex. These data agree with previously published results demonstrating that KPNA1 binds to basic segments of RAG1 spanning amino acids 222-225 and 246-250 (Cortes et al., 1994; Spanopoulou et al., 1995).
3.3 Effect of KPNA1 on RAG1 ubiquitylation
The RAG1[218-389] fragment has previously been shown to promote its own ubiquitylation primarily at lysines K233 and K257. The addition of purified MBP-KPNA1 to this reaction resulted in a mild decrease in auto-ubiquitylation of RAG1[218-389] (Fig 4A). KPNA1 binds within the same region of RAG1 that undergoes auto-ubiquitylation (Fig 3), and one of its binding sites overlaps a position of RAG1[218-389] auto-ubiquitylation. In order for auto-ubiquitylation to occur in this system, the E2 enzyme must bind the intact RING domain and access free lysine(s) onto which ubiquitin is transferred. The presence of the KPNA1 protein bound to these lysine residues might shield them from modification, which may explain the mild reduction in auto-ubiquitylation activity. Based on this, we expected that KPNA1 would not be able to bind to RAG1 that had already undergone ubiquitylation.
Figure 4.
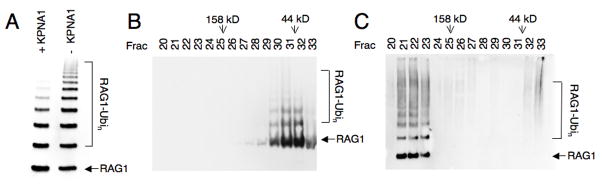
To test this hypothesis, RAG1[218-389] was allowed to undergo auto-ubiquitylation prior to being combined with KPNA1, then the complexes were analyzed by gel filtration chromatography. Auto-ubiquitylated RAG1[218-389] alone eluted as a broad peak between 13 and 14 ml, slightly earlier than unmodified RAG1[218-389] (Fig 4B). Ubiquitylated RAG1 protomers did not clearly separate from the unmodified species despite the addition of up to 48 kD in ubiquitin moieties, suggesting that both modified and unmodified RAG1 protomers exist together in the same complex. When ubiquitylated RAG1[218-389] was combined with KPNA1, both ubiquitylated and unmodified RAG1 complexes were shifted to higher molecular weight, eluting between 10.5 and 11.5 ml during gel filtration (Fig 4C). Thus there was no clear evidence that KPNA1 binding was blocked by ubiquitylation of RAG1, indicating that the KPNA1 binding site does not overlap 100% with the site of RAG1 auto-ubiquitylation. It is possible that in a RAG1 dimer, only one protomer is ubiquitylated, and KPNA1 may bind exclusively to the unmodified partner. Taken together however, these results indicate that ubiquitylation of KPNA1 and auto-ubiquitylation of RAG1 are not mutually exclusive.
4. Discussion
Much of the previous biochemical work on RAG1 has focused on the so-called core region of the protein, both because this region was relatively easy to purify and because it recapitulated many of the functions of the full length protein in vivo. There is, however, a large body of evidence to indicate that the non-core regions also include activities that are essential for normal immune development (Dudley et al., 2003; McMahan et al., 1997; Roman et al., 1997; Simkus et al., 2007). Most notably, multiple mutations and deletions within the amino terminal non-core region are associated with Omenn syndrome (Villa et al., 2001), a B-cell negative form of severe combined immune deficiency in humans, and additional immunodeficiencies associated with granulomas (Schuetz et al., 2008). Several of these mutations occur within the RING finger ubiquitin ligase domain (Schuetz et al., 2008; Villa et al., 2001), and we have previously demonstrated that at least one of them eliminates both ubiquitin ligase activity and recombination (Simkus et al., 2007). Other mutations that only partially abrogate ubiquitin ligase activity appear to have a similar effect on recombination (Simkus C. and Jones J. M. manuscript under submission). Thus there is a correlation between the ability of RAG1 to act as a ubiquitin ligase and its ability to support recombination. We have suggested that RAG1 ubiquitin ligase activity may be required to promote dissociation from a negative regulator to enable recombination (Simkus et al., 2007).
Our finding that KPNA1 is a substrate for RAG1 ubiquitin ligase activity in vitro brings together two formerly disparate lines of evidence regarding the role of the amino terminal non-core region. KPNA1 was identified as a RAG1 cohort protein nearly fifteen years ago (Cortes et al., 1994), but because it bound outside the core and was not found to be required for nuclear localization of RAG1, it received relatively little attention. Intriguingly, KPNA1 was shown to influence sub-nuclear localization of RAG1 (Spanopoulou et al., 1995). RAG1 protein bound to KPNA1 displayed a punctate or speckled pattern within the nucleus, while mutants that could not bind KPNA1 were found to be distributed diffusely throughout the nucleus (Spanopoulou et al., 1995). These speckles were too numerous to represent active recombination complexes, which are generally confined to only two per cell (Chen et al., 2000). V(D)J recombination is already know to be regulated through subnuclear localization of the recombination loci (Kosak et al., 2002). This theme could be repeated through sequestration of the recombinase itself.
We propose a model in which RAG1 is imported into the nucleus based on interaction between KPNA2 and a nuclear localization signal within the core region (Spanopoulou et al., 1995). Association of KPNA1 with binding sites upstream of the RING domain in the non-core region, while not required for import, could keep RAG1 tethered to the nuclear pore complex. Normally such complexes are dissociated after import in a manner dependent on the RAG1 GTPase. However, certain proteins such as the SUMO-1-deconjugating enzyme Ulp1 have been found to remain tethered to the nuclear pore via association with three karyopherins, Pse1, Kap95 and Kap60 (Panse et al., 2003). Continued synthesis and import of RAG1 would produce the speckled staining pattern seen previously (Spanopoulou et al., 1995). This would also be consistent with previous results from our laboratory and others that RAG1 fractionates primarily with the nuclear lamina and not strictly with the soluble nuclear fraction (Leu and Schatz, 1995; Simkus et al., 2007). RAG1-dependent ubiquitylation of KPNA1 would be required to dissociate this complex, perhaps by promoting degradation of KPNA1 by the 26S proteasome. RAG1 would then be free to travel to the site of recombination. Sequestration of RAG1 may help prevent promiscuous and inappropriate activity of the enzyme. The central domain of RAG1 within the core is capable of non-specific cleavage of single-stranded DNA (De et al., 2004). While this activity is largely suppressed by the presence of the C-terminal domain (De et al., 2004), even a low level of DNA cleavage would be deleterious particularly during S phase when large regions of single-stranded DNA would be present. RAG2, which plays a major role in regulating and directing RAG1-dependent DNA cleavage (e.g. (Akamatsu and Oettinger, 1998; De et al., 2004; Kirch et al., 1998; Swanson and Desiderio, 1999), is also absent during S and G2 (Desiderio et al., 1996).
This model suggests that ubiquitylation of KPNA1 may a be a triggered event, occurring only when the cell is primed to undergo recombination. While the nature of the trigger is unclear, accumulation of RAG2 during G1 and its binding to RAG1 is a reasonable candidate. We are currently exploring the ubiquitin ligase activity of full length RAG1, the influence of RAG2 on this activity, and the ability of RAG1 to promote ubiquitylation of KPNA1 in vivo.
We have previously found that RAG1 promotes its own ubiquitylation at conserved lysine residues upstream of the RING domain, and that RAG1 undergoes ubiquitylation in intact cells (Jones and Gellert, 2003). Our current data indicate that auto-ubiquitylation of RAG1 and ubiquitylation of KPNA1 in trans are not mutually exclusive. We see no evidence that auto-ubiquitylation promotes dissociation from KPNA1, and therefore it would not have a role in the model proposed here. However, we do not rule out the possibility that ubiquitylation of RAG1 may promote a later step of recombination.
5. Conclusion
The RAG1 RING and upstream basic domain (amino acids 218-389) together promote the poly-ubiquitylation of KPNA1 in vitro in a manner dependent on the E2 enzyme UbcH2/Rad6 or UbcH5a. KPNA1 binds to a lysine/arginine-rich region upstream of the RING domain, but does not prevent auto-ubiquitylation of this region. This finding, along with previous data that RAG1 may be required to ubiquitylate a negative regulator to activate recombination (Simkus et al., 2007) and that KPNA1 and RAG1 form punctate complexes in cells (Spanopoulou et al., 1995) suggest that KPNA1 may be a negative regulator of RAG1 and thus of V(D)J recombination.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Award Number P30CA051008. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government. This project was supported in part by a grant to J.M.J. from the National Institutes of Health (AI062854-01). The content is solely the responsibility of the authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–80. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–5. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980;77:1365–8. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes P, Ye ZS, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci U S A. 1994;91:7633–7. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Kirch SA, Gyuris J, Brent R, Oettinger MA. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci U S A. 1994;91:6156–60. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P, Peak MM, Rodgers KK. DNA cleavage activity of the V(D)J recombination protein RAG1 is autoregulated. Mol Cell Biol. 2004;24:6850–60. doi: 10.1128/MCB.24.15.6850-6860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S, Lin WC, Li Z. The cell cycle and V(D)J recombination. Curr Top Microbiol Immunol. 1996;217:45–59. doi: 10.1007/978-3-642-50140-1_4. [DOI] [PubMed] [Google Scholar]
- Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J Exp Med. 2003;198:1439–50. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–88. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier JK, Schlissel MS. Pre-BCR signals and the control of Ig gene rearrangements. Semin Immunol. 2006;18:31–9. doi: 10.1016/j.smim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Gilchrist D, Mykytka B, Rexach M. Accelerating the rate of disassembly of karyopherin. cargo complexes. J Biol Chem. 2002;277:18161–72. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- Gilchrist D, Rexach M. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J Biol Chem. 2003;278:51937–49. doi: 10.1074/jbc.M307371200. [DOI] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Gorjanacz M, Adam G, Torok I, Mechler BM, Szlanka T, Kiss I. Importin-alpha 2 is critically required for the assembly of ring canals during Drosophila oogenesis. Dev Biol. 2002;251:271–82. doi: 10.1006/dbio.2002.0827. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980;77:1783–6. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. Auto-ubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Gordon RE, Borden KL. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci U S A. 2002a;99:15404–9. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Gordon RE, Borden KL. Self-assembly properties of a model RING domain. Proc Natl Acad Sci U S A. 2002b;99:667–72. doi: 10.1073/pnas.012317299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch SA, Rathbun GA, Oettinger MA. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–62. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kussel P, Frasch M. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol Gen Genet. 1995;248:351–63. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–81. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- Leu TM, Schatz DG. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol Cell Biol. 1995;15:5657–70. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–9. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110delta is required for down-regulation of RAG expression in immature B cells. J Immunol. 2007;178:1981–5. doi: 10.4049/jimmunol.178.4.1981. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci U S A. 1995;92:7647–51. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R, Mizuta M, Araki S, Kitamura D. RAG2 is down regulated by cytoplasmic sequestration and ubiquitin-dependent degradation. J Biol Chem. 2002;277:41423–41427. doi: 10.1074/jbc.M206605200. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol. 2003;5:21–7. doi: 10.1038/ncb893. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–90. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- Roman CA, Cherry SR, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1(−/−) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Schuetz C, Huck K, Gudowius S, Megahed M, Feyen O, Hubner B, Schneider DT, Manfras B, Pannicke U, Willemze R, Knuchel R, Gobel U, Schulz A, Borkhardt A, Friedrich W, Schwarz K, Niehues T. An immunodeficiency disease with RAG mutations and granulomas. N Engl J Med. 2008;358:2030–8. doi: 10.1056/NEJMoa073966. [DOI] [PubMed] [Google Scholar]
- Showe LC, Croce CM. The role of chromosomal translocations in B- and T-cell neoplasia. Annu Rev Immunol. 1987;5:253–77. doi: 10.1146/annurev.iy.05.040187.001345. [DOI] [PubMed] [Google Scholar]
- Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci U S A. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkus C, Anand P, Bhattacharyya A, Jones JM. Biochemical and folding defects in a RAG1 variant associated with Omenn syndrome. J Immunol. 2007;179:8332–40. doi: 10.4049/jimmunol.179.12.8332. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E, Cortes P, Shih C, Huang CM, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–26. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb MM, Tongaonkar P, Vu L, Nomura M. Evidence for separable functions of Srp1p, the yeast homolog of importin alpha (Karyopherin alpha): role for Srp1p and Sts1p in protein degradation. Mol Cell Biol. 2000;20:6062–73. doi: 10.1128/mcb.20.16.6062-6073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli GE, Rathbun GA, Oltz EM, Stamato TD, Jeggo PA, Alt F. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce CM. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–3. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombinations in a cell-free system by RAG1 and RAG2 proteins. Curr Top Microbiol Immunol. 1996;217:1–10. doi: 10.1007/978-3-642-50140-1_1. [DOI] [PubMed] [Google Scholar]
- Verkoczy L, Duong B, Skog P, Ait-Azzouzene D, Puri K, Vela JL, Nemazee D. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. 2007;178:6332–41. doi: 10.4049/jimmunol.178.10.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, Cortes P, Duse M, Fasth A, Filipovich AM, Infante AJ, Jones A, Mazzolari E, Muller SM, Pasic S, Rechavi G, Sacco MG, Santagata S, Schroeder ML, Seger R, Strina D, Ugazio A, Valiaho J, Vihinen M, Vogler LB, Ochs H, Vezzoni P, Friedrich W, Schwarz K. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]