Abstract
To search for genetic variants that are associated with prostate cancer risk in the genome, we combined the data from our genome-wide association study (GWAS) in a population-based case-control study in Sweden with publicly available GWAS data from the Cancer Genetic Markers of Susceptibility (CGEMS) study. We limited the cases to those with aggressive disease, in an attempt to identify risk variants that are associated with this most clinically relevant form of the disease. Among the most likely candidate SNPs identified from the two GWAS, we sequentially confirmed one SNP at 22q13 in two independent study populations; the remaining subjects in CAPS and a hospital-based case-control study at Johns Hopkins Hospital. Association of aggressive prostate cancer with the SNP at 22q13 was also observed in the publicly available data of four additional study populations from the second stage of the CGEMS study. In all seven study populations examined, the frequency of allele ‘C’ of rs9623117 at 22q13 was consistently higher in aggressive cases than in controls. The combined allelic test was highly significant, with a P = 5.9 × 10−7. The Odds Ratio (OR) of allele ‘C’ for aggressive prostate cancer was estimated to be 1.18 (95% CI: 1.11-1.26). However, the SNP was also associated with non-aggressive prostate cancer, with an estimated OR of 1.11 (95% CI: 1.04-1.19, P = 0.003. The risk associated variants are located within the genomic region of TNRC6B, a gene involved in miRNA mediated mRNA degradation. Additional studies are warranted to further confirm the association.
Keywords: prostate cancer, association, genome-wide, 22q13, TNRC6B
Introduction
More than a dozen prostate cancer risk associated variants have been identified from genome-wide association studies (GWAS) and consistently replicated in multiple independent study populations (1-9). These risk variants, although only moderately associated with prostate cancer risk individually, have a stronger cumulative association with prostate cancer risk (10-11). In addition, most of these risk variants are located between genes or in genes not previously considered as important in prostate carcinogenesis; thus they may provide novel insight into disease etiology. It is anticipated that results from GWAS will lead to better prediction of prostate cancer for earlier detection and a better understanding of the molecular mechanisms of this disease.
However, these identified prostate cancer risk variants only account for a small fraction of the genetic variance for prostate cancer risk. For example, two highly significant and independent prostate cancer risk SNPs at 17q12, rs4430796 and rs11649743 were estimated to account for only 0.5% and 0.3% of the total genetic variance in Sweden, respectively (12). Additional prostate cancer risk variants in the genome may exist. The goal of this study is to search for these risk variants using existing data from two GWAS: one from our study in a Swedish population (CAPS) and the other from the publicly available data of the National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) Study.
To increase the likelihood of identifying risk variants that are associated with more clinically relevant, aggressive prostate cancer, we focus on comparisons between control subjects and prostate cancer patients with non-organ confined and/or high grade disease (6). Different from our previous study where only ∼60,000 single nucleotide polymorphisms (SNPs) that overlapped between the two genotyping platforms (Affymetrix 500K arrays and Illumina 317K and 240K) of the two studies were analyzed, we are now able to examine ∼2.5 million SNPs in these two studies by imputing nongenotyped SNPs from flanking genotyped SNPs. In this report, we describe a novel prostate cancer risk associated SNP that was discovered from the two GWAS and confirmed in five independent study populations.
Materials and Methods
Subjects in the two GWAS of aggressive prostate cancer
The first GWAS included 498 aggressive prostate cancer patients and 494 control subjects that matched the age distribution of case subjects from CAPS (CAncer of the Prostate in Sweden), a population-based case-control study in Sweden (6). The 498 patients with aggressive prostate cancer met at least one of the following criteria based on the biopsy specimen: clinical stage T3/T4, N+, M+, Differential Grade III, Gleason Score ≥ 8, or pre-operative serum PSA ≥ 50 ng/mL. In the second GWAS, we analyzed the data from 737 patients with aggressive prostate cancer, defined as clinical stage T3/T4 or Gleason Score ≥7 based on biopsy specimen, and 1,105 age-matched control subjects, all from the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial of the first stage CGEMS study (4,7). Individual genotype data were obtained through an approved data request application. All of the subjects selected from CGEMS were of European ancestry.
Subjects in the six confirmation study populations
The first confirmation study population consisted of the remaining subjects from CAPS, including 733 aggressive cases, 1,619 non-aggressive cases, and 1,287 controls from Sweden. The second study population was a hospital-based prostate cancer case-control study of European Americans from Johns Hopkins Hospital (JHH) (13). It included 983 aggressive cases, 527 non-aggressive cases, and 482 controls. Tumors with a Gleason score of 7 or higher or stage pT3 or higher or N+ or M1 (i.e., either high-grade or non— organ-confined disease) were defined as aggressive disease. The other four study populations were from the second stage of the CGEMS study. They were the American Cancer Society Cancer Prevention Study II (CPS-II); the Health Professionals Follow-up Study (HPFS); CeRePP French Prostate Case-Control Study (FPCC); and Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (7). Summary genotype information from these four populations was downloaded from a public CGEMS website http://cgems.cancer.gov/data/. The number of subjects in each of these study populations is described in Table 1.
Table 1.
Number of study subjects in each study population Cases
| Cases |
||||
|---|---|---|---|---|
| Study | Non-aggressive | Aggressive | Unclassified | Controls |
| CAPS | 1619 | 1231 | 49 | 1722 |
| JHH | 527 | 982 | 18 | 482 |
| ATBC | 516 | 240 | 184 | 940 |
| FPCC | 0 | 671 | 0 | 671 |
| HPFS | 405 | 123 | 91 | 620 |
| PLCO | 489 | 691 | 0 | 1105 |
| ACS | 699 | 926 | 165 | 1797 |
| ALL | 4255 | 4864 | 507 | 7337 |
Genotyping for the GWAS
Methods for the GWAS in CAPS were described in detail elsewhere (6). Briefly, the GeneChip Human Mapping 500K Array Set, supplemented with 30K genomic fill-in SNPs and 20K nonsynonymous SNPs from Affymetrix Inc. (Santa Clara, CA) were genotyped in 498 aggressive prostate cancer cases and 494 controls in CAPS. The BRLMM algorithm from Affymetrix was used to make genotype calls, and the average genotyping call rate was 99.1%. Genotype concordance for the positive controls was > 99%. The GWAS in CGEMS was performed using HumanHap300 and HumanHap240 assays from Illumina Corp. (San Diego, CA).
Genotyping for the confirmation study
A subset of SNPs was genotyped using the MassArray System from Sequenom (San Diego, CA). Polymerase chain reaction (PCR) and extension primers for these SNPs were designed using the MassARRAY Assay Design 3.0 software. PCR and extension reactions were performed according to the manufacturer’s instructions, and extension product sizes were determined by mass spectrometry using the Sequenom iPLEX system. Duplicates and water samples, to which the technician was blind, were included in each 96-well plate as PCR negative controls. The genotype call rates of these SNPs were > 98% and the average concordance rate between samples was >99%.
Statistical methods
A Hardy-Weinberg equilibrium test was performed using the Fisher’s exact test. We imputed all of the known SNPs in the genome based on the genotyped SNPs and haplotype information in the HapMap Phase II data (CEU) using a computer program, IMPUTE (14). A posterior probability of 0.9 was used as a threshold to call genotypes. Allele frequency differences between case patients and control subjects were tested for each SNP, using a chi-square test with 1 degree of freedom. The allelic odds ratio (OR) and 95% confidence interval (95% CI) were estimated based on a multiplicative model. Results from multiple case-control populations were combined using a Mantel-Haenszel model in which the populations were allowed to have different population frequencies for alleles but were assumed to have a common OR. The homogeneity of ORs among different study populations was tested using Breslow-Day chi-square test.
Results and discussion
We imputed ∼2 million SNPs across the genome among each of the two GWAS based on the genotyping data from Affymetrix 500K SNP arrays (CAPS) and Illumina 317K and 240K (CGEMS). Some quality related information on the imputed SNPs is presented in Supplementary data. Differences in the allele frequency of each genotyped and imputed SNP between aggressive prostate cancer cases and unaffected controls were tested separately in CAPS and CGEMS. A combined allelic test of these two studies was then performed for each SNP using a Mantel-Haenszel method. A set of criteria were used to select SNPs that are most likely associated with aggressive prostate cancer: 1) P < 10−5 in the combined allelic tests, 2) the direction of association is consistent in both studies, 3) the difference in allele frequencies between cases and controls is ≥ 0.02 in both studies 4) the genotype frequencies do not significantly deviate from Hardy-Weinberg equilibrium (P > 0.05) in control subjects in both studies, 5) the minor allele frequency is ≥ 0.01 in controls in both studies, 6) the call rate is > 90% for imputed SNPs in both studies, and 7) if multiple SNPs in a haplotype block met the above six criteria, we selected the most significant SNP. Twenty-one independent SNPs were found to meet these criteria (Table 2).
Table 2.
Independent SNPs that are associated with aggressive prostate cancer in CAPS and CGEMS
| CAPS-GWAS |
CGEMS |
M-H |
CAPS-500/500 |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | Position | Allele | Aggressive cases (N=498) |
Controls (N=494) |
P | OR | Type of SNPs |
% of missing data |
Aggressive cases (N=983) |
Controls (N=527) |
P | OR | Type of SNPs |
% of missing data |
P_COMBINED | Aggressive cases (N=500) |
Controls (N=500) |
P | OR |
| 1 | rs11580380 | 102,371,869 | A | 0.11 | 0.09 | 0.08251 | 1.34 | Imputed | 0.02 | 0.11 | 0.07 | 8.99E-05 | 1.58 | Imputed | 0.01 | 9.51E-05 | 0.09 | 0.12 | 0.077 | 0.77 |
| 4 | rs9684740 | 6,066,865 | A | 0.34 | 0.44 | 1.63E-05 | 0.65 | Imputed | 0.00 | 0.37 | 0.39 | 0.1283 | 0.90 | Imputed | 0.03 | 2.94E-05 | 0.40 | 0.38 | 0.582 | 1.05 |
| 4 | rs4466060 | 131,939,926 | C | 0.18 | 0.15 | 0.1065 | 1.25 | Imputed | 0.07 | 0.20 | 0.15 | 4.30E-05 | 1.45 | Imputed | 0.00 | 6.08E-05 | ||||
| 4 | rs10032223 | 190,115,687 | T | 0.23 | 0.15 | 4.05E-05 | 1.68 | Imputed | 0.01 | 0.21 | 0.19 | 0.1466 | 1.13 | Imputed | 0.02 | 7.73E-05 | 0.18 | 0.20 | 0.098 | 0.90 |
| 5 | rs2548035 | 33,853,908 | C | 0.39 | 0.49 | 2.47E-05 | 0.66 | Imputed | 0.02 | 0.45 | 0.47 | 0.1531 | 0.91 | Imputed | 0.00 | 5.11E-05 | 0.47 | 0.45 | 1.097 | 1.13 |
| 5 | rs6580317 | 143,752,911 | C | 0.19 | 0.23 | 0.02985 | 0.76 | Genotyped | 0.09 | 0.19 | 0.25 | 1.15E-04 | 0.71 | Imputed | 0.09 | 4.66E-05 | 0.25 | 0.24 | 1.074 | 0.98 |
| 5 | rs4448013 | 162,585,129 | T | 0.15 | 0.19 | 0.01802 | 0.73 | Imputed | 0.02 | 0.15 | 0.20 | 3.47E-04 | 0.71 | Genotyped | 0.05 | 8.12E-05 | 0.14 | 0.13 | 1.163 | 0.98 |
| 7 | rs221770 | 99,946,745 | A | 0.17 | 0.10 | 9.75E-05 | 1.77 | Genotyped | 0.04 | 0.13 | 0.09 | 0.00331 | 1.38 | Imputed | 0.00 | 5.15E-06 | 0.13 | 0.15 | 0.859 | 0.76 |
| 9 | rs1535453 | 5,400,624 | C | 0.05 | 0.02 | 1.84E-04 | 3.04 | Imputed | 0.06 | 0.05 | 0.03 | 0.02826 | 1.46 | Genotyped | 0.04 | 6.84E-05 | 0.07 | 0.06 | 0.37 | 1.10 |
| 9 | rs4559327 | 38,727,017 | C | 0.36 | 0.32 | 0.1135 | 1.19 | Imputed | 0.08 | 0.35 | 0.28 | 4.16E-05 | 1.37 | Imputed | 0.08 | 6.27E-05 | ||||
| 9 | rs10123553 | 89,934,974 | C | 0.40 | 0.50 | 7.57E-05 | 0.68 | Imputed | 0.00 | 0.43 | 0.46 | 0.07177 | 0.88 | Imputed | 0.01 | 7.13E-05 | 0.47 | 0.46 | 0.735 | 1.06 |
| 10 | rs1335540 | 26,767,244 | T | 0.28 | 0.37 | 1.85E-04 | 0.66 | Imputed | 0.10 | 0.35 | 0.39 | 0.01842 | 0.84 | Imputed | 0.06 | 4.64E-05 | 0.36 | 0.36 | 0.94 | 1.00 |
| 10 | rs7913838 | 36,639,228 | C | 0.07 | 0.04 | 0.00328 | 1.97 | Imputed | 0.01 | 0.08 | 0.06 | 0.00182 | 1.53 | Imputed | 0.04 | 7.76E-05 | 0.07 | 0.06 | 0.615 | 1.03 |
| 12 | rs4576883 | 118,152,179 | G | 0.10 | 0.15 | 0.00488 | 0.65 | Imputed | 0.00 | 0.10 | 0.14 | 0.00106 | 0.70 | Imputed | 0.01 | 6.79E-05 | 0.20 | 0.23 | 0.132 | 0.93 |
| 13 | rs7327461 | 39,961,028 | G | 0.19 | 0.12 | 5.20E-04 | 1.63 | Imputed | 0.06 | 0.17 | 0.14 | 0.01364 | 1.28 | Genotyped | 0.07 | 9.11E-05 | 0.19 | 0.17 | 0.503 | 0.97 |
| 14 | rs2332389 | 70,032,772 | T | 0.09 | 0.07 | 0.1694 | 1.29 | Imputed | 0.01 | 0.14 | 0.09 | 6.61E-06 | 1.63 | Imputed | 0.00 | 1.65E-05 | ||||
| 14 | rs17127296 | 90,908,001 | A | 0.06 | 0.04 | 0.02855 | 1.70 | Imputed | 0.09 | 0.06 | 0.04 | 2.45E-04 | 1.77 | Imputed | 0.01 | 9.00E-05 | 0.04 | 0.04 | 0.844 | 1.08 |
| 16 | rs743672 | 13,814,689 | G | 0.05 | 0.02 | 0.00791 | 2.05 | Imputed | 0.00 | 0.08 | 0.05 | 7.28E-05 | 1.77 | Genotyped | 0.01 | 8.84E-06 | 0.06 | 0.04 | 0.137 | 1.00 |
| 20 | rs7271186 | 37,291,509 | A | 0.09 | 0.05 | 0.00235 | 1.82 | Imputed | 0.05 | 0.09 | 0.06 | 0.0013 | 1.52 | Genotyped | 0.01 | 4.16E-05 | 0.08 | 0.08 | 0.314 | 0.89 |
| 21 | rs6586243 | 42,109,475 | T | 0.06 | 0.10 | 9.45E-04 | 0.54 | Imputed | 0.06 | 0.08 | 0.12 | 5.32E-04 | 0.66 | Imputed | 0.06 | 7.79E-06 | 0.13 | 0.12 | 0.905 | 1.02 |
| 22 | rs7291691 | 38,778,569 | G | 0.24 | 0.19 | 0.01037 | 1.37 | Imputed | 0.04 | 0.25 | 0.20 | 4.05E-04 | 1.34 | Imputed | 0.01 | 5.61E-05 | 0.23 | 0.19 | 0.043 | 1.25 |
As a first step of confirmation, we genotyped 18 SNPs among an additional 500 aggressive cases and 500 controls from CAPS (three SNPs were not genotyped because they could not be grouped into a single multiplex iPLEX group). One SNP, rs7291691 at 22q13, was confirmed using an allelic test, with a nominal P of 0.04 (Table 2). The direction of association was the same as in the two GWAS, i.e., the risk allele found in the GWAS was more frequent in aggressive cases than in controls in the confirmation study. As an additional attempt at confirmation, we genotyped this SNP in the remaining 233 aggressive cases and 726 controls of CAPS and in 982 aggressive cases and 482 control subjects of European descent from Johns Hopkins Hospital (JHH). Again, the risk allele was consistently more common in aggressive cases than controls in both groups. The difference was significant in JHH (nominal P = 0.02) but not significant in the remainder of CAPS, likely due to small sample size. The genotype counts of SNP rs7291691 in CAPS and JHH are presented in Supplementary Table 2.
To obtain additional evidence for the association from other study populations, we searched the publicly available data of four study populations from the second stage of the CGEMS study, including CPS-II, HPFS, FPCC, and ATBC. The SNP rs7291691 was not genotyped in these populations. However, a SNP (rs9623117) that is less than 2 kb from rs7291691 was genotyped. To evaluate LD between the two SNPs, we typed this new SNP (rs9623117) in all aggressive cases and controls of CAPS and JHH. These two SNPs are in strong LD, with an estimated pair-wise r2 of 0.96 and 0.98 in the control subjects of CAPS and JHH, respectively. Consequently, the SNP rs9623117 was also significantly associated with aggressive prostate cancer in CAPS (P = 0.02) and in JHH (P = 0.01) (Table 3a). The allele ‘C’ was more common in aggressive cases than in controls. Similarly, this SNP (directly genotyped) was also significantly associated with aggressive prostate cancer (P = 6.6 × 10−4) in the GWAS of CGEMS (PLCO). For the four independent study populations of the second CGEMS study, the allele ‘C’ was more common in aggressive cases than in controls in each population (Table 3a). The difference was statistically significant in ATBC (P = 0.02) and FPCC (P = 0.04). Remarkably, the frequency of allele ‘C’ was consistently higher in aggressive cases than in controls in each of the seven study populations examined. The combined allelic test in the seven populations was highly significant, with a P = 5.9 × 10−7. The Odds Ratio (OR) of allele ‘C’ for aggressive prostate cancer was estimated to be 1.18 (95% CI: 1.11-1.26).
Table 3.
Association of rsrs9623117 at 22q13 with prostate cancer risk
| Risk | Genotype counts (TT, TC, CC) |
Allele frequency |
Allelic OR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Allele | Cases | Controls | Cases | Controls | P* | P^ | OR | 95% CI | ||||
| a. Aggressive disease | |||||||||||||
| CAPS | C | 745 | 411 | 67 | 1080 | 535 | 63 | 0.22 | 0.20 | 0.016 | |||
| JHH | C | 523 | 365 | 49 | 287 | 161 | 14 | 0.25 | 0.20 | 0.01 | |||
| ATBC | C | 145 | 81 | 12 | 638 | 246 | 36 | 0.22 | 0.17 | 0.02 | |||
| FPCC | C | 369 | 244 | 43 | 409 | 207 | 39 | 0.25 | 0.22 | 0.04 | |||
| HPFS | C | 68 | 42 | 8 | 360 | 224 | 26 | 0.25 | 0.23 | 0.51 | |||
| PLCO | C | 384 | 260 | 43 | 684 | 385 | 31 | 0.25 | 0.20 | 6.60E-04 | |||
| ACS | C | 527 | 337 | 49 | 1034 | 648 | 93 | 0.24 | 0.23 | 0.79 | |||
| ALL | C | 2751 | 1735 | 269 | 4492 | 2406 | 302 | 0.24 | 0.21 | 4.96E-07 | 0.19 | 1.18 | 1.11-1.26 |
| b. Non-aggressive disease | |||||||||||||
| CAPS | C | 970 | 570 | 73 | 1080 | 535 | 63 | 0.22 | 0.20 | 0.01 | |||
| JHH | C | 287 | 203 | 22 | 287 | 161 | 14 | 0.24 | 0.20 | 0.05 | |||
| ATBC | C | 345 | 144 | 20 | 638 | 246 | 36 | 0.18 | 0.17 | 0.59 | |||
| FPCC | C | 0 | 0 | 0 | 409 | 207 | 39 | NA | 0.22 | NA | |||
| HPFS | C | 217 | 148 | 24 | 360 | 224 | 26 | 0.25 | 0.23 | 0.19 | |||
| PLCO | C | 293 | 174 | 21 | 684 | 385 | 31 | 0.22 | 0.20 | 0.25 | |||
| ACS | C | 393 | 264 | 25 | 1034 | 648 | 93 | 0.23 | 0.23 | 0.73 | |||
| ALL | C | 2486 | 1490 | 186 | 4492 | 2406 | 302 | 0.22 | 0.21 | 0.004 | 0.43 | 1.11 | 1.03-1.18 |
We also tested association of the SNP rs9623117 with non-aggressive prostate cancer. A trend of higher frequency of allele ‘C’ in non-aggressive cases than in controls was observed in all six populations (no non-aggressive cases in FPCC) (Table 3b), although a significant difference was only observed in two study populations (CAPS and JHH). The combined allelic test of the seven populations was significant at P = 0.003. The OR of allele ‘C’ for non-aggressive prostate cancer was estimated to be 1.11 (95% CI: 1.04-1.19). Although the frequency of allele ‘C’ was lower in non-aggressive cases than aggressive cases in most study populations, the difference was not statistically significant in the combined analysis, P = 0.11. Similarly, this SNP was not associated with Gleason score, pre-operative PSA, and age at diagnosis in CAPS or JHH.
The SNP rs9623117 was not significantly associated with plasma PSA levels in control subjects of CAPS (P = 0.91) and JHH (P = 0.68) using an additive model. For example, the mean PSA levels were 1.56, 1.58, and 1.57 ng/mL for men who had 0, 1, or 2 copies of the ‘C’ allele, respectively, among 1,722 control subjects in CAPS. Therefore, the association between rs9623117 at 22q13 and prostate cancer risk is unlikely to be confounded by PSA screening.
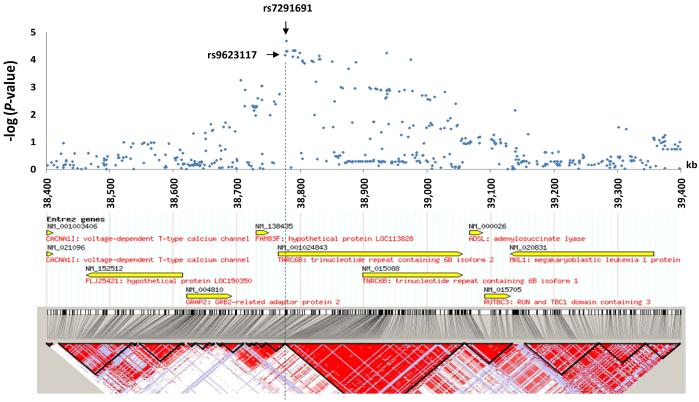
To explore the region of prostate cancer association at 22q13, we performed allelic tests for aggressive prostate cancer risk with all the genotyped and imputed SNPs in the 1 Mb-region flanking the SNP rs9623117 (38.4-39.4 Mb) based on the GWAS data from CGEMS and CAPS. Figure 1 presents the results of the combined test in these two populations using a Mantel-Haenszel model. Multiple SNPs in the region between 38.77-38.97 kb were strongly associated with aggressive prostate cancer risk; the strongest one was rs7291691. These most significant SNPs were within a large haplotype block and within the TNRC6B gene. TNRC6B (trinucleotide repeat-containing gene 6b), a RNA recognition motif (RRM)-containing protein, was localized to the mRNA-degrading cytoplasmic P bodies, and is functionally required to mediate miRNA-guided mRNA cleavage (15). TNRC6B is expressed in many normal tissues, including the prostate. In addition, TNRC6B expression was suppressed in hormone refractory metastatic prostate cancer compared to prostate carcinoma (http://www.oncomine.org). Alterations in TNRC6B gene expression due to genetic variations might perturb the levels of mRNA species normally under its control and therefore contribute to carcinogenesis. Additional fine-mapping efforts to cover the promoter region of TNRC6B are warranted.
Figure 1.
A schematic view of genetic association between SNPs at 22q13 and prostate cancer risk. (Upper panel) Combined allele tests for SNPs at 22q13 (38.4-39.4 Mb) and aggressive prostate cancer risk based on two genome-wide association studies in CAPS and CGEMS. (Lower panel) Known genes and inferred haplotype blocks of SNPs at this region based on the CEU study population HapMap data.
It is also noted that the prostate cancer association at 22q13 is also within the interval of a prostate cancer linkage at 22q12.3 reported previously in families of five or more members affected with prostate cancer (16). It is, however, ∼4 Mb telomeric to the smaller (∼2.5 Mb) consensus linkage region reported in two recent studies (17-18). Additional studies are needed to test whether this novel locus at 22q13 contributes to the observed prostate cancer linkage.
In summary, a novel prostate cancer risk locus at 22q13 was discovered in two genome-wide association studies and confirmed in six additional study populations. Our study demonstrates an efficient approach to utilize publicly available data in improving the statistical power to identify risk variants that are moderately associated with disease risk. However, there are several important caveats for the prostate cancer association at 22q13. First, although the P–value of 5.9 × 10−7 for an association between aggressive prostate cancer and rs9623117 at 22q13 in the combined analysis is highly significant, it did not reach a genome-wide significance level. A nominal P-value of 2 × 10−8 is needed to ensure a 5% study-wide false positive rate, adjusting for a total of 2.5 million tests. Therefore, additional confirmation studies are needed. Second, our study is under-powered to discover risk variants of moderate effect because of the small sample size of the GWAS in CAPS and the reliance on imputed SNPs for most of the SNPs examined. Larger sample size and higher resolution (increased number of SNPs) in GWAS are needed to reduce false negative findings. Finally, even with an effort to limit cases to aggressive prostate cancer in the GWAS, the identified novel risk variant at 22q13 was not uniquely associated with aggressive prostate cancer. This result is similar to all the prostate cancer risk variants recently identified from GWAS, including at 8q24, 17q12, 17q24, 10q11, and 11q13 (19). Considering that only a small fraction of PSA-detected prostate cancer progresses is life-threatening, it is most important to identify genetic variants that can be used to identify men at greatest risk for aggressive disease. Other study designs, including those comparing aggressive with non-aggressive prostate cancer should be used to achieve this goal.
Supplementary Material
Acknowledgements
The study is supported by National Cancer Institute CA105055, CA106523, and CA95052 to J.X., CA112517 and CA58236 to W.B.I., CA86323 to AWP, Department of Defense grant PC051264 to J.X, Swedish Cancer Society (Cancerfonden) to HG and Swedish Academy of Sciences (Vetenskapsrådet) to HG. The support of Kevin P Jaffe to W.B.I is gratefully acknowledged.
The authors thank all the study subjects who participated in the CAPS study and urologists who included their patients in the CAPS study. We acknowledge the contribution of multiple physicians and researchers in designing and recruiting study subjects, including Dr. Hans-Olov Adami (for CAPS) and Drs. Bruce J. Trock, Alan W. Partin, and Patrick C. Walsh (for JHH).
The authors also thanks for the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) for making the data available publicly.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 6.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 7.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 10.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Chang B-L, Hsu F-C, Isaacs SD, Wiley KE, Zhu Y, Wiklund F, Statin P, Tao L, Liu W, Duggan D, Carpten JC, Trock BJ, Walsh PC, Grönberg H, Isaacs WB, Xu J, Zheng SL. Cumulative effect of five genetic variants on prostate cancer risk in multiple study populations. Prostate. 2008;68:1257–62. doi: 10.1002/pros.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, Gao Z, Hsu FC, Kim ST, Liu W, Zhu Y, Stattin P, Adami H-O, Wiley KE, Dimitrov L, Sun J, Li T, Turner AR, Adams TS, Adolfsson J, Johansson JE, Lowey J, Trock BJ, Partin AW, Walsh PC, Trent JM, Duggan D, Carpten J, Chang B-L, Grönberg H, Isaacs WB, Xu J. Evidence for two independent prostate cancer risk associated loci in the HNF1B gene at 17q12. Nat Genet. 2008 doi: 10.1038/ng.214. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. JNCI. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 14.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–55. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Dimitrov L, Chang BL, et al. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am J Hum Genet. 2005;77:219–29. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp NJ, Cannon-Albright LA, Farnham JM, et al. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16:1271–8. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johanneson B, McDonnell SK, Karyadi DM, et al. Fine mapping of familial prostate cancer families narrows the interval for a susceptibility locus on chromosome 22q12.3 to 1.36 Mb. Hum Genet. 2008;123:65–75. doi: 10.1007/s00439-007-0451-y. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Isaacs SD, Sun J, Li G, Wiley KE, Zhu Y, Hsu FC, Wiklund F, Turner AR, Adams TS, Liu W, Trock BJ, Partin AW, Chang B-L, Walsh PC, Grönberg H, Isaacs WB, Zheng SL. Association of prostate cancer risk variants with clinicopathologic characteristics of the disease. Clinical Cancer Research. 2008 doi: 10.1158/1078-0432.CCR-08-0934. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.