Abstract
Resistance to the adamantane class of antiviral drugs by human A/H3N2 influenza viruses currently exceeds 90% in the United States and multiple Asian countries. Adamantane resistance is associated with a single amino acid change (S31N) in the M2 protein, which was shown to rapidly disseminate globally in 2005 in association with a genome reassortment event. However, the exact origin of influenza A/H3N2 viruses carrying the S31N mutation has not been characterized, particularly in South-East Asia. We therefore conducted a phylogenetic analysis of the HA, NA, and M1/2 segments of viral isolates collected between 1997 and 2007 from temperate localities in the Northern hemisphere (New York State, United States, 492 isolates) and Southern hemisphere (New Zealand and Australia, 629 isolates) and a subtropical locality in South-East Asia (Hong Kong, 281 isolates). We find that although the S31N mutation was independently introduced at least 11 times, the vast majority of resistant viruses now circulating globally descend from a single introduction that was first detected in the summer of 2003 in Hong Kong. These resistant viruses were continually detected in Hong Kong throughout 2003–2005, acquired a novel HA through reassortment during the first part of 2005, and thereafter spread globally. The emergence and persistence of adamantane resistant viruses in Hong Kong further supports a source-sink model of global influenza virus ecology, in which South-East Asia experiences continuous viral activity and repeatedly seeds epidemics in temperate areas.
Keywords: Human influenza A virus, Evolution, Adamantane resistance, Reassortment, Global migration
Introduction
After four decades of effective use of the adamantane class of antiviral drugs in the prophylaxis and treatment of influenza, global resistance to these drugs has increased dramatically among influenza viruses of the A/H3N2 subtype in recent years. In the vast majority of cases, the basis for resistance is a single Ser to Asn amino acid replacement (S31N) in the matrix M2 ion channel, which interferes with the drug's ability to block M2 ion channel activity and viral replication (Wang et al., 1993, Aoki, 1998). Whereas < 2% of A/H3N2 influenza viruses sampled globally were resistant to adamantane during 1995–2002, resistance was detected among 12.3% of viruses collected in 2004 (Bright et al., 2005). Sharp increases in adamantane resistance first appeared in Asia, particularly in China, where the prevalence of resistance increased from 8.2% in 2002 to 73.8% in 2004, and also in Hong Kong (69.8% resistance in 2004) (Bright et al., 2005). Recently, the S31N mutation was detected in 100% of influenza viruses sampled from multiple Asian countries, although sample sizes were relatively small (Deyde et al., 2007). In the United States, adamantane resistant influenza viruses emerged somewhat later, with prevalence increasing from < 2% in 2003 to 11% in 2004–2005, and over 90% during the 2005–2006 epidemic (Bright et al., 2005, Bright et al., 2006). Since 2006 the USA Centers for Disease Control and Prevention (CDC) has recommended the cessation of use of adamantanes for treatment or prophylaxis of influenza (CDC 2006).
Our understanding of the evolutionary basis for this dramatic global increase in adamantane resistant influenza viruses remains incomplete, particularly the role, if any, of direct selection for drug resistance. Although mutations conferring drug resistance emerge readily when adamantanes are administered to control influenza outbreaks in nursing homes and long-term care facilities (Shiraishi et al., 2003, Suzuki et al., 2003), resistant strains typically persist only transiently. Accordingly, the prevalence of resistance to adamantane at a population level has remained low for many decades (< 2%), despite periods of significant adamantane usage in certain areas, such as Japan in the late 1990s (Ziegler et al., 1999, Masuda et al., 2000).
The rise of adamantane resistance in countries where the use of adamantane drugs has not increased in recent years, such as in the United States and Australia, implicates evolutionary mechanisms other than local drug selection pressure (Simonsen et al., 2007, Barr et al., 2007a). In particular, it has been proposed that the increased global frequency of the S31N mutation in the M2 protein resulted from a chance hitchhiking event involving linkage to advantageous mutations in other segments following a major 4 + 4 reassortment event, including mutations in HA1 at positions 193 and 225 near the receptor-binding site (Simonsen et al., 2007). This reassortment event, which most likely occurred in early 2005, generated a new global lineage of adamantane resistant A/H3N2 viruses, termed the ‘N-lineage’, characterized by an antigenically A/Wisconsin/67/2005-like HA and an M2 protein bearing the S31N replacement (Simonsen et al., 2007). Further characterization of influenza viruses circulating globally identified additional independent introductions of resistant A/H3N2 influenza viruses that emerged prior to the emergence of the ‘N-lineage’, but which did not spread globally (Deyde et al., 2007). However, the specific roles of local evolution, migration, and reassortment in the genesis of adamantane resistance in years prior to the global emergence of the ‘N-lineage’ are unknown.
Although reassortment and genomic hitchhiking appear to be important in the global dissemination of A/H3N2 adamantane resistant viruses to North America and Oceania, it is possible that localized direct selection pressure for drug resistance underlies the initial emergence of adamantane resistance in South-East Asia. In particular, the over-the-counter use of adamantane drugs may have increased in some areas, including China, possibly in association with the recent outbreaks of severe acute respiratory syndrome (SARS) and H5N1 avian influenza virus (Bright et al., 2006, Deyde et al., 2007, Hayden, 2006). Furthermore, it has been proposed that South-East Asia serves as an epicenter for continuous influenza virus activity, with antigenic variants emerging in tropical South-East Asian countries before disseminating globally (Russell et al., 2008). According to the ‘source-sink’ model of influenza A virus evolution, tropical regions in general support continual viral activity, greater genetic diversity, and consequently stronger natural selection, whereas epidemics in temperate areas repeatedly die out due to strong seasonal bottlenecks and require continual re-seeding (Rambaut et al., 2008, Nelson et al., 2007, Nelson et al., 2006, Tang et al., 2008a). Overall, the seasonality of influenza in the tropics is less defined than in temperate regions but in some areas is associated with high rainfall (Shek and Lee 2003).
The city of Hong Kong has a subtropical climate with a semiannual pattern of influenza virus activity, with one peak in winter and a second peak in late spring or early summer, although in some years only one peak occurs (Wong et al., 2006, Yang et al., 2008). Given the early detection of adamantane resistant viruses in China and Hong Kong (Bright et al., 2005) and the prominence of South-East Asia in global influenza virus evolution (Russell et al., 2008), we sought to investigate the specific role of Hong Kong in the origin and rapid global dissemination of adamantane resistant viruses between 1997 and 2007. As drug selection pressure alone inadequately explains the global domination of adamantane resistant viruses in temperate countries where adamantane usage has not increased in recent years, we explored how the interaction between local evolution, genomic reassortment, and ongoing antigenic change more completely explains the rapid global dissemination of adamantane resistance. In particular, to understand the global emergence of the S31N adamantane resistance mutation within the context of antigenic evolution, we conducted a phylogenetic analysis of influenza virus sequence data for the HA, M1/2, and NA segments that was obtained through ten years of sampling in both subtropical and temperate regions.
Results
Of the 1402 viral isolates sampled between 1997 and 2007 that are included in this analysis, 119 (8.5%) exhibited the S31N mutation in the M2 protein conferring resistance to adamantane antiviral drugs. In Australia and New Zealand, 6.2% (39 / 629) of isolates were resistant to adamantane drugs, 4.5% (22 / 492) of the isolates from New York State, USA were adamantane resistant, and 20.6% (58 / 281) of Hong Kong isolates were resistant (Supplementary Tables 1–3). In the years prior to the global proliferation of adamantane resistance viruses associated the emergence of the ‘N-lineage’ in early 2005 (Simonsen et al., 2007), the overall prevalence of adamantane resistance in our data set was 3.0% (38 / 1250): 11.0% (27 / 246) in Hong Kong, 0.6% (3 / 533) in New Zealand/Australia, and 1.7% (8 / 471) in New York State. Following the emergence of the ‘N-lineage’ in 2005, the prevalence of resistance increased to 53.3% (81 / 119): 88.6% (31 / 35) in Hong Kong, 66.7% (14 / 21) in New York, and 37.5% (36 / 96) in Australia and New Zealand. The prevalence of resistance is somewhat lower in New Zealand due to the paucity of isolates from 2006 to 2007.
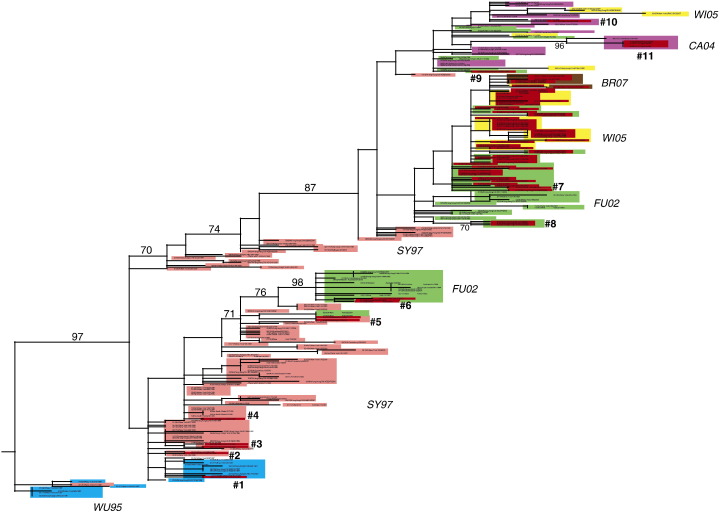
Phylogenetic analysis of the M1/2 segment of A/H3N2 influenza virus isolates collected from Hong Kong, New York State, Australia, and New Zealand reveals at least 11 independent introductions (i.e., 11 separate clades) of the S31N mutation between the years 1997 and 2007 (Fig. 1 , Table 1 ). This estimate is conservative, as clusters of genetically related resistant isolates are regarded as a single introduction of the S31N replacement even though it is possible (although less parsimonious) that the mutation arose independently among these isolates rather than through viral transmission.
Fig. 1.
Phylogenetic relationships of the M1/2 segment of 226 A/H3N2 influenza viruses sampled from Hong Kong (n = 108), New York State, USA (n = 72), and New Zealand and Australia (n = 46) between 1997 and 2007, estimated using an ML method. Isolates are shaded by antigenic characteristics (inferred from HA tree): A/Wuhan/359/1995-like (WU95) isolates are blue, A/Sydney/5/1997-like (SY97) isolates are pink, A/Fujian/411/2002-like (FU02) isolates are green, A/California/07/2004-like (CA04) isolates are purple, A/Wisconsin/67/2005-like (WI05) isolates are yellow, and A/Brisbane/10/2007-like (BR07) isolates are brown. In addition, isolates bearing the S31N mutation are shaded in red. Independent introductions of the S31N replacement are numbered #1–#11. Bootstrap values (> 70%) are shown for key nodes. The tree is rooted by the oldest isolate (A/New York/564/1997, 01/02/1997), and all horizontal branch lengths are drawn to scale.
Table 1.
Isolate name, location and date of collection, patient age, and antigenic characterization of 71 adamantane resistant A/H3N2 influenza viruses that are associated with Introductions #1–11 of the S31N mutation depicted in Fig. 1.
| Introduction | Isolate name | Location | Patient age | Date of collection | Antigenicity |
|---|---|---|---|---|---|
| #1 | A/Hong Kong/CUHK4245/1997 | HK | 1–10 y | 03/14/1997 | A/Wuhan/1995-like |
| #2 | A/New York/512/1998 | NYS | 80 y | 01/29/1998 | A/Sydney/5/1997-like |
| #3 | A/New York/315/1999 | NYS | 31 y | 01/06/1999 | A/Sydney/5/1997-like |
| A/New York/324/1999 | NYS | 84 y | 01/26/1999 | A/Sydney/5/1997-like | |
| #4 | A/New York/280/1999 | NYS | 80 y | 04/12/1999 | A/Sydney/5/1997-like |
| #5 | A/Wellington/71/2002 | AUS/NZ | 62 y | 07/15/2002 | A/Sydney/5/1997-like |
| A/Wellington/79/2002 | AUS/NZ | 11 y | 08/20/2002 | A/Sydney/5/1997-like | |
| #6 | A/Queensland/34/2003 | AUS/NZ | 11 mo | 08/25/2003 | A/Fujian/411/2002-like |
| A/New York/1/2003 | NYS | 21 y | 10/28/2003 | A/Fujian/411/2002-like | |
| A/New York/31/2004 | NYS | 66 y | 01/05/2004 | A/Fujian/411/2002-like | |
| #7 | A/Hong Kong/CUHK50895/2003 | HK | 1–10 y | 07/06/2003 | A/Fujian/411/2002-like |
| A/Hong Kong/CUHK51353/2003 | HK | 1–10 y | 07/18/2003 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52011/2003 | HK | 1–10 y | 08/05/2003 | A/Fujian/411/2002-like | |
| A/New York/204/2003 | NYS | 39 y | 08/16/2003 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52467/2003 | HK | 1–10 y | 08/18/2003 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52923/2003 | HK | 1–10 y | 08/26/2003 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK71313/2003 | HK | 1–10 y | 09/21/2003 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK24289/2004 | HK | 1–10 y | 04/02/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK33677/2004 | HK | 1–10 y | 05/04/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK33851/2004 | HK | 1–10 y | 05/08/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK35013/2004 | HK | 1–10 y | 06/01/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK50600/2004 | HK | 1–10 y | 07/06/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK50563/2004 | HK | 1–10 y | 07/06/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52274/2004 | HK | 1–10 y | 08/06/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52677/2004 | HK | 1–10 y | 08/13/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52863/2004 | HK | 1–10 y | 08/17/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK52941/2004 | HK | 1–10 y | 08/18/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK53726/2004 | HK | 1–10 y | 09/02/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK53766/2004 | HK | 1–10 y | 09/03/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK53732/2004 | HK | 1–10 y | 09/03/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK6422/2005 | HK | 1–10 y | 01/31/2005 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK6898/2005 | HK | 1–10 y | 02/12/2005 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK7221/2005 | HK | 1–10 y | 02/20/2005 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK7546/2005 | HK | 1–10 y | 02/27/2005 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK7711/2005 | HK | 1–10 y | 03/01/2005 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK7733/2005 | HK | 1–10 y | 03/02/2005 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK11949/2005 | HK | 1–10 y | 04/28/2005 | A/Wisconsin/67/05-like | |
| A/Canterbury/24/2005 | AUS/NZ | 54 y | 05/04/2005 | A/Wisconsin/67/05-like | |
| A/Otago/1/2005 | AUS/NZ | 28 y | 07/01/2005 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK42241/2005 | HK | 1–10 y | 07/02/2005 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK42644/2005 | HK | 1–10 y | 07/09/2005 | A/Wisconsin/67/05-like | |
| A/Queensland/51/2005 | AUS/NZ | 12 y | 07/12/2005 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK43751/2005 | HK | 1–10 y | 07/26/2005 | A/Wisconsin/67/05-like | |
| A/Canterbury/234/2005 | AUS/NZ | 42 y | 07/27/2005 | A/Wisconsin/67/05-like | |
| A/Western Australia/29/2005 | AUS/NZ | 1 y | 08/03/2005 | A/Wisconsin/67/05-like | |
| A/New York/923/2006 | NYS | 13 y | 02/23/2006 | A/Wisconsin/67/05-like | |
| A/New York/933/2006 | NYS | 17 y | 03/01/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK53004/2006 | HK | 1–10 y | 03/04/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK53005/2006 | HK | 1–10 y | 03/04/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK53798/2006 | HK | 1–10 y | 03/17/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK53907/2006 | HK | 1–10 y | 03/19/2006 | A/Wisconsin/67/05-like | |
| A/New York/6/2006 | NYS | 15 y | 04/06/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK60052/2006 | HK | 1–10 y | 06/29/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK64082/2006 | HK | 1–10 y | 09/10/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK65241/2006 | HK | 1–10 y | 09/28/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK66001/2006 | HK | 1–10 y | 10/13/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK45130/2006 | HK | 1–10 y | 12/04/2006 | A/Wisconsin/67/05-like | |
| A/Hong Kong/CUHK69904/2006 | HK | 1–10 y | 12/29/2006 | A/Wisconsin/67/05-like | |
| A/New York/UR06-0040/2007 | NYS | 45 y | 01/17/2007 | A/Brisbane/10/07-like | |
| A/New York/UR06-0437/2007 | NYS | 24 y | 02/27/2007 | A/Brisbane/10/07-like | |
| A/New York/UR06-0510/2007 | NYS | 10 y | 03/05/2007 | A/Wisconsin/67/05-like | |
| A/New York/UR06-0529/2007 | NYS | 34 y | 03/06/2007 | A/Brisbane/10/07-like | |
| A/New York/UR06-0607/2007 | NYS | 8 y | 03/14/2007 | A/Brisbane/10/07-like | |
| #8 | A/Hong Kong/CUHK13126/2004 | HK | 1–10 y | 02/02/2004 | A/Fujian/411/2002-like |
| A/Hong Kong/CUHK21164/2004 | HK | 1–10 y | 02/12/2004 | A/Fujian/411/2002-like | |
| A/Hong Kong/CUHK24289/2004 | HK | 1–10 y | 04/02/2004 | A/Fujian/411/2002-like | |
| #9 | A/Hong Kong/CUHK35011/2004 | HK | 1–10 y | 06/01/2004 | A/Fujian/411/2002-like |
| #10 | A/New York/391/2005 | NYS | 37 y | 01/07/2005 | A/California/7/04-like |
| #11 | A/Wellington/9/2005 | AUS/NZ | 29 y | 08/23/2005 | A/California/7/04-like |
| A/Otago/3/2005 | AUS/NZ | 19 y | 08/31/2005 | A/California/7/04-like | |
| A/Otago/4/2005 | AUS/NZ | 19 y | 08/31/2005 | A/California/7/04-like |
Viruses are sorted by isolation dates.
The first introduction of S31N that is observed in these data occurred in Hong Kong in 1997 (isolate A/Hong Kong/CUHK4245/1997, Introduction #1, Fig. 1, Table 1). Notably, this isolate is involved in a reassortment event, as A/Hong Kong/CUHK4245/1997 is related to a clade of A/Wuhan/359/1995-like isolates in the HA tree (shaded in blue, Fig. 2 ), but on the M1/2 and NA phylogenies this isolate is closely related to A/Sydney/5/97-like isolates (shaded in pink, Fig. 1, Fig. 3 ). Such phylogenetic incongruity suggests that a reassortment event involving the M1/2 and NA occurred prior to the jump between the Wuhan/95 and Sydney/97 antigenic clusters (Smith et al., 2004), such that A/Wuhan/359/1995-like viruses first acquired novel NA and M1/2 segments approximately 10–12 months before acquiring an antigenically divergent A/Sydney/5/1997-like HA.
Fig. 2.
Phylogenetic relationships of the HA gene segment of 231 A/H3N2 influenza viruses sampled from Hong Kong (n = 108), New York State, USA (n = 72), New Zealand and Australia (n = 46) between 1997 and 2007, and 5 influenza vaccine reference strains (A/Sydney/5/1997, A/Wyoming/3/2003 (A/Fujian/411/2002-like), A/California/7/2004, A/Wisconsin/67/2005, and A/Brisbane/10/2007), estimated using an ML method. Labels, shading, and rooting are the same as for Fig. 1, with Introductions #1–#11 referring to those identified in Fig. 1 and influenza vaccine reference strains shaded in dark green. Isolates labeled ‘#7F’ are associated with Introduction #7 and are A/Fujian/411/2002-like, and isolates labeled ‘7W’ are associated with Introduction #7 and are A/Wisconsin/67/2005-like.
Fig. 3.
Phylogenetic relationships of the NA gene segment of 226 A/H3N2 influenza viruses sampled from Hong Kong (n = 108), New York State, USA (n = 72), and New Zealand and Australia (n = 46) between 1997 and 2007, estimated using an ML method. Labels, shading, and rooting are the same as for Figs. 1 and 2, with Introductions #1–#11 referring to those identified in Fig. 1.
That antigenically similar isolates (based on the HA phylogeny) occupy divergent phylogenetic positions on the M1/2 and NA phylogenies indicates that reassortment involving these three segments has occurred on a regular basis over this decade of sampling (Fig. 1, Fig. 2, Fig. 3). Whereas the evolution of the HA segment is strongly linear and temporally structured (Fig. 2), reflecting the action of continual selection for antigenic novelty, multiple clades from the same time period occupy highly divergent phylogenetic positions and the NA and M1/2 trees (Fig. 1, Fig. 3), providing a clear signature of reassortment. For example, at least four distinct lineages of A/Sydney/5/1997-like isolates and four distinct lineages of A/Fujian/411/2002-like isolates co-circulate on both the M1/2 and NA phylogenies, as indicated by the phylogenetically divergent clusters of isolates shaded in pink and light green, respectively, on the M1/2 and NA trees (Fig. 1, Fig. 3). While the genetic diversity of the HA appears to be restricted at a given time, influenza viruses clearly periodically acquire novel M1/2 and NA segments through reassortment.
Notably, isolate A/Hong Kong/CUHK4245/1997 is the only S31N mutant from 1997 in our data set and is related to no other adamantane resistant isolates, compatible with no or limited spread of this virus in this viral population. The second emergence of the S31N substitution involves a A/Sydney/5/1997-like isolate from New York State (A/New York/512/1998, Introduction #2, Table 1, Fig. 1). This introduction involves a single isolate, again indicative of limited or no spread in this population at this level of detection. It is also possible that this introduction represents an instance of within-host evolution of adamantane resistance in response to drug therapy, given that the patient from whom this isolate was collected was 80 years old (Table 1) and the elderly routinely receive adamantane drugs in the United States (Cohen et al., 2008, Couch, 2000).
Introduction #3 involves two A/Sydney/5/1997-like isolates, A/New York/315/1999 and A/New York/324/1999, which were collected on January 6, 1999 and January 26, 1999, respectively (Table 1, Fig. 1). To be as conservative as possible in our estimate of independent introductions of the S31N mutation, we regard these two isolates as a single introduction, given the phylogenetic similarity of these two isolates, their geographic and temporal proximity, and the low number of S31N mutations detected at this time. However, it is possible that these S31N both arose de novo and represent independent introductions, rather than being transmitted through a network of New York State viruses not sampled in our study population.
Introduction #4 involves another New York State isolate collected in 1999 (A/New York/280/1999). However, as this introduction is associated with a single virus, we again assume that it experienced little or no spread in this population. In fact, no adamantane resistant isolates are detected in any locality during 2000–2001, a period when A/H1N1 influenza viruses were dominant (CDC 2001). In 2002 the S31N mutation is first detected in Oceania (Introduction #5), involving the two A/Sydney/5/1997-like isolates A/Wellington/71/2002 and A/Wellington/79/2002 (Fig. 1, Table 1). Again, given the geographic and temporal proximity of these phylogenetically related isolates, we infer that this occurrence of the S31N mutation has disseminated locally in New Zealand (Table 1).
The first S31N introduction that exhibits possible spread between multiple localities is Introduction #6, which is detected first in Australia on 08/25/2003 (A/Queensland/34/2003) and subsequently observed twice in New York State (A/New York/1/2003, 10/28/2003; A/New York/31/2004, 01/05/2004) (Table 1). Introduction #6 also represents the first S31N mutation observed among A/Fujian/411/2002-like viruses. Given the time and distance separating these three isolates, it is possible that each isolate represents a separate introduction of the S31N mutation, although there is insufficient phylogenetic resolution in this section of the M1/2 phylogeny to distinguish between the single versus multiple introductions of these three isolates (Fig. 1). Again, to be conservative in the number of independent introductions, we suggest that this introduction spread from the summer 2003 epidemic in Australia to the winter 2003–2004 epidemic in New York State (Table 1).
Notably, A/Queensland/34/2003, A/New York/1/2003, and A/New York/31/2004 are members of a clade of A/Fujian/411/2002-like isolates that was involved in a major reassortment event associated with the jump from Sydney/97 to Fujian/02 antigenic clusters (Holmes et al., 2005). Although these isolates cluster with A/Fujian/411/2002-like isolates on the HA tree (Fig. 2), they are members of a clade that is related to older A/Sydney/5/1997-like isolates on both the M1/2 and NA trees (Fig. 1, Fig. 3). The first A/Fujian/411/2002-like isolates observed in our data set are non-reassortant and detected during the summer of 2002 in Hong Kong (A/Hong Kong/CUHK33199/2002, 07/04/2002; A/Hong Kong/CUHK34193/2002, 08/02/2002). Non-reassortant A/Fujian/411/2002-like isolates also are detected in New York State in the winter of 2002–2003 (e.g., A/New York/485/2003, 01/06/2003). However, all A/Fujian/411/2002-like isolates from Oceania that emerge in the summer of 2003 are reassortant, with NA and M1/2 segments that derive from the major clade of A/Sydney/5/1997-like isolates. Reassortant A/Fujian/411/2002-like isolates are subsequently detected in New York State and Hong Kong during the 2003–2004 season. These reassortant M1/2 and NA lineages continue to circulate for at least a year (last detected in October 2004 in Australia) before dying off during a purging of genetic diversity associated with the selective sweep of the A/California/7/2004-like viruses (Rambaut et al., 2008).
Whereas the six previous introductions of the S31N mutation likely exhibit limited spread in time and space, Introduction #7 is continually detected among this viral population for more than 3.5 years (from 07/06/2003 to 03/14/2007) and gives rise to the majority of adamantane resistant isolates in our data set, including the major ‘N-lineage’ that is associated with the global spread of adamantane resistance that begins in 2005 (Table 1, Fig. 1) (Simonsen et al., 2007). Introduction #7 is first detected in Hong Kong on 07/06/2003 (A/Hong Kong/CUHK50895/2003) and is continually detected in Hong Kong for nearly two years (through 04/28/2005) (Table 1). A single resistant isolate is observed during this time outside of Hong Kong (A/New York/204/2003, 08/16/2003), but during the non-epidemic summer months in New York State and is likely to be a spill-over from Asia (Table 1, Fig. 1).
All resistant isolates circulating in Hong Kong from 2003 to 2004 are A/Fujian/411/2002-like in antigenicity. However, in the early spring of 2005 these resistant viruses acquire an antigenically novel HA segment described as A/Wisconsin/67/2005-like, at which point this lineage has been referred to as the ‘N-lineage’ (Simonsen et al., 2007). This reassortment event is evidenced by the phylogenetic distance on the HA tree between two sets of isolates that are both closely related on the M1/2 tree in association with Introduction #7 (Fig. 2, Fig. 1, respectively). On the HA tree, one of these sets clearly is A/Fujian/411/2002-like (‘#7F’, Fig. 2), while the other is A/Wisconsin/67/2005-like (‘#7W’, Fig. 2). In addition, the four phylogenetically distinct clusters of ‘#7F’ isolates on the HA tree suggest that A/Fujian/411/2002-like isolates may have acquired the #7 S31N introduction through multiple additional reassortment events. The phylogenetic divergence between two clusters of ‘#7F’ isolates is particularly pronounced on the NA tree (‘#7F1’ and ‘#7F2’, Fig. 3), providing clear evidence of reassortment in this case.
The first of the reassortant ‘N-lineage’ isolates is detected in Hong Kong on March 1, 2005 (A/Hong Kong/CUHK7711/2005) (Table 1). Adamantane resistant reassortant isolates rapidly proliferate globally thereafter, emerging in Oceania by the late spring of 2005 and reaching New York State during the 2005–2006 winter epidemic (Table 1). An expanded phylogenetic analysis of the M1/2 segment that includes additional adamantane resistant isolates collected globally from 1997 to 2007, mainly from South-East Asia from 2003 to 2005, indicates that the ‘N-lineage’ circulated widely in South-East Asia during the late spring and summer of 2005, including Vietnam, Malaysia, Philippines, Thailand, and Macau (Supplementary Fig. 1). Notably, none of the additional global isolates in the ‘N-lineage’ isolates was sampled prior to the original 03/01/2005 emergence date we identified in Hong Kong. Nor do any of the global background isolates associated with Introduction #7 pre-date its first detection in Hong Kong on 07/06/2003 (Supplementary Fig. 1). As expected, the majority of these global resistant isolates are associated with the main Introduction #7, and all 2005 global isolates are members of the ‘N-lineage.’
The third independent introduction of the S31N mutation among A/Fujian/411/2002-like viruses is detected in 2004 involving three isolates from Hong Kong (Introduction #8, Table 1, Fig. 1), while the fourth, Introduction #9, occurs along a phylogenetically divergent lineage of the M1/2 segment that is primarily sensitive to adamantane drugs (isolate A/Hong Kong/CUHK35011/2004) (Table 1, Fig. 1). Despite the global dominance of adamantane resistant influenza viruses from 2005 to 2007, a phylogenetically distinct lineage of the M1/2 segment that for the most part lacks the S31N substitution co-circulates from 2004 to 2007 (Fig. 1). This adamantane sensitive lineage includes all of the A/California/7/2004-like isolates, as well as small clusters of sensitive A/Fujian/411/2002-like and A/Wisconsin/67/2005-like isolates. Consequently, while the vast majority of A/Wisconsin/67/2005-like isolates are resistant to adamantanes, several A/Wisconsin/67/2005-like isolates are adamantane sensitive due to reassortment with this sensitive M1/2 lineage that co-circulates: isolates A/Canterbury/127/2005, A/Hong Kong/CUHK63957/2006, A/Hong Kong/CUHK68792/2006, A/New York/UR06-373/2007, and A/New York/UR06-515/2007 (Figs. 1 and 2).
Two additional introductions of the S31N mutation occur among this adamantane sensitive M1/2 lineage, both involving A/California/7/2004-like isolates. Introduction #10 involves a single isolate, A/New York/391/2005 (Table 1, Fig. 1), and Introduction #11 consists of a cluster of three isolates detected during the summer of 2005 in New Zealand: A/Otago/3/2005, A/Otago/4/2005, and A/Wellington/9/2005 (Table 1, Fig. 1). These three isolates are separated by only 8 days and relatively little geographic distance, suggesting limited local spread. Following this last introduction of the S31N mutation in the summer of 2005, all adamantane resistant influenza viruses hereon are associated with the ‘N-lineage’, strongly suggesting that this clade has a major fitness advantage.
Discussion
Whereas previous phylogenetic analyses of the emergence and evolution of adamantane resistant influenza viruses have been limited by the lack of data from subtropical regions, or by a primary focus on the evolution of the HA1 region (Simonsen et al., 2007, Deyde et al., 2007), this study is the first to incorporate multi-segment sequence data intensively sampled from a subtropical locality in South-East Asia. By tracing the evolution of adamantane resistance among three geographically and climatically distinct localities for the HA, M1/2, and NA segments, we are able to determine evolutionary processes operating at both regional and global scales. Given that the therapeutic usage of adamantane drugs has been at a constant and relatively low level for many decades in temperate areas, including the United States, simple models based on local drug selection pressures alone inadequately explain the rapid rise in the global prevalence of adamantane resistance in recent years. Rather, we find that the emergence and spread of adamantane resistance appears to be a complex evolutionary process that includes geographically variable selection pressures, extensive global migration, and frequent reassortment.
Specifically, our findings indicate that adamantane resistance independently emerged at least 11 times throughout 1997–2007 in all three globally dispersed localities. This estimate is also conservative, given the limitations of our sampling and our capacity to identify additional de novo introductions among genetically related resistant isolates. As several introductions of the resistant S31N mutation were detected in elderly patients in temperate regions where adamantane is more frequently administered to seniors (Cohen et al., 2008, Couch, 2000), it is possible that intra-host selection was involved in a number of these introductions. However, 10/11 of these introductions were represented by no more than three isolates in each case, suggesting that these introductions experienced only limited in spread in our studied population, and that drug selection pressure alone was insufficient to increase the prevalence of S31N.
Importantly, the vast majority of adamantane resistant influenza virus isolates in our data set descend from a single introduction (Introduction #7), which is first detected in July 2003 in Hong Kong, where it was detected continually for almost two years (and likely was present in other South-East Asian locations sparsely sampled in this study), before spreading globally following the major reassortment event in March 2005 and the emergence of the ‘N-lineage’ (Simonsen et al., 2007). Given the lack of viral sequence data from other South-East Asian countries, it is not possible to determine whether the viral lineage associated with Introduction #7 persisted in Hong Kong or was continually re-introduced into Hong Kong from neighboring South-East Asian countries over this time period, such as other parts of China.
Although Introduction #7 is present in Hong Kong for several years, it is only through reassortment with a novel HA (A/Wisconsin/67/2005-like) that this resistant M1/2 lineage is able to spread globally. Although our data set is limited to the HA, NA, and M1/2 segments, we were still able to observe a number of reassortment events among these three genome segments. While genetic diversity appears to be more limited in the HA, presumably due to strong immune selection pressures, multiple divergent lineages co-circulated on the NA and M1/2 phylogenies prior to the selective sweep associated with the emergence of A/California/7/2004-like viruses that has been described previously (Rambaut et al., 2008). It is this long-term preservation of genetic diversity in the M1/2 segment that allows for the continued circulation of both adamantane sensitive and adamantane resistant influenza viruses to the present day. Indeed, just as influenza viruses acquired adamantane resistance through reassortment in 2005, it has been observed that resistant influenza viruses may re-acquire drug sensitivity through reassortment with adamantane sensitive lineages that continue to co-circulate in both the United States (Nelson et al., 2008) and in Japan (Furuse et al., 2008).
Recent studies also suggest that South-East Asia may serve as a reservoir for influenza A virus, continually re-seeding new epidemics in temperate areas (Russell et al., 2008). Our findings provide further evidence that South-East Asia serves as an epicenter for influenza virus activity, with viral lineages such as Introduction #7 originating in South-East Asia before disseminating globally to temperate areas. Although our inclusion of background global sequences from South-East Asia in our expanded phylogenetic tree (Supplementary Fig. 1) suggests that Hong Kong is likely to be representative of the South-East Asian region for the purposes of this study, intensive sampling from additional localities in South-East Asia is greatly needed to understand the evolutionary dynamics in this critical region. Further sampling would likely be most profitable in countries such as China, where high levels of adamantane resistance were first detected, and yet publicly available influenza virus sequences are limited (Bright et al., 2005, Bright et al., 2006).
Finally, the processes of local evolution, reassortment, and global spread through genomic hitchhiking we observe in the spread of S31N among A/H3N2 viruses are also likely to be involved in the recent global proliferation of A/H1N1 influenza viruses that are resistant to adamantanes (Deyde et al., 2007, Barr et al., 2007b), as well as in the unexpected proliferation of the H274Y substitution conferring resistance to NA inhibitors (oseltamivir) (Sheu et al., 2008, Besselaar et al., 2008, Lackenby et al., 2008). Understanding the evolutionary basis for the current proliferation of drug resistant viruses will be particularly important for developing effective strategies for administering antiviral drugs in the event of a pandemic (Lackenby et al., 2008), but will require the public availability of additional whole-genome sequences from isolates collected intensively from multiple global localities from the earliest time of resistance detection.
Methods
Phylogenetic analysis of influenza A viruses from Hong Kong, New York State, USA, Australia and New Zealand from 1997 to 2007
All influenza A/H3N2 virus sequence data were collected from the National Institute of Allergy and Infectious Disease's Influenza Genome Sequencing Project (http://www.niaid.nih.gov/dmid/genomes/mscs/influenza.htm) for the period 1997–2007. A total of 485 influenza A/H3N2 viruses from 1997 to 2006 were sampled from all 11 regions in New York State, USA, and were collected by the Virus Reference and Surveillance Laboratory at the Wadsworth Center, New York State Department of Health. In addition, we included 7 A/H3N2 influenza viruses that were collected in 2007 by Surveillance Data Inc., as part of a larger 2006–2007 U.S. surveillance effort. In Oceania, a total of 629 influenza A/H3N2 viruses were sampled from both the North and South Islands of New Zealand and from four territories in Australia (Western Australia, New South Wales, South Australia, and Queensland). Influenza viruses from New Zealand were collected by Canterbury Health Laboratories in Christchurch, New Zealand. In Australia, viruses from New South Wales were collected by the Prince of Wales Hospital, New South Wales; viruses from South Australia were collected by the Institute of Medical and Veterinary Sciences, South Australia; viruses from Queensland were collected by the Queensland Health Science Services, Queensland; and viruses from Western Australia were collected by PathWest Laboratory Medicine, Western Australia. A total of 281 influenza A/H3N2 viruses from 1997 to 2006 (∼30 isolates from each year) were collected from children of ages 1–10 presenting with acute respiratory illness to the Prince of Wales Hospital in Hong Kong. In this case only the HA, NA, and M1/2 genome segments were available. Procedures for the isolation and sequencing of these isolates have been described previously (Ghedin et al., 2005, Tang et al., 2008a, Tang et al., 2008b). Accordingly, sequence data for the coding regions of the HA, NA, and M1/2 for 281 influenza isolates from Hong Kong (Supplementary Table 1), 492 influenza isolates from New York State, USA (Supplementary Table 2), and 629 influenza isolates from Australia and New Zealand (Supplementary Table 3) were downloaded from the National Center for Biotechnology Information (NCBI) Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) available on GenBank (Bao et al., 2008). Dates of sampling were available on GenBank, and information on patient age was available on GenBank for each influenza isolate collected from New York State, Australia, and New Zealand (but not for Hong Kong) (Supplementary Tables 1–3).
Phylogenetic analysis
Sequence alignments were manually constructed for the major coding regions for the HA, (1698 nt), NA (1407 nt), and M1/2 (979 nt) using the Se-Al program (Rambaut 2002). To determine the evolutionary relationships between all 1410 influenza isolates an initial phylogenetic tree was inferred using neighbor-joining methods available in PAUP⁎ (Swofford 2003), with the robustness of each node determined through a bootstrap resampling process (1000 replications). Clades of related isolates were identified by high bootstrap values (> 70%) and exceptionally long branch lengths. The occurrence of resistance to adamantane antiviral drugs was identified through a visual screening for the S31N amino acid replacement in the M2 protein using the Se-Al program (Rambaut 2002).
Due to the large number of closely related isolates that adds little to a phylogenetic analysis of circulating lineages, we selected from the original data set a representative data set of 226 influenza isolates sampled from Hong Kong (n = 108), New York State (n = 72), and Australia and New Zealand (n = 46) (Supplementary Table 4). All isolates that are resistant to adamantane drugs that emerged prior to the emergence of the ‘N-lineage’ were retained in this smaller data set, as were all isolates that occupied divergent topological positions across the HA, NA and M1/2 phylogenies, the earliest isolate that was detected in each location (Hong Kong, New York State, and Australia/New Zealand), and a maximum of four isolates for each season, each location, and each antigenic type. We also included the following five vaccine reference strains in our HA data set (n = 231) to determine antigenic groups: A/Sydney/5/1997, A/Wyoming/03/2003 (A/Fujian/411/2002-like), A/California/07/2004, A/Wisconsin/67/2005, and A/Brisbane/10/2007. To study the emergence of adamantane resistance in a broader global context, an additional 20 A/H3N2 influenza virus isolates exhibiting the S31N adamantane resistance mutation (and for which HA1 sequences were also available) were identified, and M1/2 and HA1 sequence data was downloaded from GenBank (Bao et al., 2008) and added to the original data sets (n = 246). These background isolates were collected between 2003 and 2005, mainly from South-East Asia (Macau, Malaysia, Philippines, Singapore, Taiwan, Thailand, and Vietnam) (Supplementary Table 5).
The evolutionary relationships for the HA, M1/2, and NA among these 226 influenza isolates (231 isolates for HA, including 5 vaccine reference strains) were inferred using the maximum likelihood (ML) methods available in PAUP⁎ (Swofford 2003). In each case, the best-fit model of nucleotide substitution was identified by MODELTEST (Posada and Crandall, 1998) as the general reversible (GTR+I+Γ4) model, with the frequency of each substitution type, proportion of invariant sites (I), and the gamma distribution of among-site rate variation with four rate categories (Γ4) estimated from the empirical data (parameter values available upon request). In all cases tree bisection-reconnection (TBR) branch-swapping was utilized to determine the globally optimal tree. To assess the robustness of each node on the phylogenetic tree, a bootstrap resampling process (1000 replications) using the neighbor-joining (NJ) method was used, incorporating the ML substitution model. Clades of related isolates were again identified by high bootstrap values (> 70) and exceptionally long branch lengths. Independent introductions of the S31N replacement were inferred from clade positions on the M1/2 phylogeny and the location and dates of sampling.
The antigenic characterization of each isolate was inferred from its phylogenetic position on the HA tree with respect to the five influenza vaccine strains of known antigenicity: A/Sydney/5/1997, A/Wyoming/3/2003, A/California/7/2004, A/Wisconsin/67/2005, and A/Brisbane/10/2007. Specific amino acid changes along key branches separating antigenic groups were identified using the MacClade program (Maddison and Maddison 2000) (Supplementary Table 6).
Acknowledgments
Funding support for Lone Simonsen came from the Research and Policy for Infectious Disease Dynamics (RAPIDD) joint program of the Science and Technology Directorate, Department of Homeland Security, and Fogarty International Center, National Institutes of Health.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2009.03.026.
Appendix A. Supplementary data
Phylogenetic relationships of the M1/2 segment of 246 A/H3N2 influenza viruses sampled from Hong Kong (n = 108), New York State (n = 72), New Zealand and Australia (n = 46), and a background sample of global adamantane resistant isolates (n = 20) between 1997 and 2007, estimated using an ML method. Adamantane resistant isolates from Hong Kong, New York State, and Australia and New Zealand are shaded in red, while global background isolates are shaded in orange. Rooting same as Fig. 1, Fig. 2, Fig. 3.
References
- Aoki F.Y. Amantadine and rimantadine. In: Nicholson K.G., Webster R.G., Hay A.J., editors. Textbook of Influenza. Blackwell Science; Oxford: 1998. pp. 457–476. [Google Scholar]
- Barr I.G., Hurt A.C., Iannello P., Tomasov C., Deed N., Komadina N. Increased adamantane resistance in influenza A(H3) viruses in Australia and neighboring countries in 2005. Antiviral Res. 2007;73:112–117. doi: 10.1016/j.antiviral.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Barr I.G., Hurt A.C., Deed N., Iannello P., Tomasov C., Komadina N. The emergence of adamantane resistance in influenza A(H1) viruses in Australia and regionally in 2006. Antiviral Res. 2007;75:173–176. doi: 10.1016/j.antiviral.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Bao Y., Bolotov P., Dernovoy D., Kiryutin B., Zaslavsky L., Tatusova T., Ostell J., Lipman D. The Influenza Virus Resource at the National Center for Biotechnology Information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselaar T.G., Naidoo D., Buygs A., Gregory V., McAnerney J., Manamela J.M., Blumberg L., Schoub B.D. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 2008;14:1809–1810. doi: 10.3201/eid1411.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright R.A., Medina M., Xu S., Perez-Oronoz G., Wallis R., David X.M., Povinelli L., Cox N.J., Klimov A.I. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- Bright R.A., Shay D.K., Shu B., Cox N.J., Klimov A.I. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC), 2001. 2000–2001 influenza season summary. Accessed 01/05/2009: http://www.cdc.gov/flu/weekly/weeklyarchives2000-2001/00-01summary.htm.
- Centers for Disease Control and Prevention (CDC) High levels of adamantine resistance among influenza A(H3N2) viruses and interim guidelines for use of antiviral agents — the United States, 2005–2006 influenza season. MMWR. 2006;55:44–46. [PubMed] [Google Scholar]
- Cohen N.J., Morita J.Y., Plate D.K., Jones R.C., Simon M.T., Nawrocki J., Siston A.M., Gerber S.I. Control of an outbreak due to an adamantane-resistant strain of influenza A (H3N2) in chronic care. Infection. 2008;36:458–462. doi: 10.1007/s15010-008-7295-9. [DOI] [PubMed] [Google Scholar]
- Couch R.B. Prevention and treatment of influenza. N. Engl. J. Med. 2000;343:1778–1787. doi: 10.1056/NEJM200012143432407. [DOI] [PubMed] [Google Scholar]
- Deyde V.M., Xu X., Bright R.A., Shaw M., Smith C.B., Zhang Y., Shu T., Gubareva L.V., Cox N.J., Klimov A.I. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- Furuse, Y., Suzuki, A., Kamigaki, T., Shimizu, M., Fuji, N., Oshitani, H., 2008. Reversion of influenza A (H3N2) from adamantane-resistant to adamantane-sensitive by further reassortment in Japan during the 2006–2007 season. J. Clin. Microbiol. 47 Electronic publication ahead of print. [DOI] [PMC free article] [PubMed]
- Ghedin E., Sengamalay N.A., Shumway M., Zaborsky J., Feldblyum T., Subbu V., Spiro D.J., Sitz J., Koo H., Bolotov P., Dernovoy D., Tatusova T., Bao Y., St. George K., Taylor J., Lipman D.J., Fraser C.M., Taubenberger J.K., Salzberg S.L. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- Hayden F.G. Antiviral resistance in influenza viruses — implications for management and pandemic response. New Eng. J. Med. 2006;354:785–788. doi: 10.1056/NEJMp068030. [DOI] [PubMed] [Google Scholar]
- Holmes E.C., Ghedin E., Miller N., Taylor J., Bao Y., St. George K., Grenfell B.T., Salzberg S.L., Fraser C.M., Lipman D.J., Taubenberger J.K. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005:3, e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackenby A., Thompson C.I., Democratis J. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 2008;21:626–638. doi: 10.1097/QCO.0b013e3283199797. [DOI] [PubMed] [Google Scholar]
- Maddison D.R., Maddison W.P. Sinauer Associates; Sunderland, Massachusetts: 2000. MacClade. Analysis of Phylogeny and Character Evolution, version 4.0 [computer program] [Google Scholar]
- Masuda H., Suzuki H., Oshitani H., Saito R., Kawasaki S., Nishikawa M., Satoh H. Incidence of amantadine resistant influenza A viruses in sentinel surveillance sites and nursing homes in Niigata, Japan. Microbiol. Immunol. 2000;44:833–839. doi: 10.1111/j.1348-0421.2000.tb02571.x. [DOI] [PubMed] [Google Scholar]
- Nelson M.I., Simonsen L., Viboud C., Miller M.A., Taylor J., St George KS, Griesemer S.B., Ghedin E., Sengamalay N.A., Spiro D.J., Volkov I., Grenfell B.T., Lipman D.J., Taubenberger J.K., Holmes E.C. Stochastic processes are key determinants of the short-term evolution of influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.I., Simonsen L., Viboud C., Miller M.A., Holmes E.C. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3:e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.I., Edelman L., Spiro D.J., Boyne A.R., Bera J., Halpin R., Sengamalay N., Ghedin E., Miller M.A., Simonsen L., Viboud C., Holmes E.C. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 2008;4:e1000133. doi: 10.1371/journal.ppat.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., 2002. Sequence alignment editor. Computer program downloaded from http://tree.bio.ed.ac.uk/software/seal/.
- Rambaut A., Pybus O., Nelson M.I., Viboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J., Hurt A.C., de Jong J.C., Kelso A., Klimov A.I., Kageyama T., Komadina N., Lapedes A.S., Lin T.P., Mosterin A., Obuchi M., Odagiri T., Osterhaus A.D., Rimmelzwaan G.R., Shaw M.W., Skepner E., Stohr K., Tashiro M., Fouchier R.A., Smith D.J. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- Shek L.P.C., Lee B.W. Epidemiology and seasonality of respiratory tract infections in the tropics. Paediatr. Respir. Rev. 2003;4:105–111. doi: 10.1016/s1526-0542(03)00024-1. [DOI] [PubMed] [Google Scholar]
- Sheu T.G., Deyde V.M., Okomo-Adhiambo M., Garten R.J., Xu X., Bright R.A., Butler E.N., Wallis T.R., Klimov A.I., Gubareva L.V. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004–2008. Antimicrob. Agents. Chemother. 2008;52:3284–3292. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K., Mitamura K., Sakai-Tagawa Y., Goto H., Sugaya N., Kawaoka Y. High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J. Infect. Dis. 2003;188:57–61. doi: 10.1086/375799. [DOI] [PubMed] [Google Scholar]
- Simonsen L., Viboud C., Grenfell B.T., Dushoff J., Jennings L., Smit M., Macken C., Hata M., Gog J., Miller M.A., Holmes E.C. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol. Biol. Evol. 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Saito R., Masuda H., Oshitani H., Sato M., Sato I. Emergence of adamantane-resistant influenza A viruses: epidemiological study. J. Infect. Chemother. 2003;9:195–200. doi: 10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, Massachusetts: 2003. PAUP⁎: Phylogenetic Analysis Using Parsimony (⁎and Other Methods) Version 4.0 [Computer Program] [Google Scholar]
- Tang J.W., Ngai K.L.K., Lam W.Y., Chan P.K.S. Seasonality of influenza A(H3N2) virus: a Hong Kong perspective. PLoS One. 2008;3:e2768. doi: 10.1371/journal.pone.0002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Ngai K.L.K., Wong C.L., Lam W.Y., Chan P.K.S. Emergence of adamantane-resistant influenza A(H3N2) viruses in Hong Kong between 1997–2006. J. Med. Virol. 2008;80:895–901. doi: 10.1002/jmv.21155. [DOI] [PubMed] [Google Scholar]
- Wang C., Takeuchi K., Pinto L.H., Lamb R.A. Ion channel activity of influenza A virus A virus M2 protein: characterization of the amantadine block. J. Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.H., Yang L., Chan K.P., Leung G.M., Chan K.H., Guan Y., Lam T.H., Hedley A.J., Peiris J.S.M. Influenza-associated hospitalization in a subtropical city. PLoS Med. 2006;3:e121. doi: 10.1371/journal.pmed.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wong C.M., Lau E.H.Y., Chan K.P., Ou C.Q., Peiris J.S.M. Synchrony of clinical and laboratory surveillance for influenza in Hong Kong. PLoS One. 2008;3:e1399. doi: 10.1371/journal.pone.0001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T., Hemphill M.L., Ziegler M.L., Perez-Oronoz G., Klimov A.I., Hapson A.W., Regnery H.L., Cox N.J. Low incidence of rimantadine resistance in field isolates of influenza A viruses. J. Infect. Dis. 1999;180:935–939. doi: 10.1086/314994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships of the M1/2 segment of 246 A/H3N2 influenza viruses sampled from Hong Kong (n = 108), New York State (n = 72), New Zealand and Australia (n = 46), and a background sample of global adamantane resistant isolates (n = 20) between 1997 and 2007, estimated using an ML method. Adamantane resistant isolates from Hong Kong, New York State, and Australia and New Zealand are shaded in red, while global background isolates are shaded in orange. Rooting same as Fig. 1, Fig. 2, Fig. 3.