Abstract
A series of hydroxamic acid based histone deacetylase inhibitors 6–15, containing an isoxazole moiety adjacent to the Zn-chelating hydroxamic acid, is reported herein. Some of these compounds showed nanomolar activity in the HDAC isoform inhibitory assay and exhibited micro molar inhibitory activity against five pancreatic cancer cell lines.
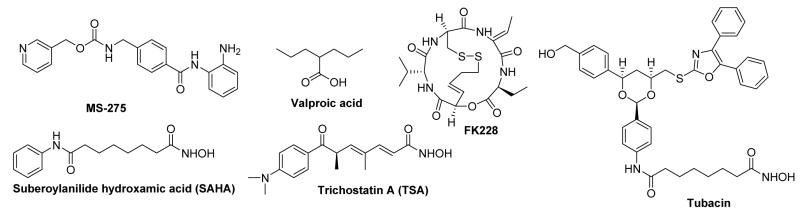
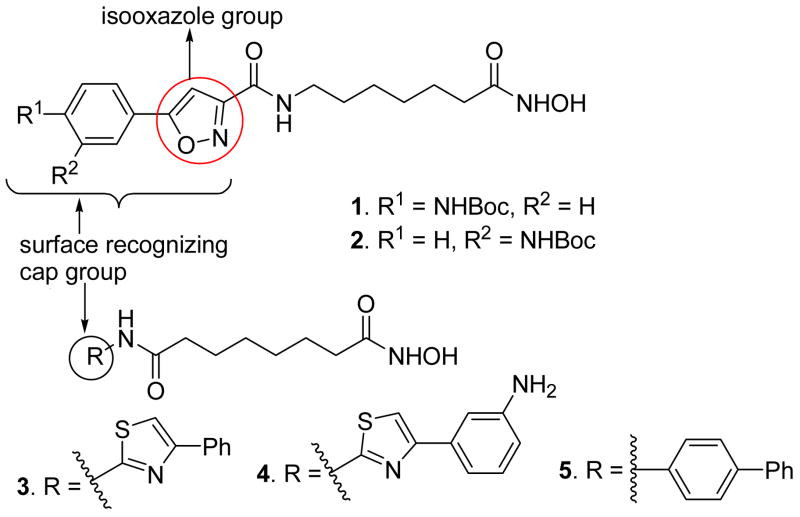
Epigenetic regulation of gene expression is partly controlled by chromatin remodeling which is initiated either by post-translational modification of histone proteins1,2 or DNA methylation.3 Several different modifications that occur in histone amino termini are defined as acetylation, methylation, ubiquitinylation, and glycosylation. Histone acetylation-deacetylation is a reversible phenomenon and is tightly regulated by two competing enzymes known as histone acetyltransferases (HATs) and histone deacetylases (HDACs).4 In general, HATs catalyse the acetylation of N-terminal lysine ε-amino groups in nuclear histones, resulting in neutralization of the positive charges on the histones and a more open, transcriptionally active chromatin structure while the HDACs catalyze the reverse reaction and suppress transcription. Altered HDAC activity is seen to be associated with many cancers, validating HDACs as promising targets for cancer therapy. Several small molecule HDAC inhibitors5–7 (Figure 1) such as suberoylanilide hydroxamic acid (SAHA), depsipeptide FK228, MS-275, and valproic acid have been found to have promising antitumor activities in clinical studies and in 2006, the US FDA approved SAHA for the treatment of a rare cancer, cutaneous T-cell lymphoma (CTCL).8 These compounds owe their antiproliferative action to their ability to allow the transcription and expression of repressed genes including tumor suppressor genes. Beside their antitumor activity, HDAC inhibitors are also found to have potential applications in the treatment of neurodegenerative disorders such as Parkinson’s and Huntington’s diseases, as well as maleria.9 Human HDACs are divided into four classes based on their function and structural homology. Class I (HADC1, 2, 3, and 8), class II (HDAC4, 5, 6, 7, 9 and 10), and class IV (HDAC11) HDACs are zinc-dependent enzymes while class III HDACs are sirtuin related proteins and require the cofactor NAD+ for their deacetylase function.7 To learn more about the role the individual HDACs play in cell growth and differentiation, neuronal protection, and apoptosis, it is important to develop agents that show selectivity for individual isoforms or a small subset of these isoforms. To date, a large number of structurally diverse, natural and synthetic, hydroxamic acid based HDAC inhibitors (Figure 1) have been reported which include trichostatin A (TSA), SAHA, depsipeptide, and tubacin etc., but the number of isoform selective HDAC inhibitors are very limited.5,6 Recently, we have reported hydroxamate-based HDAC inhibitors bearing substituted aryl-isoxazoles (Figure 2) as the cap groups. Some of these compounds were found to be highly potent and selective at HADC6 as well as HDAC3 (compound 1 and 2, Table 1).10
Figure 1.
Structures of some known HDAC inhibitors
Figure 2.
HDAC inhibitors containing a substituted isoxazole, a substituted thiazole, or a biphenyl group as the cap
Table 1.
In vitro HDAC Inhibitory Activities of Compounds 1–5.
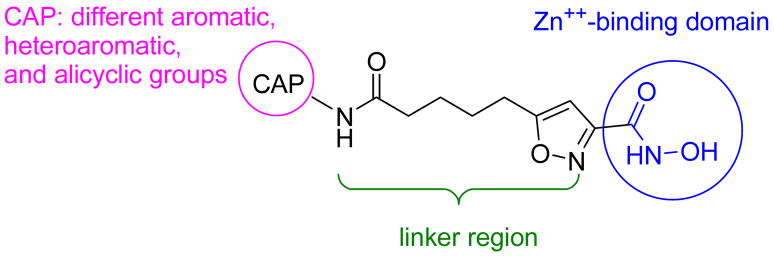
We anticipated that the exceptional potency and selectivity of those compounds were due to the presence of the isoxazole moiety whose mode of interaction with surface residues is still under investigation. In this paper, we report a series of hydroxamic acid-based HDAC inhibitors with an isoxazole moiety in the linker region adjacent to the hydroxamic acid group (Figure 3). These compounds were designed with the idea that the more rigid and bulkier zinc binding group might allow for a different selectivity. The choice of the cap groups was based on the HDAC inhibitors previously explored by us11,12 (Figure 2, Table 1) and Miyata et al.13 In this context, the hydroxamic acid was suitably replaced by the other functionalities like alcohol, carboxylic acid, and amide to explore their Zn-binding capabilities. All the synthesized compounds were subjected to in vitro HDAC inhibition study by using catalytically active recombinant HDACs, and their antiproliferative activities were tested on various pancreatic cancer cell lines as well.
Figure 3.
General structural of HDAC inhibitors 6–15
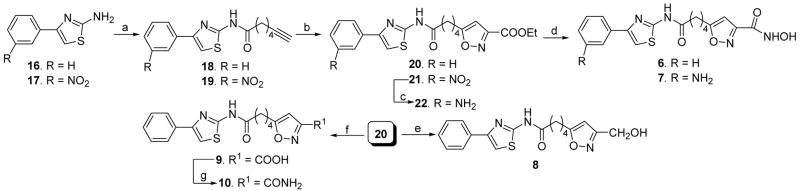
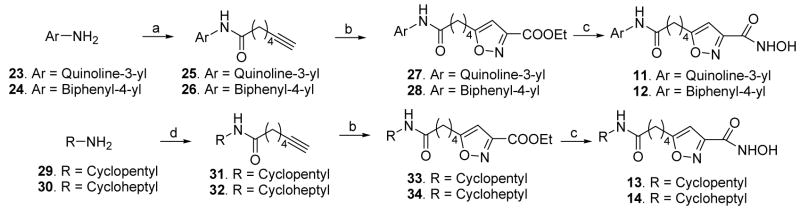
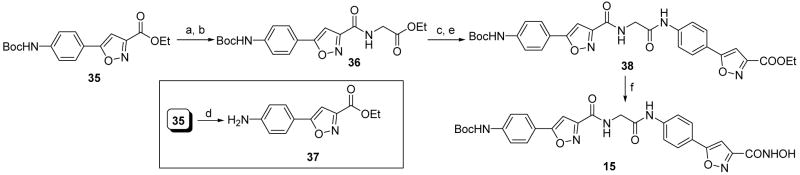
Different heteroaromatic, aromatic and alicyclic amino cap groups, namely 2-amino-4-phenylthiazole (16), 4-(3-nitrophenyl)thiazol-2-ylamine (17), 3-aminoquinoline (23), 4-aminobiphenyl (24), cyclopentylamine (29), and cycloheptylamine (30) were coupled with 6-heptynoic acid under peptide coupling conditions to yield terminal acetylenic compounds 18, 19, 25, 26, 31, and 32, respectively. Subsequently the terminal acetylenic group of these compounds was converted by the use of nitrile oxide cycloaddition chemistry14 to the substituted-isoxazole carboxylic acid ethyl ester (20, 21, 27, 28, 33, and 34). Lastly, these ester compounds were reacted with hydroxylamine hydrochloride in the presence of potassium hydroxide in accord with a known literature procedure (Schemes 1 and 2)15 to provide hydroxamic acids 6, 7, and 11 – 14. In case of the synthesis of compound 7, the intermediate 21 bearing a nitro cap group was reduced to the aniline derivative 22 by catalytic hydrogenation (Scheme 1), followed by hydroxamic acid formation. Sodium borohydride reduction of compound 20 led to alcohol 8. The carboxylic acid 9 was synthesized by saponification (Scheme 1) of the corresponding ester 20. The amide 10 was obtained from 9 by an active ester method (Scheme 1). A different strategy was applied to synthesize compound 15 as shown in Scheme 3. Sequential saponification and amide coupling10 of 35 gave the intermediate 38 via the ester 36. The ester 38 was converted to hydroxamic acid 15 following the same general method used to prepare previous hydroxamic acid compounds.
Scheme 1.
Synthesis of compounds 6–10. Reagents and conditions: (a) 7-Heptynoic acid, POCl3, pyridine, −13 °C to rt, 30 min. (b) Ethyl chlorooxamidoacetate, triethylamine, THF, rt, 16 h. (c) H2, 10 % Pd/C, EtOAC, EtOH, 60 °C, 30 min. (d) NH2OH·HCl, KOH, MeOH, rt, 15 min. (e) NaBH4, MeOH, 0 °C to rt, 2 h. (f) LiOH, THF:MeOH:H2O (3:1:1), 0 °C, 30 min. (g) Ethyl chloroformate, triethylamine, THF, 0 °C, 30 min, then aqueous NH3, rt, 12 h.
Scheme 2.
Synthesis of compounds 11–14. Reagents and conditions: (a) 7-Heptynoic acid, POCl3, pyridine, −13 °C to rt, 30 min. (b) Ethyl chlorooxamidoacetate, Et3N, THF, rt, 16 h. (c) NH2OH·HCl, KOH, MeOH, rt, 15 min. (d) EDCI, HOBT, DIEA, amine, DMF, 0 °C to rt, 12 h.
Scheme 3.
Synthesis of compound 15. Reagents and conditions: (a) LiOH, THF:MeOH:H2O (3:1:1), 0 °C, 30 min. (b) EDCI, HOBT, DIEA, glycine ethyl ester hydrochloride, DMF, 0 °C to rt, 12 h. (c) LiOH, THF:MeOH:H2O (3:1:1), 0 °C, 30 min. (d) TFA, CH2Cl2, 0 °C to rt, 2 h. (e) 37, POCl3, pyridine, −13 °C to rt, 30 min. (f) NH2OH·HCl, KOH, MeOH, rt, 15 min.
All the compounds 6–15 were tested against both the class I (1, 2, and 3) and class II (6 and 10) HDACs and their IC50 values are shown in Table 2. Compound 6, 7, and 11, with a phenylthiazole, an m-aminosubstituted phenylthiazole, and a quinoline moiety as cap groups, were the most active HDAC inhibitors in this series. These compounds 6, 7, and 11 were comparatively more active at HDAC3 and HDAC6 than HDAC1, HDAC2, and HDAC10. The hydroxamic acids 12–15 are very poor HDAC inhibitors except for compound 12, which was weakly active at HDAC1 and HDAC6. The inhibitors 6, 7 and 11 were less potent than those compounds with an isoxazole moiety in the cap region10 (Table 1) and also SAHA. Placement of the isoxazole group in the linker region decreased the inhibitory activity of compounds 6 and 7, when compared to those hydroxamic acids 3 and 4 bearing the same cap groups and a linear linker (Figure 2). At the same time, these structural alterations made 6 and 7 relatively more selective as HDAC3 and HDAC6 inhibitors, respectively (6 vs 3, and 7 vs 4, Table 1 and Table 2). Attempted replacement of the hydroxamate group by the other functional groups such as alcohol, carboxylic acid, and amide, failed to provide any significant HDAC inhibitory activity (Compound 8, 9, and 10). This is not surprising in light of other publications.16,17
Table 2.
In vitro HDAC inhibitory assay results for compounds 6–15.
| IC50 (nM) |
|||||
|---|---|---|---|---|---|
| Compds | HDAC1 | HDAC2 | HDAC3 | HDAC6 | HDAC10 |
| 6 | 303 ± 22 | 430 ± 37 | 30 ± 1 | 68 ± 5 | 254 ± 25 |
| 7 | 139 ± 4 | 164 ± 4 | 25 ± 1 | 82 ± 5 | 250 ± 6 |
| 8 | >30,000 | >30,000 | >30,000 | >30,000 | >30,000 |
| 9 | >30,000 | >30,000 | 10,600 ± 829 | >30,000 | >30,000 |
| 10 | >30,000 | >30,000 | >30,000 | >30,000 | >30,000 |
| 11 | 328 ± 10 | 469 ± 18 | 102 ± 3 | 51 ± 0.9 | 471 ± 20 |
| 12 | 885 | 3,750 | 7,410 | 885 | NDa |
| 13 | 17,100 | > 30,000 | nda | 17,100 | NDa |
| 14 | 12,300 | > 30,000 | > 30,000 | 12,300 | NDa |
| 15 | 9,200 ± 670 | >30,000 | 1,920 ± 73 | 1710 ± 82 | 13,300 ± 560 |
| SAHAb | 96 | 282 | 17 | 14 | 72 |
ND = Not Detected.
SAHA data provided by Amphora Inc.
We also investigated the ability of this set of compounds 6–15 to inhibit cancer cell growth using different pancreatic cancer cell lines. Pancreatic cancer is the fourth leading cause of cancer death in the United States, and essentially remains an incurable disease with a five-year survival rate of < 5%.18 This cancer fails to respond to widely-used chemotherapeutic drugs like 5-fluorouracil and gemcitabine,19 and thus there is an urgent need for more efficient drugs. All HDAC inhibitors 6–15, along with the currently marketed drug SAHA were tested on five pancreatic cancer cell lines using the MTT assay. These data are provided in the Table 3. As is apparent from these data, compound 6 nicely inhibited all the five transformed cell lines and its IC50 values were similar to those of SAHA. The relatively weak HDAC inhibitor 12 showed only moderate activity against three of the five transformed cell lines, namely, BxPC-3, HuPT3 and Su.86.86. In contrast, the most potent HDAC inhibitor in this series, compound 7, only moderately inhibited the five pancreatic cancer cell lines. Compound 11 failed to show similar antiproliferative activity compared to compound 6, although its IC50 values in the in vitro HDAC inhibitory assay are very similar to those of 6. The weak inhibitory activities of compound 7 and 11 against different pancreatic cancer cell lines might be due to their poor cell permeability. Compounds 8–10 and 13–15 exhibited poor antiproliferative activities against all five pancreatic cancer cell lines, which are consistent with their poor HDAC inhibitory activities.
Table 3.
In vitro growth inhibition assay results against pancreatic cancer cell lines for compounds 6–15.
| IC50 (μM) |
|||||
|---|---|---|---|---|---|
| Compds | BxPC-3 | HupT3 | Mia Paca-2 | Panc 04.03 | Su.86.86 |
| 6 | 3 | 1 | 4 | 4 | 1 |
|
| |||||
| 7 | 10.5 | 28.4 | 9 | 14.4 | 15 |
| 8 | > 50 | > 50 | > 50 | > 50 | > 50 |
| 9 | > 50 | > 50 | > 50 | > 50 | > 50 |
| 10 | > 50 | > 50 | > 50 | > 50 | > 50 |
| 11 | 36 | 49 | 24 | 44 | 25 |
| 12 | 9 | 7 | 30 | > 50 | 9 |
| 13 | > 50 | > 50 | 24 | 30 | 43 |
| 14 | > 50 | > 50 | > 50 | > 50 | > 50 |
| 15 | > 50 | > 50 | > 50 | > 50 | > 50 |
| SAHA | 5 | 0.8 | 1 | 1 | 1 |
In summary, we synthesized a series of hydroxamic acid based HDAC inhibitors wherein the isoxazole moiety was embedded in the linker region and directly attached to the Zn-chelating hydroxamic acid group. The present findings suggest that no significant isozyme selectivity is gained using the more rigid isoxazole hydroxamate as zinc binding group.
Acknowledgments
This work was supported in part by gift funds from an anonymous donor, by a grant (no. 271210) from the ADDF/Elan, and by the Mayo Foundation and the Pancreatic Cancer SPORE P50 CA102701.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Grunstein M. Nature. 1997;389:349. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe AP, Guschin D. J Struct Biol. 2000;129:102. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 3.Abreu PA, Dellamora-Ortiz G, Leao-Ferreira LR, Gouveia M, Braggio E, Zalcberg I, Santos DO, Bourguinhon S, Cabral LM, Rodrigues CR, Castro HC. Expert Opin Ther Targets. 2008;12:1035. doi: 10.1517/14728222.12.8.1035. [DOI] [PubMed] [Google Scholar]
- 4.Marks PA, Richon VM, Rifkind RA. J Natl Cancer Inst. 2000;92:1210. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 5.Butler KV, Kozikowski AP. Curr Pharm Des. 2008;14:505. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 6.Paris M, Porcelloni M, Binaschi M, Fattori D. J Med Chem. 2008;51:1505. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 7.Bolden JE, Peart MJ, Johnstone RW. Nat Rev Drug Discov. 2006;5:769. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 8.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. Oncologist. 2007;12:1247. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 9.Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, D’Mello SR. Mol Cell Biol. 2006;26:3550. doi: 10.1128/MCB.26.9.3550-3564.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozikowski AP, Tapadar S, Luchini DN, Kim KH, Billadeau DD. J Med Chem. 2008;51:4370. doi: 10.1021/jm8002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozikowski AP, Chen Y, Gaysin AM, Savoy DN, Billadeau DD, Kim KH. ChemMedChem. 2008;3:487. doi: 10.1002/cmdc.200700314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B, Petukhov PA, Jung M, Velena A, Eliseeva E, Dritschilo A, Kozikowski AP. Bioorg Med Chem Lett. 2005;15:1389. doi: 10.1016/j.bmcl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Itoh Y, Suzuki T, Kouketsu A, Suzuki N, Maeda S, Yoshida M, Nakagawa H, Miyata N. J Med Chem. 2007;50:5425. doi: 10.1021/jm7009217. [DOI] [PubMed] [Google Scholar]
- 14.Simoni D, Invidiata FP, Rondanin R, Grimaudo S, Cannizzo G, Barbusca E, Porretto F, D’Alessandro N, Tolomeo M. J Med Chem. 1999;42:4961. doi: 10.1021/jm991059n. [DOI] [PubMed] [Google Scholar]
- 15.Gediya LK, Chopra P, Purushottamachar P, Maheshwari N, Njar VC. J Med Chem. 2005;48:5047. doi: 10.1021/jm058214k. [DOI] [PubMed] [Google Scholar]
- 16.Wahhab A, Smil D, Ajamian A, Allan M, Chantigny Y, Therrien E, Nguyen N, Manku S, Leit S, Rahil J, Petschner AJ, Lu AH, Nicolescu A, Lefebvre S, Montcalm S, Fournel M, Yan TP, Li Z, Besterman JM, Deziel R. Bioorg Med Chem Lett. 2009;19:336. doi: 10.1016/j.bmcl.2008.11.081. [DOI] [PubMed] [Google Scholar]
- 17.Hanessian S, Vinci V, Auzzas L, Marzi M, Giannini G. Bioorg Med Chem Lett. 2006;16:4784. doi: 10.1016/j.bmcl.2006.06.090. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Xie K, Wolff R, Abbruzzese JL. Lancet. 2004;363:1049. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 19.Haller DG. Semin Oncol. 2003;30:3. doi: 10.1016/s0093-7754(03)00296-3. [DOI] [PubMed] [Google Scholar]