Abstract
Topoisomerase enzymes regulate superhelical tension in DNA resulting from transcription, replication, repair, and other molecular transactions. Poxviruses encode an unusual type IB topoisomerase that acts only at conserved DNA sequences containing the core pentanucleotide 5′-(T/C)CCTT-3′. In X-ray structures of the variola virus topoisomerase bound to DNA, protein-DNA contacts were found to extend beyond the core pentanucleotide, indicating that the full recognition site has not yet been fully defined in functional studies. Here we report quantitation of DNA cleavage rates for an optimized 13 bp site and for all possible single base substitutions (40 total sites), with the goals of understanding the molecular mechanism of recognition and mapping topoisomerase sites in poxvirus genome sequences. The data allow a precise definition of enzyme-DNA interactions and the energetic contributions of each. We then used the resulting “action matrix” to show that favorable topoisomerase sites are distributed all along the length of poxvirus DNA sequences, consistent with a requirement for local release of superhelical tension in constrained topological domains. In orthopox genomes, an additional central cluster of sites was also evident. A negative correlation of predicted topoisomerase sites was seen relative to early terminators, but no correlation was seen with early or late promoters. These data define the full variola virus topoisomerase recognition site and provide a new window on topoisomerase function in vivo.
Introduction
Many DNA transactions, including transcription, DNA replication, and DNA repair, introduce superhelical tension into DNA. Topoisomerase enzymes release DNA supertwists and are required to facilitate these processes (Kornberg and Baker, 1991; Wang, 1996; Wang, 2002). The type IB class of topoisomerases act by introducing transient nicks in DNA which allow supercoil release by swiveling of the continuous DNA strand around the nick site. The mechanism involves initial binding of the topoisomerase, formation of a 3′ phospho-tyrosine covalent intermediate, supercoil release by rotation, religation to restore DNA continuity, and product release (Champoux, 2001; Shuman, 1998).
All poxviruses encode an unusual type IB topoisomerase, which is a member of a family that is now known to include enzymes from mimivirus and many bacteria (Benarroch et al., 2006; Gubser et al., 2004; Hwang, Wang, and Bushman, 1998; Krogh and Shuman, 2002; Shuman, 1998; Upton et al., 2003). At about 33 kDa in size, these are by far the smallest topoisomerases known. All the poxvirus enzymes studied have been found to act at the core sequence 5′-(T/C)CCTT-3′ (base pairs +5 to +1; see Figure 1 for numbering of the DNA) together with optimal flanking sequences (Hwang, Burgin, and Bushman, 1999; Hwang, Wang, and Bushman, 1998; Shuman and Prescott, 1990; Tian et al., 2004). Purified enzymes have been studied from vaccinia, variola virus, molluscum contagiosum (MCV), leporipox (Palaniyar et al., 1996), and two entomopox viruses (MSV and AMV)(Bauer et al., 1977; Hwang, Rhodes, and Bushman, 2000; Hwang et al., 1999b; Hwang, Wang, and Bushman, 1998; Krogh et al., 1999; Perry et al., 2006; Petersen et al., 1997; Shuman, 1998). These studies have shown that phylogenetically diverse poxviruses encode enzymes with simliar specificities. Studies of sequence specific binding and DNA cleavage indicate that sequence-specific recognition is not important just for binding, but also for a conformational step after binding that activates the enzyme for transesterification to form the phosphotyrosine intermediate (Koster et al., 2005; Nagarajan et al., 2005; Shuman, 1998; Stivers, Shuman, and Mildvan, 1994a; Stivers, Shuman, and Mildvan, 1994b).
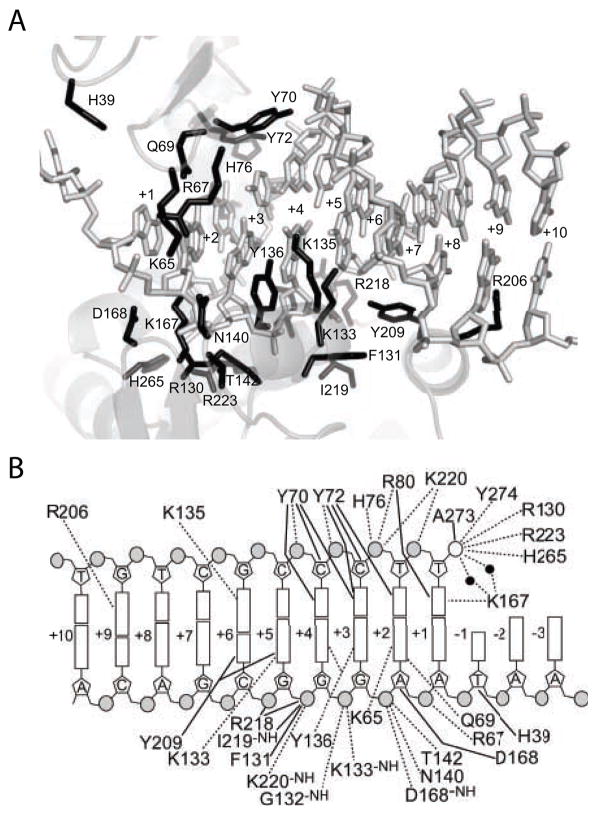
Figure 1.
The variola virus topoisomerase/DNA interface. A) Structure of the topoisomerase/DNA interface. DNA contact residues are shown in black. B) Diagram of the topoisomerase/DNA contacts. Dotted lines indicate hydrogen bonds of less than or equal to 3.3 Angstroms. Solid lines indicate Van der Waals contacts of less than or equal to 4.0 Angstroms.
The elucidation of X-ray structures of the variola virus toposiomerase bound to DNA in covalent and non-covalent complexes has clarified the contacts at the enzyme-DNA interface and suggested mechanisms for activation of catalysis (Perry et al., 2006). The topoisomerase is composed of two domains that wrap around the core 5′-(T/C)CCTT-3′ recognition sequence, forming a C-shaped clamp (Cheng et al., 1998; Cheng and Shuman, 1998; Hwang et al., 1999a; Perry et al., 2006; Sekiguchi and Shuman, 1994). A network of contacts extending from −1 to the +9 region stabilize the catalytic domain in an active conformation (Perry et al., 2006). This is consistent with biochemical studies, which indicated that the topoisomerase when bound covers a considerably larger site than just the conserved 5′-(T/C)CCTT-3′ (Hwang, Burgin, and Bushman, 1999; Hwang et al., 2006; Hwang, Wang, and Bushman, 1998; Sekiguchi and Shuman, 1994). Comparison of the structure of the catalytic domain in the presence and absence of DNA indicates that DNA contacts stabilize a major conformational change which assembles the active site (Cheng et al., 1998; Perry et al., 2006). A particularly notable feature in the topoisomerase-DNA complex is the formation of a new alpha-helix in the presence of DNA, which makes sequence specific contacts to the DNA major groove to base pairs +3 to +6 and thereby helps deliver one of the catalytic residues (R130) to the enzyme active site (Perry et al., 2006).
Previous studies have elucidated many key features of the DNA recognition sequence (Hwang, Burgin, and Bushman, 1999; Hwang, Wang, and Bushman, 1998; Shuman, 1991; Shuman and Prescott, 1990; Tian et al., 2004), but a complete functional characterization of the full site has not been reported. Such data is important both for clarifying the contributions of observed topoisomerase-DNA contacts and allowing analysis of topoisomerase sites in poxvirus genomes. Here we report a systematic study of the energetic contribution of each base in the variola virus topoisomerase DNA recognition site from position +10 to −3. As mentioned above, following DNA binding, the topoisomerase must undergo a conformation step to activate the cleavage reaction, and correct DNA contacts are required for activation. For this reason, we measured the role of DNA sequence not in binding assays, but in the forward rate of cleavage, which includes both specific binding and subsequent catalysis.
Results and Discussion
Measuring the rates of covalent complex formation on suicide substrates
The rate of the initial cleavage step of covalent complex formation can be determined using synthetic oligonucleotide substrates. In these assays, a short partially double stranded oligonucleotide duplex (16-mer annealed to a complementary 18-mer) was used as substrate. This DNA contained an optimal topoisomerase cleavage site or derivatives with single base-pair substitutions (Table 1). This sequence was derived from a favored cleavage site in the cloning vector pUC19 (Hwang, Burgin, and Bushman, 1999; Shuman and Prescott, 1990) and was referred to as sub a in (Hwang, Burgin, and Bushman, 1999). The oligonucleotide DNAs were synthesized so that only four bases extended 3′ of the scissile phosphate on the cleaved strand (Figure 2). After cleavage, the four base strand does not remain stably base paired, and so dissociates and is lost by dilution, thereby trapping the covalent complex (Figure 2A). To visualize reaction products, the 5′ end of the cleaved strand was end-labeled with 32P, the reactant DNA separated from covalent complexes by SDS-PAGE, and the products quantified by PhosphorImager. Time course studies were carried out to compare different DNA substrates (Figure 2B and 2C).
Table 1.
Rates of cleavage by the smallpox topoisomerase
Table 1. Positional weight matrix for DNA recognition by the variola virus topoisomerase derived from measurements of the forward rate of DNA cleavage.
| +10 | +9 | +8 | +7 | +6 | +5 | +4 | +3 | +2 | +1 | −1 | −2 | −3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T | C | G | C | C | C | T | T | A | T | T | |
| G | 35.6% | 100% | 66.7% | 222.2% | 100% | 0.43% | 0.18% | 0.55% | 0.04% | 0.75% | 214.8% | 527.8% | 47.2% |
| A | 36.1% | 12.7% | 105.6% | 183.3% | 66.7% | 0.31% | 0.015% | 0.63% | 0.69% | 1.4% | 100% | 83.3% | 94.4% |
| T | 100% | 9.4% | 100% | 77.8% | 13.3% | 9.4% | 0.89% | 0.45% | 100% | 100% | 0.72% | 100% | 100% |
| C | 83.3% | 2.1% | 38.9% | 100% | 10.5% | 100% | 100% | 100% | 1.3% | 0.44% | 51.1% | 9.4% | 88.9% |
A rate of 100% = 1.9 × 10−2/sec.
The sequence along the top is the reference sub a substrate. Single base substitutionsare as shown in the left column.
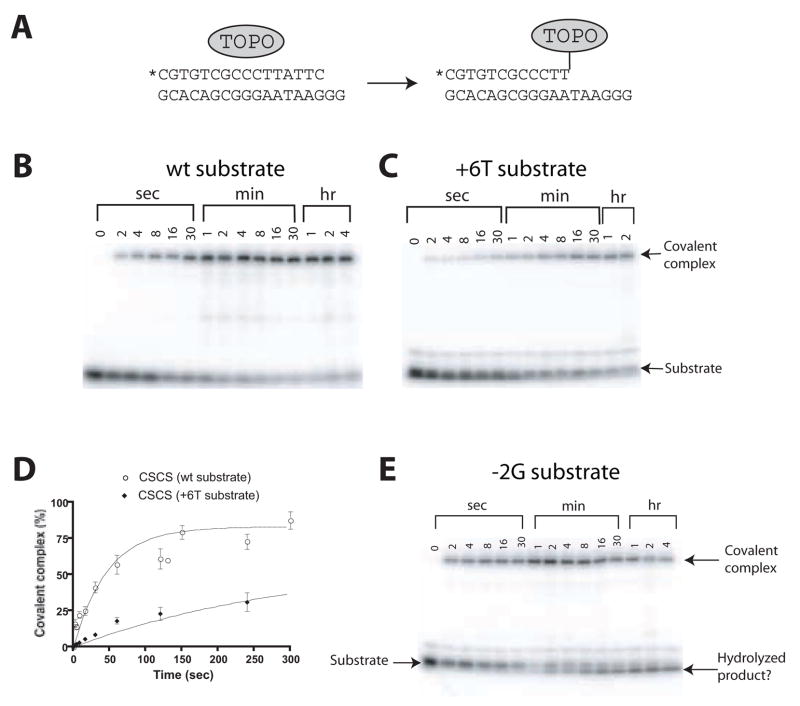
Figure 2.
Kinetic assays of single cycle DNA cleavage by the variola virus topoisomerase. A) Diagram of the assay method used, illustrating trapping of the covalent intermediate using the suicide substrate. The asterisk indicates the position of a 32P label on the 5′ end of the cleaved strand. B) Kinetic analysis of cleavage reactions in vitro. Covalent DNA-topoisomerase complexes were separated from unreacted DNA on SDS PAGE gels. C) Cleavage rate data for a DNA substrate with a Thy base at position +6. D) Example of quantitation of the rate of covalent complex formation. E) Breakdown of the topoisomerase-DNA covalent intermediate promoted by the -2G substituted suicide substrate.
As a source of topoisomerase activity, the variola virus topoisomerase derivative used in the DNA co-crystallization study was employed in the kinetic analysis to facilitate comparison with structural data. The enzyme contains two substitutions, C100S and C211S, that were found to be required for crystallization. The C211S-substituted enzyme was reduced in the forward rate of cleavage about 10-fold compared to the variola virus wild-type enzyme (Hwang et al., 2006), which is convenient here for allowing more accurate measurement of the faster rates. Comparison to previous data with vaccinia virus and MCV topoisomerase indicates that the relative rates, where tested, were comparable with the variola virus enzyme used here (data not shown). This supports the idea that the base substitutions in the variola virus topoisomerase studied, which are far from the DNA interface, had no influence on DNA recognition. The amount of topoisomerase used was shown in preliminary experiments to be sufficient for saturation on the wild-type substrate (data not shown).
Rates were determined by fitting curves to exponential models using non-linear regression. The experiment was carried out for each of the three possible nucleotide substitutions at positions +10 to −3, for a total of 39 sequence variants in addition to the optimized sub a sequence (Table 1). Most rates showed good fits to the exponential model (Figure 2D). All rate measurements were repeated at least twice, and errors were generally modest, though uncertainty was greater for the most defective substrates. The free energy change resulting from each base pair substitution (kcal/mol) could be calculated from
where km is the mutant rate and ks the rate for the reference sub a substrate (Table 2).
Table 2.
Free energy changes accompanying each base pair substitution (Kcal/mol)
| +10 | +9 | +8 | +7 | +6 | +5 | +4 | +3 | +2 | +1 | −1 | −2 | −3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T | C | G | C | C | C | T | T | A | T | T | |
| G | 0.61 | 0.00 | 0.24 | −0.47 | 0.00 | 3.23 | 3.74 | 3.08 | 4.64 | 2.90 | −0.45 | −0.99 | 0.44 |
| A | 0.60 | 1.22 | −0.03 | −0.36 | 0.24 | 3.42 | 5.22 | 3.00 | 2.95 | 2.53 | 0.00 | 0.11 | 0.03 |
| T | 0.00 | 1.40 | 0.00 | 0.15 | 1.20 | 1.40 | 2.80 | 3.20 | 0.00 | 0.00 | 2.92 | 0.00 | 0.00 |
| C | 0.11 | 2.29 | 0.56 | 0.00 | 1.34 | 0.00 | 0.00 | 0.00 | 2.57 | 3.21 | 0.40 | 1.40 | 0.07 |
However, for substrates containing a Gua base at −1 or −2 on the scissile strand, a different behavior was seen (Figure 2E). The covalent complex formed, then decayed, so that the topoisomerase became released from DNA without religation. The mechanistic basis for this reaction is under investigation. To extract the forward rates for the −1 Gua and −2 Gua substrates, only the early part of the time course was used, before extensive breakdown of the covalent complex took place.
Effects of substrate sequence on cleavage rate
The effects of each substitution, and their structural interpretations, are summarized below.
Position +10
The strongest effects of substitution at this position was about 3-fold, which corresponds to less than 0.6 kcal/mol, and no direct contacts were seen to this base in the topoisomerase-DNA complex structures. We conclude that this base is outside of the protein-bound region, and so maps an upstream boundary.
Position +9
A Gua base is strongly favored at +9. Substitution of Ade at this position reduced the rate about 8-fold (up to 2.29 kcal/mol), and the pyrimidine substitutions showed greater reductions. In the X-ray structures of the variola virus topoisomerase-DNA complex, the Gua base is contacted by R206, which makes a canonical bidentate hydrogen bond in the major groove. A functional study of the R206 side chain reveals that it is responsible for sequence discrimination at this position (Hwang et al., 2006).
Positions +8 and +7
Substitutions at +8 and +7 had no strong effects, and no contacts to these bases were evident in the topoisomerase-DNA structures.
Position +6
The Gua at position +6 in the structure is contacted from the major groove side by residues K135 and Y209. At this position, both Gua and Ade are well tolerated (Shuman, 1991). The two pyrimidine substitutions resulted in a reduction in cleavage of at least 7.5-fold. Y209 interacts via a Van der Waal’s contact with the Cyt, and may be able to interact similarly with the Thy. K135 contacts the Gua at the N7 position in the structure, and the same contact is possible to Ade, explaining why this substitution is well tolerated. The K135 side chain is in different positions in the two available topoisomerase-DNA complexes (Perry et al., 2006), also suggesting flexible recognition.
Position +5
At +5, substituting the C with T resulted in a 11-fold reduction in activity, while the purine substitutions were both down at least 200-fold. The Gua on the uncleaved strand at +5 is contacted by both the Y209 and K133, in part by hydrogen bonding of K133 to the N7 position. Again similar contacts are possible at the Ade and Gua N7 bases at +5, and the K133 interaction may be flexible as judged by comparing covalent and non-covalent topoisomerase-DNA structures, explaining why Ade is tolerated.
Position +4
All substitutions at +4 are down 110-fold (up to 5 kcal/mol). Base pairs +1 to +4 are closely approached by the topoisomerase on both strands of the DNA helix due to closure of the topoisomerase clamp around the DNA. Y70 lies flat along the Cyt and hydrogen bonds to the phosphate backbone. Thy may not be allowable at this position due to steric clashes with the Thy methyl group. This base is also contacted from the catalytic domain by Y136, and a network of hydrogen bonds, some water mediated, are present in this region.
Position +3
Substitutions at +3 are all down at least 158-fold. The Cyt at +3 is contacted from the amino-terminal domain by Y70 and Y72, and the sugar-phosphate backbone on the other strand is contacted from the catalytic domain by Y136, G132, and K133. Y136 also contacts the Gua base via its OH group.
Position +2
Substitutions at +2 are all down at least 77-fold. Residues K65 and Q69 hydrogen bond to the Ade base, and Y72 forms a Van der Waals interaction with Thy +2. Several further residues contact the sugar (D168) and phosphates (K220, H76, R80, R67) at +2.
Position +1
Substitutions at +1 are all down at least 71-fold. This base pair is adjacent to the scissile phosphate, and engages in a network of interactions with the topoisomerase. The +1 Thy makes Van der Waals interactions with R80 and a hydrogen bond with the active site residue K167, and the phosphate 5′ of the Thy +1 forms a hydrogen bond with K220. A study of substrates containing 5-fluro-2′-deoxyuridine substituted for +1 Thy showed that these were well accommodated by the toposiomerase, and that the 5-F-dUrd was in a dynamic equilibrium between stacked and unstacked conformations in both the noncovalent and covalent complexes (Kwon et al., 2002). The phosphate 3′ of Thy +1 participates in the phospho-tyrosine linkage with Y274, and in the structures this phosphate is contacted by the active site residues R130, K167, R223, and H265.
Position −1
A substitution of Thy on the top strand at −1 reduced the rate 139-fold, but other substitutions were well tolerated. For DNA positions −1 to −3, the nature of protein contacts is less certain because the topoisomerase-DNA structures lack the DNA 3′ of the scissile phosphate on the cleaved-strand (top strand in Table 1). A backbone contact is donated by H39 to the uncleaved strand, but no further contacts are evident from the structure, and the mechanism of discrimination against Thy (on the top strand) is not clear. It is likely that during religation the toposiomerase contacts the returning strand to align the 5′-hydroxyl with the phosphotyrosine for transesterification (i. e. −1 to −3 on the cleaved strand).
Position −2
At position −2, substitution of Cyt for Thy reduced the cleavage rate 11-fold, but Ade and Gua were well tolerated. In fact, the Gua substitution increased the cleavage rate 5-fold. No contacts to the uncleaved strand are evident at this position in the structures (the cleaved strand is missing). The mechanism of sequence-specific recognition at −2 is uncertain.
Position −3
Changes at −3 have a 2-fold effect at most (0.44 kcal/mol at most), and there were no obvious contacts to the structure in this region, indicating that −3 is the down stream boundary of the DNA region recognized in a sequence-specific fashion.
In summary, the mutagenesis study generally indicated that protein-DNA interactions identified as important in the structures scored as sensitive to base-pair substitutions. Positions that were not obviously contacted in the structure (+10, +8, +7, and −3) did not show strong responses to base pair substitutions. Several examples of flexible recognition were noted, including position +5 and +6. Both of these two interactions appear to be mediated at least in part by contacts to the purine N7 position which is shared between Gua and Ade.
Calculating a topoisomerase “action matrix” based on cleavage rates
A second goal in carrying out the comprehensive study of DNA recognition was to map topoisomerase sites in poxvirus genomes. Topoisomerase sites could then be tested for association with other types of genomic annotation, allowing investigation of the biological role of the topoisomerase.
To accomplish this, we used the rate measurements in Table 1 to generate a matrix describing the relative favoring of each base within the 13 bp sequence studied. We then transformed the rate matrix into a probability matrix by dividing the rate for each nucleotide substitution by the sum of activity levels at that position (Table 3). The sum of values at each base position was constrained to add up to 1. Such probability matrices are commonly used to represent the DNA binding specificity of a transcription factor. Typically several binding sites are aligned and for each position the fraction of each of the 4 observed bases is used in the probability matrix. All binding sites are treated equally because the binding affinity is usually unknown. Our approach instead takes advantage of the cleavage rate measurements to generate a quantitative “action matrix”.
Table 3.
Probability matrix based on the cleavage rate data
| +10 | +9 | +8 | +7 | +6 | +5 | +4 | +3 | +2 | +1 | −1 | −2 | −3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T | C | G | C | C | C | T | T | A | T | T | |
| G | 0.140 | 0.805 | 0.214 | 0.381 | 0.525 | 0.004 | 0.0018 | 0.005 | 0.000 | 0.007 | 0.586 | 0.733 | 0.143 |
| A | 0.142 | 0.102 | 0.339 | 0.314 | 0.350 | 0.003 | 0.0001 | 0.006 | 0.007 | 0.014 | 0.273 | 0.116 | 0.286 |
| T | 0.392 | 0.076 | 0.321 | 0.133 | 0.070 | 0.085 | 0.0088 | 0.004 | 0.980 | 0.975 | 0.002 | 0.139 | 0.303 |
| C | 0.327 | 0.017 | 0.125 | 0.171 | 0.055 | 0.908 | 0.9893 | 0.984 | 0.013 | 0.004 | 0.139 | 0.013 | 0.269 |
Given the action matrix describing topoisomerase activity, we can computationally estimate the activity level of any 13 nucleotide long DNA sequence. For instance, to estimate the activity of DNA sequence 5′-AAAAAAAAAAAAA-3′, we multiply the probability of A at each position using data from Table 3, i.e., 0.142*0.102*0.339*….*0.286 = 8.6e-19. More formally, the activity of a 13 nt long DNA sequence is estimated by the probability that the given sequence S is generated by the probability matrix M.
Next we calibrate the score to a value between 0 and 1 as follows. For the matrix we compute the minimum score (MIN) achievable by the matrix by multiplying together each of the smallest values in each column. We also compute the maximum score, MAX, achievable by the matrix by multiplying together the largest values in each column. The calibrated score is computed as:
We score every 13 nt long window in the viral genome on both strands and output the location of the windows that achieve a threshold score.
Poxvirus genome sequences were downloaded from Poxvirus Bioinformatics Resource Center (http://www.poxvirus.org/index.asp) and analyzed. For the comparative study, we selected a collection of orthopox viruses (variola virus, vaccinia, cowpox, camelpox, ectromelia, and monkeypox), all of which encode closely related topoisomerase proteins, and representatives of two other poxvirus groups, AME (a Betaentomopoxvirus) and MCV (a Molluscipoxvirus), for which in vitro studies have confirmed that the topoisomerase is active on the 5′-(T/C)CCTT-3′ sequence (Hwang, Burgin, and Bushman, 1999; Petersen et al., 1997). In carrying out this analysis, we make the assumption that the site specificity on oligonucleotide substrates is meaningful in the context of viral replication in vivo. The topoisomerase action matrix was scanned along each genome and matches above 0.85 recorded. Inspection of the sequences returned indicated that sites achieving 0.85, while close in sequence to highly active sites, nevertheless often contained significantly suboptimal bases that would be expected from data in Table 1 to reduce activity. Sequences at 0.88 or higher were generally free of adverse bases. A table of all sites scoring above 0.85 in the genomes studied can be found in Supplementary Material S1.
Experimental validation of predicted topoisomerase sites
Studies of DNA relaxation by poxvirus topoisomerases in vitro have often used the bacterial plasmid pUC19 as a substrate, and in the course of this work the topoisomerase recognition sites have been carefully quantified (Hwang, Wang, and Bushman, 1998; Shuman, 1991; Shuman and Prescott, 1990). To assess whether the PWM determined above is a reliable predictor of favored sites, we used it to predict sites in pUC19, then compared the calls to experimental results.
A single site was strongly favored in pUC19 by the vaccinia virus and MCV topoisomerases (named sub a in (Hwang, Burgin, and Bushman, 1999)), which was mapped by direct analysis of in vitro cleavage products. Less frequently cleaved minor sites were also identified by synthesizing oligonucleotides matching candidate regions of pUC19 and testing their activity as substrates in vitro. Under standard reaction conditions, six oligonucleotides matching minor pUC19 cleavage sites formed covalent complexes with efficiencies ranging from 2% to 0.2% that of sub a (Hwang, Wang, and Bushman, 1998; Shuman, 1991; Shuman and Prescott, 1990). Upon analysis of pUC19 using the above PWM, the sub a site received the highest score (0.94). The second most active site in vitro received the second highest score of 0.86 (named sub g in (Hwang, Burgin, and Bushman, 1999)). Other minor sites identified computationally in the 0.86-0.85 range did not match experimentally determined minor sites. We conclude that the computational analysis accurately identifies highly favored sites and is partially successful in the 0.85-0.86 range.
Distribution of topoisomerase sites in poxvirus genomes
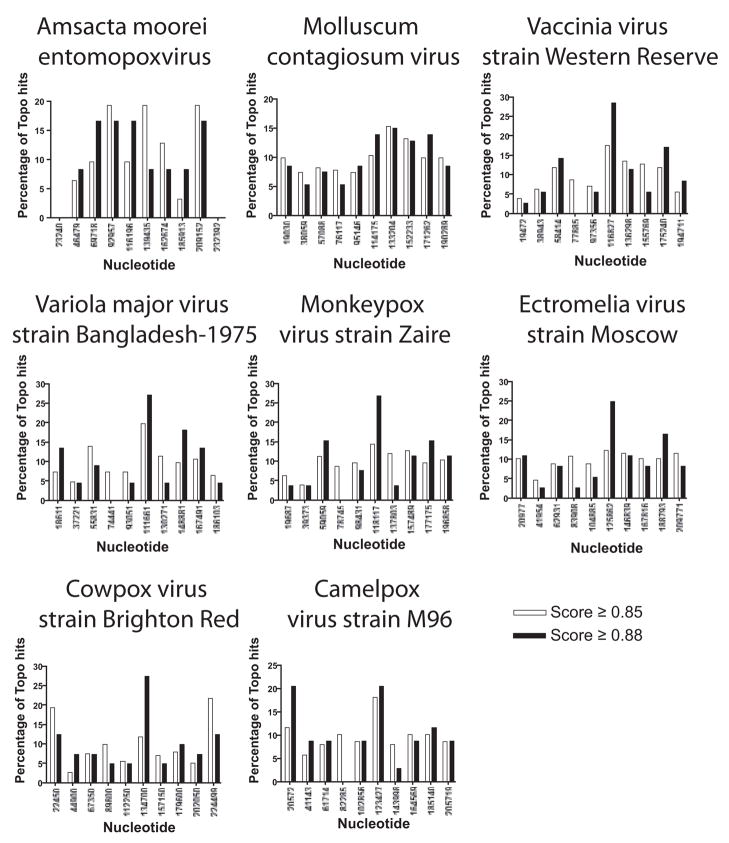
As a first step in investigating the distribution of topoisomerase sites, the poxvirus genomes were each divided into 10 intervals, and the proportions of sites scoring >0.85 or >0.88 were plotted (Figure 3). Most intervals in each genome contained at least one topoisomerase site scoring 0.88 or better, indicating that highly active sites could be found all along the genomes, consistent with a requirement for local release of superhelical tension.
Figure 3.
Locations of matches to the topoisomerase action matrix in poxvirus genomes. Each genome was divided into 10 bins and the proportion of all sites scored for each bin. Data for matches to the PWM achieving 0.85 are shown in white, matches achieving 0.88 are shown black. For statistical comparison, 2,500 random sites were generated in each poxvirus genome, and the distribution of topoisomerase sites in each bin was compared to the distribution of random sites using Chi Square, which showed significant differences from random distribution for vaccinia and cowpox.
A test of the overall frequency of sites in poxvirus genomes showed no statistically significant enrichment compared to a control DNA not exposed to the poxvirus topoisomerase. Specifically, analysis of phage T4 DNA using the above action matrix showed no statistically significant difference in the number of sites per kilobase compared to the poxvirus genomes (0.7 sites per kilobase, P=0.3; Wilcoxon Signed Rank test for the T4 value of sites/kb versus the collection of poxvirus site/kb values).
We next investigated whether topoisomerase sites were enriched in protein coding regions, since these regions will likely be more actively transcribed on average than noncoding regions. No consistent pattern was seen. Vaccinia virus and ectromelia virus showed a significant surplus of topoisomerase sites in coding regions compared to random distribution, but cowpox virus showed a surplus outside coding regions, and the other five showed no significant trend (Binomial test, analyzed for sites scoring 0.85 and above, data not shown). We conclude that there is no conserved relationship of topoisomerase sites to protein coding regions.
Notably, for all the poxviruses, genomic intervals with surplus density of high-scoring sites were present near the center of the genomes. This was particularly clear-cut for the orthopox viruses (cowpox, camelpox, vaccinia, variola, monkeypox, and ectromelia). The predicted distribution of sites was significantly different from a computationally generated random set of 2500 sites for the vaccinia and cowpox genomes (P<0.05 by Chi Square).
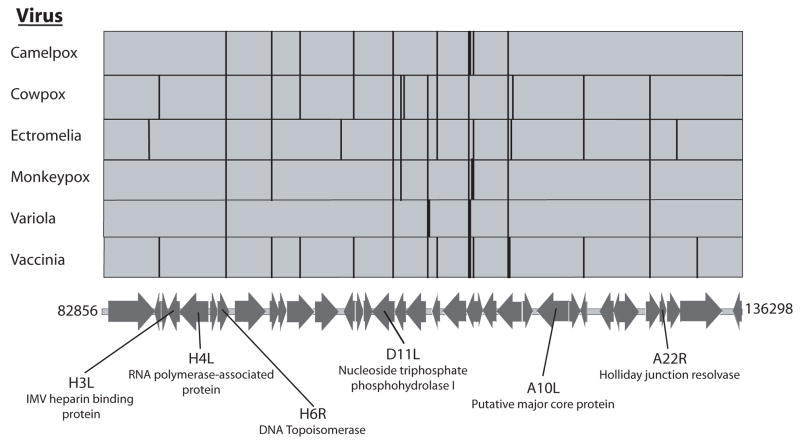
Figure 4 shows the location of the central cluster of predicted topoisomerase sites relative to conserved orthopox genes. Predicted sites are found both within genes and in intergenic regions. For sites within genes, no bias was detected for orientation of the topoisomerase site with respect to the orientation of transcription. Many sites are identical with respect to their DNA sequence and position relative to the underlying coding sequences among multiple orthopox genomes.
Figure 4.
Genetic map of the central region of the vaccinia virus genome annotated with the locations of high-scoring topoisomerase sites for several orthopoxes. The vertical lines above the genetic map show the locations of the sites. The base pair coordinates refer to the vaccinia genome (strain Copenhagen). There are slight differences in the sequences among the genomes in the figure. To emphasize the similarities, topoisomerase sites that were of identical sequence and identical placement within a gene were aligned by hand. Only the major open reading frames are shown.
The placement of the topoisomerase sites were then compared to predictions for early promoters, early terminators, and late promoters in the vaccinia virus genome (Copenhagen strain). Vaccinia was chosen as the reference genome since the most experimental data for promoter motifs was available for this virus. Vaccinia virus promoters were predicted using a newly developed algorithm based on interpolated context models (ICMs) (Delcher et al., 1999). These models take into account not only compositional features of known promoters, but also inter-dependencies between base pairs. ICM models derived from experimentally verified early or late promoters were used to predict promoters for vaccinia virus genes (Wang, C., Hendrickson, R.C., and Lefkowitz, E.J., manuscript in preparation). We predicted 127 topoisomerase sites (0.85 criteria), 128 early promoters, 143 late promoters, and 685 early termination sites. For terminators we used the sequence TTTTTNT, and did not take into account possible effects of sequence context. For 60 genes, both early and late promoters were predicted.
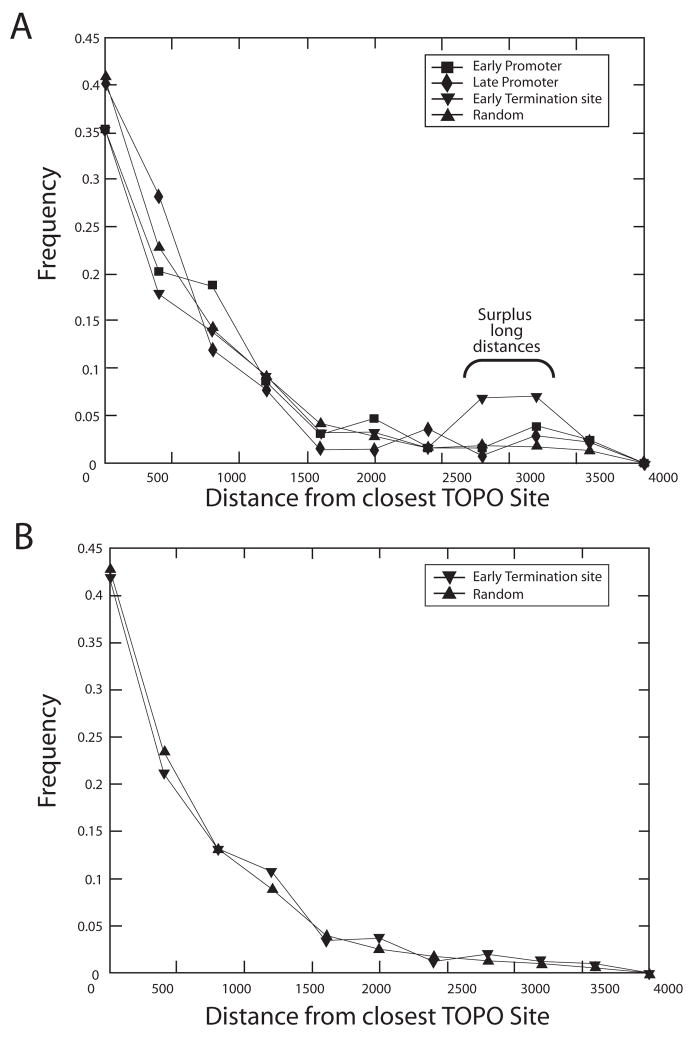
We then analyzed the relationship of topoisomerase sites to promoters and terminators. For each promoter or termination site, we computed the distance to the closest topoisomerase site. Additionally, we generated 10,000 random locations on the vaccinia virus (Copenhagen strain) genome and measured the distance to the closest topoisomerase site for each. Figure 5A plots the collection of lengths to the nearest topoisomerase site for the early and late promoters, the late terminators, and the random sites. The collection of distances between topoisomerase sites and the promoters were not significantly different from the distances for the random sites. However, the distances between topoisomerase sites and early termination sites showed a peak at longer lengths (Mann-Whitney U test based P-value ~ 0). The analysis was repeated for the vaccinia virus WR strain and essentially identical results obtained (data not shown).
Figure 5.
Frequency histograms analyzing lengths of DNA intervals between predicted topoisomerase sites and predicted early promoters, early terminators, and late promoters in the vaccinia genome (strain Copenhagen). A) The plot shows the distance to the closest topoisomerase site with a match of 0.88 or better. There is no significant favoring or disfavoring of early or late promoters. There is a significant disfavoring of early terminators (P=0.000; Mann-Whitney U test), indicated by the surplus long distances (bracket on the figure). B) The same analysis as above but repeated on vaccinia COP genome with the 20 kb removed from each end. The terminal region is A/T-rich and rich and densely populated with early terminators. Note that the long intervals between early terminators and topoisomerase sites are no longer present, indicating anticorrelation between the two at the genome termini.
The ends of the vaccinia virus-COP genome are AT-rich and thus the termination site signal (TTTTTNT) tends be enriched in these regions. An explanation for the apparent longer distances to the topoisomerase site is that the highly AT-rich character of the region near the ends of the genome causes an apparent enrichment for the TTTTTNT site. To test this, we repeated our analysis after removing 20kb of DNA from each end, and found that the collection of distances from topoisomerase sites to termination sites became indistinguishable from that for distances between topoisomerase sites and random sites (Figure 5B).
The overall abundance of topoisomerase sites in the terminal 10 kb regions of the eight genomes studied in Figure 3 was compared to the abundance in the more central regions. There was no significant difference in the mean number of sites per kilobase in the terminal versus central regions (Mann-Whitney U Test comparison of means; analyzed for sites scoring 0.85 and above, data not shown).
Regions of convergent transcription have the potential to accumulate superhelical stress, so we investigated possible enrichment of topoisomerase sites in such regions in the vaccinia virus (strain WR) using the above promoter predictions. Predicted convergent promoters were cataloged, and the frequency of topoisomearse sites compared in such regions to genome regions that were not subject to convergent transcription. A few genomic regions were judged to be ambiguous and not included in the analysis. No statistically significant enrichment was detected (Binomial test on sites scoring 0.85 and above, data not shown).
Of possible interest is the finding that many poxvirus genomes have a topoisomerase recognition site within the topoisomerase gene itself. Of the eight genomes analyzed above, seven have topoisomerase sites scoring 0.85 or better within the topoisomerase coding region. Transcription of the bacterial gyrase gene is controlled by a feedback loop in which changes in superhelical density influence the transcription rate (Menzel and Gellert, 1983). The finding of topoisomerase sites in the topoisomerase coding region raises the question of whether a related system might operate in poxviruses.
Thus the main findings from this analysis were that 1) predicted topoisomerase recognition sites are fairly evenly distributed along poxvirus genomes, 2) some clustering is seen in the central region, 3) there was no obvious clustering relative to early or late promoters, and 4) predicted vaccinia virus topoisomerase sites were anti-correlated with early termination sequences at the edges of the genome, probably a result of enrichment of terminator sequences in these regions.
Models for the function of poxvirus topoisomerases in vivo
The poxvirus genome is linear, and so could potentially relieve superhelical tension generated during transcription by revolving around the long axis of the DNA. However, the viral DNA is likely bound to proteins that would inhibit free rotation, necessitating action of a topoisomerase to relieve superhelical stress in constrained topological domains. Predicted highly active topoisomerase sites were found distributed along the length of all the poxvirus sequences analyzed, consistent with a requirement for local release of superhelical tension at many points along the genome.
Deleting the topoisomerase gene selectively impairs early transcription (Da Fonseca and Moss, 2003). During the early phase of infection the viral genome is tightly packaged in factory centers, so that topological constraints to rotation may be particularly strict at this time. Thus the finding of a cluster of topoisomerase sites in late genes in the center of the genome was unexpected. Possibly it is important to have a “swivel point” located near the center of the genome to allow release of superhelical tension, and so the relationship with late genes is only a result of their conserved central position. However, it is not ruled out that the topoisomerase sites are preserved among poxviruses in late genes because some other function of the DNA primary sequence, for example, the amino-acid coding capacity.
Going forward, the predicted topoisomerase sites can be used by members of the poxvirus field for comparison to new tracks of genomic analysis as they are generated. This will allow investigation of topoisomerase function in light of ongoing genomic annotation.
Materials and Methods
Topoisomerase protein
The gene encoding the variola virus topoisomerase protein used here was initially generated by introducing three nucleotide changes resulting in three amino acid substitutions into a gene encoding the closely related vaccinia virus topoisomerase (D24N, E47G, anda E159K). The variola virus topoisomerase coding region was further modified to encode the C100S and C211S substitutions which facilitated protein crystallization as described in (Perry et al., 2006). The CSCS variola virus topoisomerase was purified as described in (Perry et al., 2006).
Cleavage assay with suicide substrates
The 16-mer oligonucleotide shown in Figure 2 was end labeled by treatment with 32P gamma-ATP and T4 polynucleotide kinase. After purification over a G50 spin column, the oligonucleotide was annealed to the 18-mer complementary bottom strand also shown in Figure 2A. Cleavage reaction mixtures contained (per 20 μl) 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 8 pmol of 16-mer/18-mer DNA, and 80 pmol topoisomerase. After incubation at 25°C, reactions were stopped by addition of 2% SDS. Reconstruction experiments were carried out to verify that the reactions were indeed stopped by this treatment. Reaction mixtures were subjected to electrophoresis through a 12% polyacrylamide gel containing 0.1% SDS. The extent of covalent adduct formation was quantitated using a STORM PhosphoImager. The cleavage rate was extracted by fitting the measured product decrease as a function of time to the exponential decay equation (100 - % of cleaved product) = 100e−kt using non-linear regression with GraphPad PRIZM version 4 software.
Bioinformatic analysis
The methods used were as described in the text and in (Delcher et al., 1999). The poxvirus sequences analyzed were obtained from http://www.poxvirus.org/index.asp. The identification and analysis of vaccinia early promoters, early terminators, and late promoters will be reported elsewhere by R.C.H. and E.J.L.
Supplementary Material
Acknowledgments
We thank members of the Bushman and Van Duyne laboratories for helpful discussions. This work was supported by a grant from the Mid-Atlantic Regional Center of Excellence for Biodefense Research to F. D. B., and an NIH/NIAID Bioinformatics Resource Center Contract HHSN266200400036C to E.J.L. G.V. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Data
Location of predicted topoisomerase sites in poxvirus genomes and pUC19 (0.85 cut-off).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauer WR, Ressner EC, Kates J, Patzke JV. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc Natl Acad Sci U S A. 1977;74(5):1841–5. doi: 10.1073/pnas.74.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch D, Claverie JM, Raoult D, Shuman S. Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria. J Virol. 2006;80(1):314–21. doi: 10.1128/JVI.80.1.314-321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA Topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- Cheng C, Shuman S. A Catalytic Domain of Eukaryotic DNA Topoisomerase I. J Biol Chem. 1998;273:11589–11595. doi: 10.1074/jbc.273.19.11589. [DOI] [PubMed] [Google Scholar]
- Da Fonseca F, Moss B. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proc Natl Acad Sci U S A. 2003;100(20):11291–6. doi: 10.1073/pnas.1534874100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nuc Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser C, Hue S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. 2004;85(Pt 1):105–17. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- Hwang Y, Burgin A, Bushman FD. DNA Contacts Stimulate Catalysis by a Poxvirus Topoisomerase. J Biol Chem. 1999;274:9160–9168. doi: 10.1074/jbc.274.14.9160. [DOI] [PubMed] [Google Scholar]
- Hwang Y, Minkah N, Perry K, Van Duyne GD, Bushman FD. Regulation of catalysis by the smallpox virus topoisomerase. J Biol Chem. 2006 doi: 10.1074/jbc.M608858200. Epub. [DOI] [PubMed] [Google Scholar]
- Hwang Y, Park M, Fischer WH, Bushman F. Domain structure of the type-1B topoisomerase encoded by molluscum contagiosum virus. Virology. 1999a;262:479–491. doi: 10.1006/viro.1999.9920. [DOI] [PubMed] [Google Scholar]
- Hwang Y, Rhodes D, Bushman FD. Rapid Microtiter Assays for Poxvirus Topoisomerase, Mammalian Type IB Topoisomerase, and HIV Integrase: Application to Inhibitor Isolation. Nuc Acids Res. 2000;28:4884–4892. doi: 10.1093/nar/28.24.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Rowley D, Rhodes D, Gertsch J, Fenical W, Bushman FD. Mechanism of inhibition of a poxvirus topoisomerase by the marine natural product sansalvamide A. Mol Pharmacol. 1999b;55:1049–1053. doi: 10.1124/mol.55.6.1049. [DOI] [PubMed] [Google Scholar]
- Hwang Y, Wang B, Bushman FD. Molluscum contagiosum virus topoisomerase: purification, activities and response to inhibitors. J Virol. 1998;72:3401–3406. doi: 10.1128/jvi.72.4.3401-3406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker T. DNA Replication. W. H. Freeman and Company; New York: 1991. [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434(7033):671–4. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Cheng C, Burgin A, Shuman S. Melanoplus sanguinipes entomopox DNA topoisomerase: site-specific DNA transesterification and effects of 5 ′ bridging phosphorothiolates. Virology. 1999;25:441–451. doi: 10.1006/viro.1999.0022. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Shuman S. A poxvirus-like type IB topoisomerase family in bacteria. Proc Natl Acad Sci U S A. 2002;99(4):1853–8. doi: 10.1073/pnas.032613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, Jiang YL, Song F, Stivers JT. 19F NMR studies of vaccinia type IB topoisomerase. Conformational dynamics of the bound DNA substrate. J Biol Chem. 2002;277:353–358. doi: 10.1074/jbc.M109450200. [DOI] [PubMed] [Google Scholar]
- Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34(1):105–13. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Nagarajan R, Kwon K, Nawrot B, Stec WJ, Stivers JT. Catalytic phosphoryl interactions of topoisomerase IB. Biochemistry. 2005;44:11476–11485. doi: 10.1021/bi050796k. [DOI] [PubMed] [Google Scholar]
- Palaniyar N, Fisher C, Parks R, Evans DH. SFV topoisomerase: sequence specificity in a genetically mapped interval. Virology. 1996;221(2):351–4. doi: 10.1006/viro.1996.0385. [DOI] [PubMed] [Google Scholar]
- Perry K, Hwang Y, Bushman FD, Van Duyne G. Structural basis for specificity in the poxvirus topoisomerase. Mol Cell. 2006;23:343–354. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Hall RL, Moyer RW, Shuman S. Characterization of a DNA Topoisomerase Encoded by Amsacta moorei Entomopoxvirus. Virology. 1997;230:197–206. doi: 10.1006/viro.1997.8495. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S. Vaccinia Topoisomerase Binds Circumferentially to DNA. J Biol Chem. 1994;269:31731–31734. [PubMed] [Google Scholar]
- Shuman S. Site-specific DNA Cleavage by Vaccinia Virus DNA Topoisomerase I: Role of nucleotide sequence and secondary structure. J Biol Chem. 1991;266:1796–1803. [PubMed] [Google Scholar]
- Shuman S. Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim Biophys Acta. 1998;1400:321–339. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- Shuman S, Prescott J. Specific DNA Cleavage and Binding by Vaccinia Virus DNA Topoisomerase I. J Biol Chem. 1990;265:17826–17836. [PubMed] [Google Scholar]
- Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA Topoisomerase I: Single-Turnover and Steady-State Kinetic Analysis of the DNA Strand Cleavage and Ligation Reactions. Biochemistry. 1994a;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA topoisomerase: kinetic evidence for general acid-base catalysis and a conformational step. Biochemistry. 1994b;33:15449–15458. doi: 10.1021/bi00255a027. [DOI] [PubMed] [Google Scholar]
- Tian L, Sayer JM, Jerina DM, Shuman S. Individual nucleotide bases, not base pairs, are critical for triggering site-specific DNA cleavage by vaccinia topoisomerase. J Biol Chem. 2004;279(38):39718–26. doi: 10.1074/jbc.M407376200. [DOI] [PubMed] [Google Scholar]
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J Virol. 2003;77(13):7590–600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Annu Rev Biochem. Vol. 65. Annual Reviews, Inc.; New York: 1996. DNA Topoisomerases; pp. 635–692. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.