Abstract
The flap conformations of two drug resistant HIV-1 protease constructs were characterized by molecular dynamic (MD) simulations and distance measurements with pulsed electron paramagnetic resonance (EPR) spectroscopy. MD simulations accurately regenerate the experimentally determined distance profiles and provide structural interpretations of the EPR data. The combined analyses show that the average conformation of the flaps, the range of flap opening and closing and the flexibility of the flaps differ markedly in HIV-1PR as multiple mutations arise in response to antiviral therapy, providing structural insights into the mechanism of inhibitor resistance.
Human Immunodeficiency Virus Type 1 protease (HIV-1PR) is an enzyme responsible for gag-pol processing, an essential step in viral maturation and the lifecycle of HIV-1. Inhibition of the activity of HIV-1PR results in immature virus particles that are non-infectious.1 As such, this protein represents a major target of AIDS antiviral therapy. Emerging resistance to antiviral inhibitor cocktails, due to high viral mutation rates, represents a significant challenge in AIDS treatment.2 Analysis of data from the Stanford Drug Resistance Database3 shows that while polymorphisms in the sequence of HIV-1PR naturally occur, there are regions in the protein sequence that appear invariant under normal evolutionary pressures. These invariant regions coincide with the structural elements of the dimer interface, the active site floor, the P3-P3′ substrate binding region and the β-hairpin loops (AKA, flaps). Strikingly, upon exposure to protease inhibitor (PI) cocktail treatment, numerous mutations develop, with high occurrences near residues 40–56 and 80–90, which correspond to the hairpin flaps and the P3-P3′ substrate binding cleft; respectively (data shown in Supp Info). Amino acid substitutions arise in these regions of the protein from random mutations that alter the ability of a given inhibitor to bind as tightly to the active site pocket, allowing for effective protease function with subsequent viral maturation and proliferation of the mutation. Many of these mutations also alter the kinetics of the protease for the multiple polypeptide cleavage sequences in the gag-pol polypeptide.4–7
It can be readily understood how mutations within the active site pocket reduce inhibitor effectiveness considering that many of the current PIs have been specifically designed to bind tightly to the shape of the active site cavity. However, the mechanism by which mutations that are NOT within the active site cavity modulate PI efficiency remains uncertain. It has been hypothesized that mutations in the elbow and flap regions (residues 36 to 58) may alter either the conformation of the flaps defined as the degree of flap opening or closing, or the mobility of the flaps, or both.4–6
We have previously shown that site-directed spin labeling (SDSL) and pulsed double electron-electron resonance (DEER) electron paramagnetic resonance (EPR) spectroscopy can distinguish between conformations of the flaps in the inhibitor bound “closed” state and the apo-state of HIV-1PR.8 Molecular dynamics (MD) simulations of flap motion in the LAI' (the prime indicates K55C-MTSL and active site mutation D25N, further details given in Supp Info) sequence have reproduced the DEER-based distance distribution profiles and provide a necessary link that correlates the EPR distances to structural and dynamic features of the flaps.9 From the most probable distance and the distance distribution profiles, information about the ensemble flap conformations in solution is obtained.
Here, we show that mutations that arise in response to PI treatment alter the flap conformations in the apo-state, defined as the conformations sampled in the absence of substrate/inhibitor. Specifically, the V6 and MDR769 drug-resistance constructs were investigated.6,10 Figure 1 shows the location of mutations in these constructs relative to the LAI sequence. Because HIV-1PR is a homodimer, generation of a single cysteine mutant for spin labeling provides a pair of spin labels for DEER measurements, where the magnitude of the magnetic dipolar coupling of the unpaired nitroxide electrons, which scales as 1/r3, is detected from analysis of the modulation of the spin echo amplitude.11–13 Shown in Fig. 2a are the DEER echo curves and corresponding distance distribution profiles for spin labeled constructs of LAI', V6' and MRD769'.
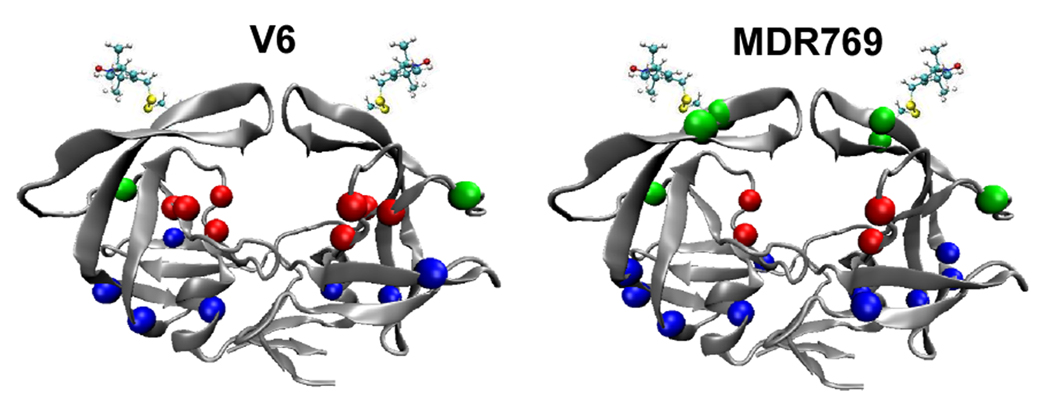
Figure 1.
Ribbon diagrams of HIV-1PR in the semi-open conformation (1HHP) with the nitroxide spin probe, MTSL, appended at site K55C. Colored spheres represent the Cα position of mutations relative to LAI in V6 (left) and MDR769 (right) in the active site cavity, non-active site region, and flaps/elbows shown in red, blue and green; respectively. Diagrams were rendered with VMD.
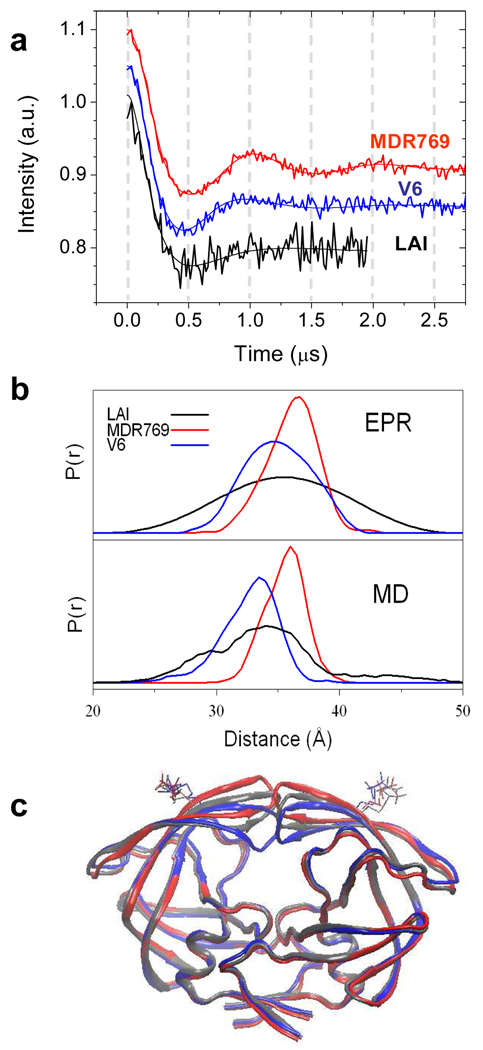
Figure 2.
(a) DEER echo curves (vertically offset by 0.05 a.u.) and best solution from Tikhonov regularization (TKR) methods11,13 for HIV-1PR samples labeled at site K55C with MTSL. (b) Distance distribution profiles from TKR of EPR data (upper) and N-N distance profiles from MD (lower). (c) Average structures sampled during MD simulations of the 3 sequences. For clarity, only the backbone ribbon and Cys-MTSL side chain are shown. KEY: LAI' “wild-type” (black); V6' (blue); MDR769' (red).
It is apparent from the distance distribution profiles (Fig. 2b) that the conformation of the flaps in V6' and MDR769' vary from those of LAI'. The most probable distance between spin labels in V6' is 34.9±0.7 Å, which is slightly shorter than that determined for LAI' (35.5±1 Å). On the other hand, for MDR769', the most probable distance is slightly larger than in LAI', and is found to be 36.4±0.5 Å. A more open structure for MDR769' than LAI' agrees well with the reported crystal structure.10 However, for the three constructs, a striking difference is observed in the breadth of the distance distribution profiles, which reflects the range of opening and conformational flexibility of the flaps. In both V6' and MDR769', analysis of the DEER data shows that the flaps do not span the full range of distances seen for LAI'. For V6', the flaps span distances of 28 to 42 Å, for MDR769', a distance breadth of 31 to 42 Å is obtained. Both of these ranges are narrower than the 23 to 48 Å seen for LAI' (From the experimental S:N, errors for distance distribution profile breadths are estimated to be ± 2Å for V6'/MRD769' and ± 4Å for LAI'.)
In order to obtain insight into structural conformations that correspond to the experimentally determined distance profiles, MD simulations in explicit solvent using the ff99SB force field14 were performed for V6' and MRD769' following the protocol we published for the MTSL-labeled LAI' sequence.9 Results are shown in Fig. 2b, and overall, the trends are in excellent agreement with the profiles derived from the DEER data. The MD reconstructed N-N distance distribution profiles for both V6' and MDR769' are narrower than for LAI', with both lacking long distances that correspond to a wide-open conformation of the flaps.9 Furthermore, the MD results predict the same shift of the most probable distance seen between nitroxide spin labels; V6' < LAI' and MDR769' > LAI'. In addition, the simulation data reveal that the distance distributions between Cα pairs for residues 55/55’ have the same trend in most probable distances as those observed for the nitroxide nitrogen pairs (Supp Info.); implying that the changes seen in the EPR data analysis for V6' and MDR769' are dominated by changes in the protein backbone positions, with only minor contributions from altered conformations of the spin label.
We calculated the average conformations sampled during the MD simulations (Fig. 2c). The flaps in the LAI' simulation adopted a degree of closure in excellent agreement with the semi-open crystal structure of apo HIV-PR (pdb code 2G69). In contrast, but also consistent with the EPR data analysis, the flaps of the MDR769' mutant adopt a more open conformation than those of LAI', while the flaps in V6' are more closed relative to LAI' (we note that all structures adopt the semi-open flap handedness.15) Analysis of the entire range of flap to active site distances sampled during the simulations of each sequence confirms the conclusions obtained from the average structures (Supp Info.).
The DEER results show that mutations linked to function and inhibitor resistance can alter flap conformations in HIV-1PR. In addition, the MD simulations of the flap motion provide a structural interpretation of the EPR data. Based upon the combined analysis, we see that both the breadth of the flap distance distribution profile and the average conformation are altered in the mutants, providing valuable insight into the coupling of drug resistance and protein backbone conformational flexibility. Perhaps the limited conformational opening of the flaps in V6' alters the ability of the inhibitor, and possibly substrate, to enter into the active site cavity; whereas in MDR769' the longer average semi-open distance might increase the free energy cost for the flaps closing tightly in the presence of inhibitor or substrate.
Supplementary Material
Further experimental details of protein expression, purification, spin labeling, circular dichroism spectra of spin labeled constructs, computational methods and additional data analyses. This material is available free of charge via the Internet at http://pubs.acs.org
ACKNOWLEDGMENT
This work was supported by AHA (Predoctoral Fellowship to LG), NHMFL-IHRP and UF-DSR seed money to GEF and support to CS by NIH (GM6167803), NCSA (NPACI MCA02N028) and DOE (DE-AC02-98CH10886) and the State of New York. We thank Ben Dunn for helpful discussions and Ralph Weber for initial DEER data collection of V6' and MDR769'.
Contributor Information
Carlos Simmerling, Email: carlos.simmerling@stonybrook.edu.
Gail E. Fanucci, Email: fanucci@chem.ufl.edu.
REFERENCES
- 1.Ashorn P, McQuade TJ, Thaisrivongs S, Tomasselli AG, Tarpley WG, Moss B. Proc Natl Acad Sci U S A. 1990;87:7472–7476. doi: 10.1073/pnas.87.19.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walensky RP, Paltiel AD, Losina E, Mercincavage LM, Schackman BR, Sax PE, Weinstein MC, Freedberg KA. J Infect Dis. 2006;194:11–19. doi: 10.1086/505147. [DOI] [PubMed] [Google Scholar]
- 3. http://hivdb.stanford.edu/ is “A curated public database designed to represent, store and analyze the divergent forms of data underlying HIV drug resistance”.
- 4.Clemente JC, Coman RM, Thiaville MM, Janka LK, Jeung JA, Nukoolkarn S, Govindasamy L, Agbandje-McKenna M, McKenna R, Leelamanit W, Goodenow MM, Dunn BM. Biochemistry. 2006;45:5468–5477. doi: 10.1021/bi051886s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemente JC, Hemrajani R, Blum LE, Goodenow MM, Dunn BM. Biochemistry. 2003;42:15029–15035. doi: 10.1021/bi035701y. [DOI] [PubMed] [Google Scholar]
- 6.Clemente JC, Moose RE, Hemrajani R, Whitford LR, Govindasamy L, Reutzel R, McKenna R, Agbandje-McKenna M, Goodenow MM, Dunn BM. Biochemistry. 2004;43:12141–12151. doi: 10.1021/bi049459m. [DOI] [PubMed] [Google Scholar]
- 7.Mahalingam B, Boross P, Wang YF, Louis JM, Fischer CC, Tozser J, Harrison RW, Weber IT. Proteins. 2002;48:107–116. doi: 10.1002/prot.10140. [DOI] [PubMed] [Google Scholar]
- 8.Galiano L, Bonora M, Fanucci GE. J Am Chem Soc. 2007;129:11004–11005. doi: 10.1021/ja073684k. [DOI] [PubMed] [Google Scholar]
- 9.Ding F, Layten M, Simmerling C. J Am Chem Soc. 2008;130:7184–7185. doi: 10.1021/ja800893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. Structure. 2005;13:1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl. Mag. Reson. 2006;30:473–498. [Google Scholar]
- 12.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 13.Chiang YW, Borbat PP, Freed JH. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornak V, Okur A, Rizzo RC, Simmerling C. Proc Natl Acad Sci U S A. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further experimental details of protein expression, purification, spin labeling, circular dichroism spectra of spin labeled constructs, computational methods and additional data analyses. This material is available free of charge via the Internet at http://pubs.acs.org