Abstract
Models that deal with the individual level of populations have shown the importance of stochasticity in ecology, epidemiology and evolution. An increasingly common approach to studying these models is through stochastic (event-driven) simulation. One striking disadvantage of this approach is the need for a large number of replicates to determine the range of expected behaviour. Here, for a class of stochastic models called Markov processes, we present results that overcome this difficulty and provide valuable insights, but which have been largely ignored by applied researchers. For these models, the so-called Kolmogorov forward equation (also called the ensemble or master equation) allows one to simultaneously consider the probability of each possible state occurring. Irrespective of the complexities and nonlinearities of population dynamics, this equation is linear and has a natural matrix formulation that provides many analytical insights into the behaviour of stochastic populations and allows rapid evaluation of process dynamics. Here, using epidemiological models as a template, these ensemble equations are explored and results are compared with traditional stochastic simulations. In addition, we describe further advantages of the matrix formulation of dynamics, providing simple exact methods for evaluating expected eradication (extinction) times of diseases, for comparing expected total costs of possible control programmes and for estimation of disease parameters.
Keywords: stochasticity, disease dynamics, time to extinction, total costs, Kolmogorov forward equations, parameter estimation
1. Introduction
The foundations of ecological and epidemiological modelling are largely based upon deterministic equations for dynamics of populations. However, a growing body of research suggests that demographic stochastic effects, due to the random nature of population events, can cause dramatic deviations from this deterministic ideal (Bartlett 1956; Rand & Wilson 1991; Fox 1993; Grenfell et al. 1998; Keeling et al. 2000; Spagnolo et al. 2003; Coulson et al. 2004). Three notable elements separate stochastic from deterministic dynamics. The first and most obvious is the random nature of the dynamics leading to differences between replicates and temporal variation, where the deterministic model will often predict an unvarying equilibrium solution (Taylor 1961; Hanski & Woiwod 1993; Keeling & Grenfell 1999). The second is the possibility of stochastic extinction, where, by chance, individuals may fail to ‘reproduce’ and the species dies out (Bartlett 1956; Lande 1993; Grenfell et al. 1995). This is important in most avenues of population modelling as recovery from extinction can usually only occur due to interactions with an external population. Finally, stochasticity induces fluctuations away from the deterministic equilibrium (attractor), so that realizations of the stochastic process often ‘track’ the deterministic transient path. This can often lead to stochastic resonance where stochasticity causes the population to oscillate at or near its natural frequency (Renshaw 1991; Blarer & Doebeli 1999; McKane & Newman 2005; Alonso et al. 2006; Ross 2006b).
Commonly, modelling of stochastic populations is performed using integer-based event-driven models (Gillespie 1976; Renshaw 1991), mimicking the supposed behaviour of the real system. Such methods of simulation generally allow a range of complex biologically realistic behaviour to be incorporated in the underlying model and offer an intuitive modelling framework to many applied researchers. However, given a single simulation, it is not clear whether the simulated dynamics are representative of average behaviour or merely a chance outlier due to a rare combination of events. Therefore, large numbers of replicate simulations are required to establish confidence in results. The same is true in the situation where our interest is in a rare event, such as occasional extinctions or unusually large epidemics (although methods have been developed to improve efficiency, e.g. importance sampling and the cross-entropy method; Rubinstein & Kroese 2004). In essence, event-driven models are in silico experiments and therefore the results must be subject to the same statistical treatments as would be employed for any experimental observation.
A number of approximation methods exist, which overcome the requirement of a large number of simulations by providing analytical approximations for expected behaviour of the population and often approximations for variability in this behaviour. Two widely used methods are moment closure techniques (Keeling et al. 2000; Nåsell 2001, 2002, 2003a,b) and diffusion approximations (Kurtz 1970, 1971; Pollett 1990; van Kampen 1992; Jacquez & Simon 1993; Andersson & Britton 2000; Ross 2006a,b). However, such methods are generally most accurate when the population size is sufficiently large. Since we are often interested in stochastic effects when population sizes are small, and thus stochasticity has a relatively large impact on dynamics, it would be desirable to have similar methods that operate effectively when population sizes are small. We present such a method for Markov processes. Markov processes are a class of stochastic process in which the future state of the population is determined solely by the current state—the system has no memory (Norris 1997; Andersson & Britton 2000); the vast majority of event-driven stochastic models used in ecology and epidemiology are Markov processes.
The basic methodology presented here pre-dates both simulation and analytical approximations; it allows the complete ensemble of stochastic behaviour to be predicted precisely by a (very) large set of deterministic equations (e.g. for susceptible–infectious–susceptible (SIS) dynamics, N+1 differential equations are required and for susceptible–infectious–recovered (SIR) dynamics, (1/2)(N+1)(N+2) differential equations are required, where N is the number of individuals). In essence, the deterministic ‘Kolmogorov forward equation’ (also called the ensemble or master equation) contains a single equation for the probability of being in each possible state, with the dynamics governed by the rates of transition between states (Norris 1997). Therefore, by solving one set of differential equations, we can obtain a complete description of all possible behaviours of the stochastic system. While these methods have existed for many years, they appear not to be widely known or used by many applied practitioners. However, with continuing advances in computing power, these techniques are becoming increasingly applicable to real-world problems. In addition, these ensemble equation methods can be used to test the validity of analytical approximation methods (such as diffusion approximations and moment closure techniques) and provide a richer understanding of the dynamics of stochastic event-driven simulations.
The solution of large numbers of differential equations generated by this method is relatively straightforward and fast on modern computers, and has been exploited by a number of researchers (Dieckmann & Law 1996; Keeling 2000; Keeling et al. 2000; Stollenwerk & Briggs 2000; Alonso & McKane 2002; Stollenwerk & Jansen 2003; Viet & Medley 2006). However, a careful examination of the Kolmogorov forward equation shows that it is linear in terms of the probabilities of being in each state—the complex nonlinearities often associated with population dynamics are simply absorbed into the matrix terms. This observation allows the equations to be recast in terms of simpler matrix and vector operations, which greatly speed computation, and provides more insight into dynamics. This paper is not intended to provide detailed computational methodology for handling the necessary matrix operations (such as finding eigenvalues, solving systems of linear equations or calculating matrix exponentials), instead readers are recommended to read one of the various textbooks on the subject (e.g. Golub & van Loan 1996) or to use one of the many computational packages that are available (e.g. Matlab, Lapack, R or Mathematica). Instead, using simple epidemiological examples, we show the power and advantages of using Kolmogorov's forward equations and the associated matrix methods, in particular when very precise results are required for relatively small populations. We hope this will motivate researchers to explore the potential that these matrix methods have to offer.
Throughout this paper, attention is focused on dynamics of infectious diseases; this is for two main reasons. First, the study of infectious diseases has been at the forefront of research into the behaviour of stochastic population processes—generally because the prevalence of infection is often low and therefore stochastic effects are felt strongly (Bartlett 1956; Grenfell 1992). Second, for disease dynamics, the number of events is strictly limited (birth, death, infection and recovery) and their nature well defined, whereas for ecological models there is far more ambiguity (e.g. consider the number of ways in which density dependence can be modelled). Three distinct epidemiological models are considered to exemplify the use of ensemble equations: the endemic SIS model with infectious imports (i.e. infection resulting from a source external to the population being modelled); the endemic SIR model with infectious imports; and the simple SIR model without births, deaths or imports. Throughout the paper, deterministic ensemble equation results are compared with results from the corresponding event-based stochastic simulations and the additional insights that come from the matrix formulation are discussed. Understanding the stochastic behaviour of diseases in small populations is a vital step towards the control of infection in subdivided communities with metapopulation-like structure (Grenfell & Harwood 1997). In particular, the methods outlined in this paper are ideal for studying infection within families (Hope Simpson 1952; Melegaro et al. 2004; Verver et al. 2004), farms (Woolhouse et al. 1996; Nodelijk et al. 2000; Stark et al. 2000; Viet & Medley 2006) and hospitals (Dooley et al. 1992; Austin et al. 1999; Cooper et al. 1999; Cooper & Lipsitch 2004). The continual improvement in computational power means these techniques will be of increasing importance and applicability in the near future as it becomes practical to deal with ever larger population sizes.
2. The SIS model
The SIS model is an accurate representation of the population dynamics for many sexually transmitted infections (Anderson & May 1992), where following treatment an infectious individual recovers from the disease but is once again susceptible to infection. We take the standard differential equation for the number of infectious individuals in a deterministic population (noting that in such formulations, population numbers are assumed continuous)
| (2.1) |
where S=N−1 is the number of susceptibles; N is the population size (assumed constant); β is the contact rate; ϵ captures the import of infection from an external source (assumed to equal 0 if the population is isolated); and g is the rate at which individuals are treated and return to the susceptible class. In this basic formulation, the natural processes of births and deaths have been ignored, as these demographic events often occur at a much slower rate than infection. A more realistic model of sexually transmitted infections would include the age, gender, sexual preference and number of partners of an individual; however, the simple one-dimensional model (equation (2.1)), in which the only events are infection and recovery (back to susceptibility), is ideal for illustrating the power of Kolmogorov's forward equations. In fact, the SIS model has many similarities with the Levins metapopulation model (Levins 1969; Alonso & McKane 2002; Ross 2006a,b), classifying hosts as either empty or occupied with infection.
For this SIS disease model, there are N+1 different states that the population can be in, corresponding to I=0, 1, …, N. We let pn(t) be the probability that there are n infectious individuals at time t and construct a set of differential equations for these state probabilities, the so-called Kolmogorov forward equations
| (2.2) |
where I=n=−1 and I=n=N+1 are not feasible states and therefore p−1 and pN+1 are set to zero. The first two terms on the right-hand side of equation (2.2) deal with ways in which an I=n population state can arise: the first term corresponds to ‘creating’ a new infectious individual from the I=n−1 state, and the second term corresponds to recovery from when I=n+1. The final term deals with ways in which an I=n population can be lost, either through gaining an extra case or through recovery of an infectious individual. While it would be relatively straightforward to numerically evolve this set of equations, more insight can be gained by changing to a vector notation.
We set p to be the row vector of the N+1 probabilities. In this vector notation, the Kolmogorov forward equation becomes
| (2.3) |
where the matrix Q is tridiagonal and consists of the transition rates
It is interesting to realize that all of the complex nonlinear dynamics associated with disease transmission (or other processes) are absorbed into the terms of the matrix, and therefore the dynamics of p are purely linear. In general, the eigenvalues (λ1, λ2, …, λN+1) and left eigenvectors of Q specify the entire dynamics. (For later use, we also specify that the right eigenvectors are , which are associated with the same set of eigenvalues.) Throughout this paper, we assume that the eigenvalues are ordered such that 0=λ1≥Re(λ2)≥⋯≥Re(λN+1), so that the first eigenvector(s) dominates the long-term dynamics. In general, the sparse nature of Q (here only 3N+1 terms out of (N+1)2 are non-zero) means that we can exploit a range of powerful numerical techniques to find dominant eigenvalues (Golub & van Loan 1996; Trefethen & Bau 1997).
Considering the Kolmogorov forward equation in general for any Markov process model (given that the system is finite—as is frequently the case), there exist two ways in which we can write the full dynamics of the ensemble for all time, hence providing a single equation that specifies the exact probabilistic dynamics of the disease. The simplest formulation extending from the differential equation (2.3) is
| (2.4) |
where the exponential of a matrix can be computed with relative ease (Moler & van Loan 1979; Sidje 1998). As an alternative representation, we can define the solution as
| (2.5) |
where the coefficients qn are determined by the inner product of the right eigenvectors of Q with the initial distribution , , and the right eigenvectors are normalized so that x. (The precise normalization of left eigenvalues is irrelevant for this formulation, although throughout we have assumed that the terms of l1 sum to 1 such that it is a valid probability distribution. We also note that for equation (2.5) to hold we have assumed that repeated eigenvalues have equal algebraic and geometric multiplicities, which is generally the case.)
For the SIS model, in particular, λ1=0 and all other eigenvalues are real and negative. Hence, from equation (2.5), we find that (normalized to sum to 1) is the final state of the ensemble equation and hence the long-term distribution of the stochastic SIS model. Figure 1 shows the exact probability distribution given by , and the error at each point when the distribution is estimated from a stochastic time series (simulation of SIS model) of length 100 and 1000 time units. The inset graph shows how the total (root mean square) error decreases as the length of the stochastic time series increases. As expected from standard statistics, the error scales inversely with the square root of the length of the time series (error∝(length)−1/2). So, either very long or very many stochastic simulations have to be performed to obtain an accurate prediction of the distribution of disease prevalence. In contrast, a single eigenvalue calculation produces the answer to very high accuracy and extremely quickly. We return to this question of computational efficiency later.
Figure 1.
Comparison between the ensemble equation solution and the average of stochastic simulations for the SIS disease model in a population of 100 individuals. The grey curve is the equilibrium distribution from the ensemble equation, calculated as the dominant eigenvector, . As a comparison, the average distribution is calculated from stochastic simulations of length 100 and 1000 time units, once transient dynamics have died away. The thick and thin black lines show the difference between these stochastically derived distributions and the ideal from the ensemble equation. The inset graph gives the root mean square error between the ensemble and stochastic distributions as the length of the simulation is varied (g=1, , ϵ=0.01, N=100).
Other eigenvalues (and the associated eigenvectors) provide additional information; in particular, the real part of λ2, λ3, … (noting again that for the SIS model, the eigenvalues are all real) informs about the rate of convergence to the equilibrium distribution. For the example given in figure 1, λ2≈−0.0098 while λ3≈−0.8725, and the entry of the eigenvector corresponding to the disease-free state I=0 is significantly larger than the other entries. This suggests that, in general, convergence from any infected state to the equilibrium distribution is swift, as contributions from the eigenvectors to decay rapidly; however, escape from disease extinction (governed by parameter ϵ and captured by λ2) is much slower.
In addition, we note that if ϵ=0, the equilibrium distribution is degenerate as I=0 is an absorbing state. In such cases, we may be interested in the long-term distribution of the process conditioned on non-absorption; in epidemiological terms, this is the prevalence distribution, given that the disease has not died out. This is precisely the information conveyed by the quasi-stationary distribution, which may once again be evaluated efficiently using a matrix formulation (Pollett 2006; Ross 2006a). We perform the same calculations as before, but for the reduced matrix Q0 derived by removing the first row and first column (rows and columns corresponding to absorbing states) from the matrix Q. The first (normalized left) eigenvector of this matrix, , is the quasi-stationary distribution of the SIS model without imports of infection. Once again, the eigenvalues provide us with additional information; the real part of the first eigenvalue tells us about the long-term rate of extinction (decay of the quasi-stationary distribution or convergence to the absorbing state I=0), and the difference between the real parts of the first and second eigenvalues (the spectral gap) tells us about the rate of convergence to the quasi-stationary distribution (Dambrine & Moreau 1981). Furthermore, for the quasi-stationary distribution to be of practical interest, the difference between the first and second eigenvalues must be substantially larger than the magnitude of the first eigenvalue.
Finally, the matrix formulation also allows exact evaluation of the likelihood for a sequence of data, and thus provides a framework for parameter estimation (Pelupessy et al. 2002; Cooper & Lipsitch 2004; Ross et al. 2006). Suppose there is a parameter (or set of parameters) θ, we wish to estimate from some given data. We allow the dependence on θ to be made explicit in our model notation by writing Q(θ) and ; we also define to be the indicator vector that corresponds to being precisely in state i, and set [·]i to be the element of a vector corresponding to state i, while [·]i,j is defined to be the i,jth element of a matrix. Suppose we now have n observations at times t1<⋯<tn, when it is recorded that the process is in states i1, …, in. The likelihood of observing these states is
| (2.6) |
This likelihood is the product of the probability of being in state ik at time tk, given that the system was in state ik−1 at time tk−1, all multiplied by the probability of observing the system in state i1 at time t1. Using the exponential matrix formulation, the probability of moving from state ik−1 to state ik in time tk−tk−1 can be explicitly calculated. Any one of a range of numerical optimization techniques can then be used to find the value of θ, which maximizes the likelihood (2.6) over the range of parameter space (Ross et al. 2006). It should be emphasized that this method of parameter estimation uses the exact likelihood of observing the given data—assuming the model is an accurate description of disease dynamics—and also incorporates dependency between observations. Although the form of the likelihood (2.6) is greatly simplified by the assumption of an accurate record of prevalence, a similar approach is possible when only partially observed reporting data are available. We note that if the data are recorded periodically (such that tk−tk−1 is constant), then the matrix exponential only needs to be calculated once, greatly improving computational efficiency.
It is informative to contrast this estimation method with MCMC techniques (Gamerman 1997). The estimation method based on the matrix approach calculates the exact probability of moving between two states (say ik−1 and ik); in contrast, MCMC techniques generally calculate the likelihood by ‘averaging’ over plausible transitions between states (Gamerman 1997). Therefore, for small population sizes and limited number of states, this matrix approach may be faster and more accurate; however, MCMC techniques can generally deal with higher-dimensional systems with much larger state spaces.
3. Endemic SIR diseases
While the SIS model provides a simple test case for the ensemble equation, the one-dimensional nature of the problem means the equilibrium solution to the ensemble equation (2.3) can be found explicitly by balancing transitions between states (pnβ(N−n)n=pn+1g(n+1); Keeling 2000; Alonso & McKane 2002). In fact, explicit expressions exist for many quantities of interest for such one-dimensional birth-and-death processes (Norris 1997), where the only transitions possible are either increase or decrease the state by 1.
When considering a disease with SIR dynamics, such that there are (at least) two independent variables, the problem is more complex and the matrix approach becomes even more appealing. In the SIR disease model, when individuals recover, they are assumed to be immune from further infection; this gives rise to the standard deterministic model for this type of disease (Anderson & May 1992)
| (3.1) |
where the birth and death rates are typically assumed to be equal (B=d) and again imports from an external source (governed by the parameter ϵ) have been included. Here six distinct events can occur (birth, death of a susceptible individual, death of an infected individual, death of a recovered individual, infection and recovery), which modify the state of the process. This means, in general, there are six ways in which the probability density for a particular state can increase and six ways in which it can decrease:
| (3.2) |
where pS,I,N is the probability of having S susceptible individuals, I infectious individuals and a total population of size N.
Unfortunately, due to the way births are modelled, a strict upper limit cannot be placed on the population size for the stochastic model, which is necessary to formulate a finite matrix expression. Hence, we require a mechanism of bounding the population. One approach to this, which should provide the most accurate approximation to the dynamics of equation (3.2), is to truncate the process at some large population size N. The influence of truncating the process at various population sizes may then be examined numerically, and for some models analytically (Cairns & Pollett 2005). In practice, one would usually choose the largest population size N for which computations remain feasible. An alternative way to bound the population size is to fix the population size N as a constant, such that the death of a susceptible, infectious or recovered individual is immediately compensated by the birth of a susceptible. This assumption has three additional advantages. First, it reduces the number of independent variables to two (just S and I). Second, it circumvents the necessity to choose between density- and frequency-dependent transmission (Begon et al. 2002). Finally, a constant population size is reasonable for many applied problems, such as modelling the transmission of infection upon farms or within hospital wards, where there is generally little variation in the number of individuals in the population (Pelupessy et al. 2002; Cooper & Lipsitch 2004; Viet & Medley 2006). It should be noted that while assuming a constant population size does not change the ODEs (3.1) (if, as is typical, we assume B=d), however, it does change the Kolmogorov forward equations
| (3.3) |
where pS,I is the probability of having S susceptible individuals and I infective individuals. This highlights an additional use of the Kolmogorov forward equations: the Kolmogorov forward equations uniquely specify the full stochastic dynamics, whereas there are multiple interpretations of the ordinary differential equations (Keeling 2000). For this model, there are possible disease states (0≤S≤N, 0≤1≤N−S), and so C different pS,I variables. To formulate a matrix expression, we need to find a method of mapping the two-dimensional pS,I into a one-dimensional vector, p. An efficient means of achieving this is to set the probability pS,I as the th element of the vector.
Now, as before, we can compute the distribution of states as , where the matrix Q is constructed from the transition rates between states; again Q is sparse (a very small proportion of non-zero terms). If we are only interested in long-term behaviour, considering the eigenvalues again provides an efficient method of calculating the equilibrium distribution. For this SIR model, the first eigenvalue, λ1, is zero while all the others have negative real parts, and thus there is again a unique equilibrium distribution. Hence, from equation (2.5),
so the entire long-term behaviour of the full stochastic system can be found by simply computing the dominant (left) eigenvector, which can be done relatively efficiently (Pollett & Stewart 1994; Golub & van Loan 1996). Figure 2a compares the distribution from the first eigenvector (contours) with the results of a long stochastic simulation (dots). As expected, there is good agreement between the two methods, although the Kolmogorov forward equation approach provides far more detail, allowing evaluation of the exact (to numerical precision) distribution of states—which can be a highly efficient way of calculating extremes of behaviour such as the 99 or 99.5% contours.
Figure 2.
Dynamics of the full stochastic SIR equation with very rapid ‘births’ and ‘deaths’, which could be compared with movements on and off farms or hospital wards. (a) The solid lines show the 10, 50, 90, 95, 99 and 99.5% level curves for the distribution from the first eigenvalue of the ensemble approach. The dots show the number of susceptible and infectious individuals from a long stochastic simulation (10 000 points each separated by 100 events); the most frequently visited points lead to a darker coloration. The diagonal line corresponds to S+I=N and is therefore the limit of possible values. (b) The extinction rate from the equilibrium distribution for a range of population sizes. The inset graph shows the nonlinear increase in the mean number of susceptibles and infecteds when the population size is small; solid line, infecteds; dashed line, susceptibles (d=0.5, g=1, , ϵ=0.1; in (a), N=100).
Looking at the other eigenvalues and eigenvectors again provides a deeper understanding of the dynamics. For the parameters used in figure 2a, once again, the second eigenvalue and eigenvector describe the slow escape from the disease-free state (S=N, I=0), with λ2≈−0.3468. Thus, if the initial distribution begins at the disease-free state, it will escape relatively slowly at rate exp(−0.3468t). This is in direct contrast to the strong instability of the disease-free state in the standard ODE model (equation (3.1)) for which I(t) grows at approximately the rate exp(2.333t) for I(0) small; this difference is because in the stochastic model, an import is required for the infection to escape zero and there is the possibility of stochastic extinctions, both of these are captured by the Kolmogorov forward equation. The third and fourth eigenvalues are a complex conjugate pair (approx. −0.7607±0.9576i) and correspond to the dominant oscillations around the equilibrium distribution. These oscillations have a slightly longer period and a slower decay than predicted by the standard ODE model (Anderson & May 1992, where the eigenvalues are approx. −0.8401±1.0287i) due to the effects of stochasticity and nonlinearities around the fixed point.
Finally, the equilibrium solution can also tell us about the long-term rates of temporary extinction and subsequent reinfection of the population. As we are at equilibrium, these two rates must be equal and can be calculated as either the rate at which infection is lost from or the rate that reinfection occurs from
Figure 2b shows the extinction rates from the equilibrium distribution, and average numbers of susceptible and infectious individuals, for a range of population sizes and with β=5/N. As expected, the extinction rate decreases exponentially with increasing population size (extinction rate≈11.13 exp(−0.0789N)), and for population sizes above 100 the mean number of susceptibles and infecteds increases linearly.
4. The simple SIR epidemic
As a further example of the power of ensemble equations, we consider the simple SIR model, without births, deaths or imports of infection
| (4.1) |
The Kolmogorov forward equations are defined as in equation (3.3), but with d=ϵ=0. For this model, we observe a single epidemic that eventually dies out as the level of susceptibles becomes too low to support the infection and cannot be replenished. While generally considering the simplest form of the deterministic SIR model, in this stochastic setting the dynamics are more complex as there are multiple equilibrium (absorbing) distributions. For a population of size N, there are N+1 equilibria corresponding to I=0, S=0, …, N. Faced with this inevitable extinction, there are two applied questions that can be addressed using the ensemble model: what is the expected time to extinction, and how many individuals escape infection, and the influence of the initial conditions on both of these quantities is of applied interest. These questions are once again readily calculated using a matrix formulation.
The expected time to extinction, , when starting in state i, is evaluated as the solution to
where 1 is a vector of 1s, and Q0 is again the matrix Q with the rows and columns corresponding to the (N+1) absorbing states removed (Mangel & Tier 1994; Norris 1997). Additionally, if costs are involved (such as treatment and isolation costs), we can replace 1 by a vector f, where the entry is the cost per unit time associated with a population in state j. The solution now corresponds to the expected total cost over the lifetime of the disease starting in state i (Norris 1997; Pollett & Stefanov 2002; Pollett 2003). This formulation highlights that the effect of initial conditions may be rapidly evaluated once τ is calculated.
In the deterministic model, the final size of the epidemic, R∞, is defined as the proportion of a totally susceptible population that become infected (and eventually recover) following the introduction of a small amount of infection (Kermack & McKendrick 1927). In this context, the final size is determined by the relationship
We now wish to consider how this theoretical quantity for a large deterministic population compares with results for small stochastic populations. Using the matrix formulation, the average final size of the epidemic () can be calculated from the number of susceptibles ‘surviving’ in the final probability distribution
We now use equation (2.5), but note that there are N+1 zero eigenvalues and that eigenvectors associated with zero eigenvalues span the null space of the matrix. The null-space calculation provides a computationally efficient means of finding the eigenvectors and hence calculating the final state of the system. (For simplicity, we can set the first N+1 left eigenvectors to be (where is a vector of length equal to the size of the state space and having a 1 in the entry corresponding to the state (S, 0) and zeros elsewhere), the right eigenvectors can then be found such that they span the null space of Q and satisfy .) From equation (2.5), the long-term distribution is given by
allowing us to compute . Hence, the influence of initial distributions on the size of the epidemic may again be computed very quickly. (We note that alternative methods exist for evaluating this quantity, as described for instance by Bailey (1953), Daley & Gani (1999) and Diekmann & Heesterbeek (2000).)
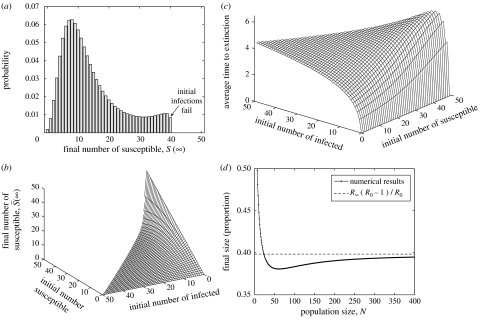
Figure 3a shows the long-term distribution ps,0(∞) in a population of 50 individuals, starting from S(0)=40 and I(0)=5. As expected, the model predicts a range of epidemic sizes, from failure of the initial cases to generate any secondary infections to epidemics infecting the entire population. Once the left and right eigenvectors have been found corresponding to the null space of the matrix, extending the calculation to include the complete range of possible initial conditions is computationally efficient (figure 3b). Figure 3c shows the corresponding average times to extinction. Several facets are clear from these results: the average time to extinction increases with both the initial number of susceptibles and the number of infecteds; in contrast, the number of individuals escaping infection decreases with the initial number infected but increases with the number that are initially susceptible.
Figure 3.
Results for the SIR ensemble equation without births or deaths. (a) For a population size of N=50, and starting with S(0)=40, I(0)=5, shown are the distribution of the number of susceptible individuals remaining once the infection has died out given by . (b) The extension of this result to the entire range of initial conditions and, of necessity, the extraction of the average number of susceptible individuals from the final distribution (the initial conditions used in (a) are shown by a dot). (c) The average time to extinction, again for the entire range of possible initial conditions. Note that for (b) and (c), only a triangular set of initial conditions are feasible as S+1 must be less than or equal to N. (d) The examination of the so-called ‘final size’ of an epidemic; starting with S(0)=N−1, I(0)=1, we show the average proportion of the population infected during the epidemic, 1−S(∞)/N. We compare the numerical results with simple theory (R∞(R0−1)/R0), where R∞ is the expected proportion infected in a deterministic model and 1−1/R0 is the probability that a stochastic invasion caused a major epidemic (throughout g=1, ).
Finally, in figure 3d, we consider how numerical calculation of final size (proportion of a totally susceptible population that becomes infected) in a stochastic population (starting with S(0)=N−1 and I(0)=1) compares with theoretical predictions of Kermack & McKendrick (1927). The theoretical value of R∞ has to be modified by a factor 1−1/R0, which accounts for failure of the initial infection to cause a major epidemic (Bartlett 1956). Although the theoretical value may be appropriate in the limit of large population sizes, understanding the deviation from this ideal for small populations is highly informative. Two conflicting elements contribute to the pattern. First, for small population sizes, the initial case is a significant proportion of the population size, hence the true final size is greater than the theoretical prediction. However, for larger population sizes, the theoretical prediction is an overestimate primarily due to underestimating the extinction risk. While these conclusions are intuitive, and could be determined by repeated simulation, the results from the ensemble equations provide an extremely accurate description of the behaviour and are computationally efficient to generate.
5. Computational efficiency
Throughout this paper, we have alluded to the computational efficiency of using the Kolmogorov forward equation when dealing with Markov models. We now wish to make these assertions more definite by providing some examples of the types of computational demands required by the various problems discussed in this paper (table 1). It is clear that some calculations, such as computing the transition matrix Q or calculating the first few dominate eigenvalues and eigenvectors, are relatively fast—and therefore applications which use this information may have substantial advantages over event-driven stochastic simulations. However, other calculations, such as finding exponentials and null spaces, are associated with significant computational overheads and therefore repeated event-driven simulations may be preferable.
Table 1.
Times required to calculate various quantities from the Kolmogorov forward equation for the SIS and SIR models. (Only the calculations that complete within a reasonable time frame are shown. It should be noted that for the SIR model, the number of states increases like 1/2N2. Calculations are performed on a 2.4 GHz PC using Matlab.)
| calculation of | time to perform calculation with population size (N) | ||
|---|---|---|---|
| 100 | 100 | 10 000 | |
| Q matrix (SIS) | 0.0005 s | 0.0024 s | 0.024 s |
| dominant eigenvalue and eigenvector (SIS) | 0.0295 s | 0.0372 s | 0.282 s |
| four dominant eigenvalues and eigenvectors (SIS) | 0.0232 s | 0.0754 s | 0.728 s |
| exp (0.1Q) (SIS) | 0.0743 s | 180 s | 6.4 h |
| exp (10Q) (SIS) | 0.0622 s | 5.3 min | — |
| Q matrix (full SIR) | 0.390 s | 4.742 s | — |
| dominant eigenvalue and eigenvector (full SIR) | 0.2785 s | 67.1 s | — |
| four dominant eigenvalues and eigenvectors (full SIR) | 0.5359 s | 107 s | — |
| Q matrix (simple SIR) | 0.023 s | 2.99 s | — |
| null space (simple SIR) | 0.376 s | 2.45 h | — |
To provide a relative comparison with the times in table 1, we shall consider what can be achieved with a similarly parametrized event-driven stochastic model in one second. Both SIS- and SIR-type models can perform approximately 2.7 million events per second using Gillespie's direct algorithm (Gillespie 1976)—the comparable speed of both SIS and SIR models is because the rate-determining step is the calculation of two random numbers per event. For the SIS model, illustrated in figure 1, we find that in the time it takes to calculate the matrix, dominant eigenvalue and associated eigenvector, the equilibrium distribution can be calculated with an error of approximately 1% for both N=100 and 1000. Hence, for this problem, the Kolmogorov forward equations have substantial benefits. However, for the simple SIR model without births or deaths (figure 3), Gillespie's direct algorithm allows the simulation of approximately 170 complete epidemics per second for a population size of N=10 000, which is just sufficient to calculate the mean final epidemic size to within 1%. In contrast, the null-space calculation for N=10 000 is computationally infeasible, so multiple stochastic simulations are the only suitable methodology.
Given the time scales and computational overheads involved with the matrix formulation of SIS and SIR models relative to event-driven simulations, we can come to the following general conclusions. Problems that involve large populations (and many possible states) are best tackled using event-driven stochastic simulations; this is because although simulation time generally increases proportional to the population size, the computational time associated with the matrix operations increases proportional to the number of states (or even faster). The advantages of the matrix-based approach are most evident when answers (or distributions) are required to high precision and multiple sets of initial conditions need to be considered. For example, the calculation of the null space for the simple SIR model is a computationally intensive process, but once found the effect of different initial conditions can be calculated using a single matrix–vector multiplication; in contrast, with event-driven stochastic simulations, each set of initial conditions requires a new set of replicate simulations. As an illustrative example, consider a set of stochastic (event-driven) epidemics for the entire range of possible initial conditions, which take the same amount of time as the associated null-space-based calculation in table 1; for N=100, the stochastic simulations could calculate the mean final epidemic size to an accuracy of just 10% while for N=1000 the accuracy increases to approximately 2.5%. Therefore, even though calculating null spaces is computationally demanding, it can be an efficient approach if multiple initial conditions need to be considered.
There are still many situations where it is currently infeasible to consider the Kolmogorov equations. For a population of N individuals, where each individual can be in one of n states, the number of possible states of the process grows like . As such, a disease with SIR dynamics has states , whereas if the dynamics are assumed to be SEIR (susceptible–exposed–infectious–recovered), the number of states increases to C=1/6((N+1)(N+2)(N+3)) (N=100⇒N=176851). Hence, although increasing computational power will allow us to deal with Kolmogorov's forward equations for ever larger population sizes, increasing the biological realism, and thus the number of states, can readily overtake any gains in processing speed.
6. Discussion
Understanding the dynamics of stochastic populations, and how they deviate from the deterministic ideal, is being viewed with increasing importance by ecologists and epidemiologists. In particular, the stochastic behaviour of diseases in small (well-mixed) populations is vital for a better understanding of control. For such situations, where the number of possible states is not prohibitively large, ensemble equations provide a very powerful and efficient alternative to replicate stochastic simulations.
In this paper, it has been shown that the natural matrix formulation (due to the linear nature of the ensemble equation) allows us to specify the dynamics in a concise form, which in turn allows a wealth of sophisticated computational approaches to be used. Four considerable benefits are gained from this approach. First, only a single calculation needs to be performed to describe the dynamics of an infinite ensemble of stochastic realizations. Second, and as a corollary to the first point, the results of the ensemble equations are exact—so the probability of rare events (which could have a large impact) can be calculated precisely. Third, once the initial calculation is performed, considering a range of initial conditions is generally highly efficient. Finally, the fact that the ensemble equations are deterministic means that a wide variety of tools from dynamical systems can be used; for example, it is possible to assess the rate of convergence to an equilibrium distribution.
The use of the Kolmogorov forward equation clearly has many benefits over more commonly used event-driven stochastic simulations; although we will never be able to completely replace the need for simulation in applied modelling. At the moment, Kolmogorov's forward equations are best used on small populations (N<1000) with SIR- or SIS-type dynamics, which complements the need for precise study of small populations where the effects of stochasticity are the greatest. We therefore expect this approach to have maximum benefit when examining the spread of infection within families, farms and hospitals and when needing to obtain precise estimates of rare events.
Acknowledgments
This research was supported by the Royal Society and Leverhulme Trust. We thank Mike Tildesley, Jon Read and David Sirl for their helpful comments.
References
- Alonso D, McKane A. Extinction dynamics in mainland–island metapopulations: an N-patch stochastic model. Bull. Math. Biol. 2002;64:913–958. doi: 10.1006/bulm.2002.0307. [DOI] [PubMed] [Google Scholar]
- Alonso D, McKane A.J, Pascual M. Stochastic amplification in epidemics. J. R. Soc. Interface. 2006;14:575–582. doi: 10.1098/rsif.2006.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1992. Infectious diseases of humans. [Google Scholar]
- Andersson H, Britton T. Springer lecture notes in statistics. vol. 151. Springer; New York, NY: 2000. Stochastic epidemic models and their statistical analysis. [Google Scholar]
- Austin D.J, Bonten M.J.M, Weinstein R.A, Slaughter S, Anderson R.M. Vancomycin-resistant enterococci in intensive-care hospital settings: transmission dynamics, persistence, and the impact of infection control programs. Proc. Natl Acad. Sci. USA. 1999;96:6908–6913. doi: 10.1073/pnas.96.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey N.T.J. The total size of a general stochastic epidemic. Biometrika. 1953;40:177–185. [Google Scholar]
- Bartlett M.S. Deterministic and stochastic models for recurrent epidemics. Proc. Third Berkley Symp. Math. Stats. and Prob. 1956;4:81–108. [Google Scholar]
- Begon M, Bennett M, Bowers R.G, French N.P, Hazel S.M, Turner J. A clarification of transmission terms in host—microparasite models: numbers, densities and areas. Epidemiol. Infect. 2002;129:147–153. doi: 10.1017/S0950268802007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blarer A, Doebeli M. Resonance effects and outbreaks in ecological time series. Ecol. Lett. 1999;2:167–177. doi: 10.1046/j.1461-0248.1999.00067.x. [DOI] [Google Scholar]
- Cairns B.J, Pollett P.K. Approximating measures of persistence in a general class of population processes. Theor. Popul. Biol. 2005;68:77–90. doi: 10.1016/j.tpb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Cooper B, Lipsitch M. The analysis of hospital infection data using hidden Markov models. Biostatistics. 2004;5:223–237. doi: 10.1093/biostatistics/5.2.223. [DOI] [PubMed] [Google Scholar]
- Cooper B.S, Medley G.F, Scott G.M. Preliminary analysis of the transmission dynamics of nosocomial infections: stochastic and management effects. J. Hosp. Inf. 1999;43:131–147. doi: 10.1053/jhin.1998.0647. [DOI] [PubMed] [Google Scholar]
- Coulson T, Rohani P, Pascual M. Skeletons, noise and population growth: the end of an old debate? Trends Ecol. Evol. 2004:359–364. doi: 10.1016/j.tree.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Daley D.J, Gani J. Cambridge University Press; Cambridge, UK: 1999. Epidemic modelling. [Google Scholar]
- Dambrine S, Moreau M. Note on the stochastic theory of a self-catalytic chemical reaction I. Physica A. 1981;106:559–573. doi: 10.1016/0378-4371(81)90126-6. [DOI] [Google Scholar]
- Dieckmann U, Law R. The dynamical theory of coevolution: a derivation from stochastic ecological processes. J. Math. Biol. 1996;34:579–612. doi: 10.1007/BF02409751. [DOI] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek J.A.P. Wiley; Chichester, UK: 2000. Mathematical epidemiology of infectious diseases: model building, analysis and interpretation. [Google Scholar]
- Dooley S.W, Villarino M.E, Lawrence M, Salinas L, Amil S, Rullan J.V, Jarvis W.R, Bloch A.B, Cauthen G.M. Nosocomial transmission of tuberculosis in a hospital unit for HIV-infected patients. J. Am. Med. Assoc. 1992;267:2632–2634. doi: 10.1001/jama.267.19.2632. [DOI] [PubMed] [Google Scholar]
- Fox G.A. Life-history evolution and demographic stochasticity. Evol. Ecol. 1993;7:1–14. doi: 10.1007/BF01237731. [DOI] [Google Scholar]
- Gamerman D. Chapman & Hall; London, UK: 1997. Markov chain Monte Carlo: stochastic simulation for bayesian inference. [Google Scholar]
- Gillespie D.T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976;22:403–434. doi: 10.1016/0021-9991(76)90041-3. [DOI] [Google Scholar]
- Golub G.H, van Loan C.F. Johns Hopkins University Press; Baltimore, MD: 1996. Matrix computation. [Google Scholar]
- Grenfell B.T. Chance and chaos in measles dynamics. J. R. Stat. Soc. B. 1992;54:383–398. [Google Scholar]
- Grenfell B.T, Bolker B.M, Kleczkowski A. Seasonality and extinction in chaotic metapopulations. Proc. R. Soc. B. 1995;259:97–103. doi: 10.1098/rspb.1995.0015. [DOI] [Google Scholar]
- Grenfell B, Harwood J. (Meta)population dynamics of infectious diseases. Trends Ecol. Evol. 1997;12:395–399. doi: 10.1016/S0169-5347(97)01174-9. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T, Wilson K, Finkenstädt B.F, Coulson T.N, Murray S, Albon S.D, Pemberton J.M, Clutton-Brock T.H, Crawley M.J. Noise and determinism in synchronized sheep dynamics. Nature. 1998;394:674–677. doi: 10.1038/29291. [DOI] [Google Scholar]
- Hanski I, Woiwod P. Mean-related stochasticity and population variability. Oikos. 1993;67:29–39. doi: 10.2307/3545092. [DOI] [Google Scholar]
- Hope Simpson R.E. Infectiousness of communicable diseases in the household. Lancet. 1952;2:549–554. doi: 10.1016/S0140-6736(52)91357-3. [DOI] [PubMed] [Google Scholar]
- Jacquez J.A, Simon C.P. The stochastic SI model with recruitment and deaths I. Comparison with the closed SIS model. Math. Biosci. 1993;117:77–125. doi: 10.1016/0025-5564(93)90018-6. [DOI] [PubMed] [Google Scholar]
- Keeling M.J. Simple stochastic models and their power-law type behaviour. Theor. Popul. Biol. 2000;58:21–31. doi: 10.1006/tpbi.2000.1475. [DOI] [PubMed] [Google Scholar]
- Keeling M.J, Grenfell B.T. Stochastic dynamics and a power law for measles variability. Phil. Trans. R. Soc. B. 1999;354:769–776. doi: 10.1098/rstb.1999.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, Wilson H.B, Pacala S.W. Re-interpreting space time-lags and functional responses in ecological models. Science. 2000;290:1758–1761. doi: 10.1126/science.290.5497.1758. [DOI] [PubMed] [Google Scholar]
- Kermack W.O, McKendrick A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. A. 1927;115:700–721. doi: 10.1098/rspa.1927.0118. [DOI] [Google Scholar]
- Kurtz T. Solutions of ordinary differential equations as limits of pure jump Markov processes. J. Appl. Prob. 1970;7:49–58. doi: 10.2307/3212147. [DOI] [Google Scholar]
- Kurtz T. Limit theorems for sequences of jump Markov processes approximating ordinary differential processes. J. Appl. Prob. 1971;8:344–356. doi: 10.2307/3211904. [DOI] [Google Scholar]
- Lande R. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull. Entomol. Soc. Am. 1969;15:237–240. [Google Scholar]
- Mangel M, Tier C. Four facts every conservation biologist should know about persistence. Ecology. 1994;75:607–614. doi: 10.2307/1941719. [DOI] [Google Scholar]
- McKane A.J, Newman T.J. Predator–prey cycles from resonant amplification of demographic stochasticity. Phys. Rev. Lett. 2005;94:218–102. doi: 10.1103/PhysRevLett.94.218102. [DOI] [PubMed] [Google Scholar]
- Melegaro A, Gay N.J, Medley G.F. Estimating the transmission parameters of pneumococcal carriage in households. Epidemiol. Infect. 2004;132:433–441. doi: 10.1017/S0950268804001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moler C.B, van Loan C.F. Nineteen dubious ways to compute the exponential of a matrix. SIAM Rev. 1979;20:801–836. doi: 10.1137/1020098. [DOI] [Google Scholar]
- Nåsell I. Extinction and quasi-stationarity in the Verhulst logistic model. J. Theor. Biol. 2001;211:11–27. doi: 10.1006/jtbi.2001.2328. [DOI] [PubMed] [Google Scholar]
- Nåsell I. Stochastic models of some endemic infections. Math. Biosci. 2002;179:1–19. doi: 10.1016/S0025-5564(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Nåsell I. Moment closure and the stochastic logistic model. Theor. Popul. Biol. 2003a;63:159–168. doi: 10.1016/S0040-5809(02)00060-6. [DOI] [PubMed] [Google Scholar]
- Nåsell I. An extension of the moment closure method. Theor. Popul. Biol. 2003b;64:233–239. doi: 10.1016/S0040-5809(03)00074-1. [DOI] [PubMed] [Google Scholar]
- Nodelijk G, de Jong M.C.M, van Nes A, Vernooy J.C.M, van Leengoed L.A.M.G, Pol J.M, Verheijden J.H.M. Introduction, persistence and fade-out of porcine reproductive and respiratory syndrome virus in a Dutch breeding herd: a mathematical analysis. Epidemiol. Infect. 2000;124:173–182. doi: 10.1017/S0950268899003246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris J.R. Cambridge University Press; Cambridge, UK: 1997. Markov chains. [Google Scholar]
- Pelupessy I, Bonten M.J, Diekmann O. How to assess the relative importance of different colonization routes of pathogens within hospital settings. Proc. Natl Acad. Sci. USA. 2002;99:5601–5605. doi: 10.1073/pnas.082412899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollett P.K. On a model for interference between searching insect parasites. J. Aust. Math. Soc. Ser. B. 1990;31:133–150. [Google Scholar]
- Pollett P.K. Integrals for continuous-time Markov chains. Math. Biosci. 2003;182:113–225. doi: 10.1016/S0025-5564(02)00161-X. [DOI] [PubMed] [Google Scholar]
- Pollett, P. K. 2006 Quasi-stationary distributions: a bibliography. See http://www.maths.uq.edu.au/∼pkp/papers/qsds/qsds.html.
- Pollett P.K, Stefanov V.T. Path integrals for continuous-time Markov chains. J. Appl. Prob. 2002;39:901–904. doi: 10.1239/jap/1037816029. [DOI] [Google Scholar]
- Pollett P.K, Stewart D.E. An efficient procedure for computing quasistationary distributions of Markov chains with sparse transition structure. Adv. Appl. Prob. 1994;26:68–79. doi: 10.2307/1427580. [DOI] [Google Scholar]
- Rand D.A, Wilson H.B. Chaotic stochasticity—a ubiquitous source of unpredictability in epidemics. Proc. R. Soc. B. 1991;246:179–184. doi: 10.1098/rspb.1991.0142. [DOI] [PubMed] [Google Scholar]
- Renshaw E. Cambridge University Press; Cambridge, UK: 1991. Modelling biological populations in space and time. [Google Scholar]
- Ross J.V. A stochastic metapopulation model accounting for habitat dynamics. J. Math. Biol. 2006a;52:788–806. doi: 10.1007/s00285-006-0372-8. [DOI] [PubMed] [Google Scholar]
- Ross J.V. Stochastic models for mainland-island metapopulations in static and dynamic landscapes. Bull. Math. Biol. 2006b;68:417–449. doi: 10.1007/s11538-005-9043-y. [DOI] [PubMed] [Google Scholar]
- Ross J.V, Taimre T, Pollett P.K. On parameter estimation in population models. Theor. Popul. Biol. 2006;70:498–510. doi: 10.1016/j.tpb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Rubinstein R.Y, Kroese D.P. Springer; Berlin, Germany: 2004. The cross-entropy method: a unified approach to combinatorial optimization Monte-Carlo simulation, and machine learning. [Google Scholar]
- Sidje R.B. Expokit. A software package for computing matrix exponentials. ACM Trans. Math. Softw. 1998;24:130–156. doi: 10.1145/285861.285868. [DOI] [Google Scholar]
- Spagnolo B, Fiasconaro A, Valenti D. Noise induced phenomena in Lotka–Volterra systems. Fluct. Noise. Lett. 2003;3:L177–L185. doi: 10.1142/S0219477503001245. [DOI] [Google Scholar]
- Stark K.D.C, Pfeiffer D.U, Morris R.S. Within-farm spread of classical swine fever virus—a blueprint for a stochastic simulation model. Vet. Quart. 2000;22:36–43. doi: 10.1080/01652176.2000.9695021. [DOI] [PubMed] [Google Scholar]
- Stollenwerk N, Briggs K.M. Master equation solution of a plant disease model. Phys. Lett. A. 2000;274:84–91. doi: 10.1016/S0375-9601(00)00520-X. [DOI] [Google Scholar]
- Stollenwerk N, Jansen V.A.A. Meningitis, pathogenicity near criticality: the epidemiology of meningococcal disease as a model for accidental pathogens. J. Theor. Biol. 2003;222:347–359. doi: 10.1016/S0022-5193(03)00041-9. [DOI] [PubMed] [Google Scholar]
- Taylor L.R. Aggregation, variation and the mean. Nature. 1961;189:732–735. doi: 10.1038/189732a0. [DOI] [Google Scholar]
- Trefethen L.N, Bau D. Society for Industrial and Applied Mathematics; Philadelphia, PA: 1997. Numerical linear algebra. [Google Scholar]
- van Kampen N.G. Noth-Holland; Amsterdam, The Netherlands: 1992. Stochastic processes in physics and chemistry. [Google Scholar]
- Verver S, Warren R.M, Munch Z, Richardson M, van der Spuy G.D, Borgdorff M.W, Behr M.A, Beyers N, van Helden P.D. Proportion of tuberculosis transmission that takes place in households in a high-incidence area. Lancet. 2004;363:212–214. doi: 10.1016/S0140-6736(03)15332-9. [DOI] [PubMed] [Google Scholar]
- Viet A, Medley G.F. Stochastic dynamics of immunity in small populations: a general framework. Math. BioSci. 2006;200:28–43. doi: 10.1016/j.mbs.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E.J, Haydon D.T, Pearson A, Kitching R.P. Failure of vaccination to prevent outbreaks of foot-and-mouth disease. Epidemiol. Infect. 1996;116:363–371. doi: 10.1017/s0950268800052699. [DOI] [PMC free article] [PubMed] [Google Scholar]