Abstract
Natural variation in the absolute and relative size of different parts of the human brain is substantial, with a range that often exceeds a factor of two. Much of this variation is generated by the cumulative effects of sets of unknown gene variants that modulate the proliferation, growth, and death of neurons and glial cells. Discovering and testing the functions of these genes should contribute significantly to our understanding of differences in brain development, behavior, and disease susceptibility. We have exploited a large population of genetically well-characterized strains of mice (BXD recombinant inbred strains to map gene variants that influence the volume of the dorsal striatum (caudate-putamen without nucleus accumbens). We used unbiased methods to estimate volumes bilaterally in a sex-balanced sample taken from the Mouse Brain Library (www.mbl.org). We generated a matched microarray data set to efficiently evaluate candidate genes (www.genenetwork.org). As in humans, volume of the striatum is highly heritable, with greater than two-fold differences among strains. We mapped a locus that modulates striatal volume on chromosome (Chr) 6 at 88 ± 5 Mb. We also uncovered an epistatic interaction between loci on Chr 6 and Chr 17 that modulates striatal volume. Using bioinformatic tools and the corresponding expression database, we have identified positional candidates in these QTL intervals.
Keywords: Striatum, Quantitative Trait Loci, Systems Genetics, Stereology, Point Counting
The volume of neuroanatomical structures varies widely in human populations (Blinkov & Glezer, 1968). For example, the volume of the basal ganglia and its major components, including the caudate nucleus, the putamen, and the globus pallidus, are highly variable: putamen volume in a control population ranged from 6.2–9.7 ml (Harris et al., 1999), and Rosas et al., (2001) reported a 1.4 fold difference in caudate volume in healthy controls. Importantly, individual differences in striatal volume are associated with differential susceptibility to several diseases including autism (Voelbel et al., 2006), schizophrenia (Beckmann & Lauer, 1997, Kreczmanski et al., 2007, Lauer & Beckmann, 1997), and other psychiatric disorders (Reiss et al., 1993).
In the present study we focus attention on the genetic basis of variation in the size of the dorsal striatum (caudate-putamen without nucleus accumbens) in a population of BXD recombinant inbred (RI) strains of mice that incorporates a level of genetic diversity roughly equivalent to that of many human populations (~2 million SNPs and CNVs). This set of genetically well-characterized BXD strains provides a system to experimentally test relations and interactions between normal variation in CNS neuroanatomical traits, differences in behavior, and differences in disease onset and severity (Rosen et al., 2003). The main advantage of using sets of genetically diverse but isogenic strains, as opposed to unique individuals, is that one can systematically accumulate a vast amount of diverse data for each strain. This enables a community of scientists with different interests and expertise to test and retest entire systems of traits, their covariance, and their genetic causes (Chesler et al., 2005, Chesler et al., 2004, Mozhui et al., 2007, Yang et al., 2008).
In a previous study using an F2 intercross between A/J and BXD5 mice, we identified a quantitative trait locus (QTL) on the distal end of chromosome (Chr) 10 that modulates striatal volume (Rosen & Williams, 2001). This prior study was constrained by the relatively small number of strains and cases being investigated. The recent addition of many new BXD strains and cases (Peirce et al., 2004) has significantly increased the power of this RI set and improved the precision of QTL mapping. In this experiment we have estimated striatal volume in both the right and left hemispheres in 53 BXD RI lines and both parental strains using unbiased stereology. We have also generated gene expression data for striatum for the same stains of mice to evaluate candidate genes within the major QTLs.
METHODS
Subjects
All histologic data for this study were obtained from The Mouse Brain Library (MBL)—a physical and Internet resource that contains high-resolution images of histologically processed slides from over 2600 adult mouse brains (www.mbl.org) with roughly balanced numbers of male and female specimens (Rosen et al., 2003). The age of most cases ranged between 50 and 100 days (young adult). Mice were obtained from either the Jackson Laboratory (Bar Harbor, ME) or the University of Tennessee Health Science Center (UTHSC) as detailed previously (Rosen & Williams, 2001). All procedures were approved by animal care and use committees and conform to NIH guidelines for humane treatment of animals. Mice were deeply anesthetized with Avertin (0.8 ml i.p.) and transcardially perfused with saline, followed by fixative (glutaraldehyde/paraformaldehyde), and their brains removed and weighed. After variable post-fixation times, the brains were embedded in 12% celloidin and sliced in either a coronal or horizontal plane at a width of approximately 30 μm. Actual section thickness was determined by direct examination of 10 sections for each brain using an x100 oil immersion objective and a z-axis micrometer.
Estimation of striatal volume
We estimated the volume of the dorsal striatum (equivalent to the human caudate nucleus and putamen) of 342 mice (176 female and 166 male) belonging to 53 BXD RI strains and the parental strains (C57BL/6J and DBA/2J, abbreviated B6 and D2). The volume of the right and left striatum was estimated by five individuals (GDR, CJP, CBO, JC, HK) using a computer controlled microscope (Nikon E800, Nikon, Inc., Melville, NY) and Stereo Investigator (MBF Biosciences, Williston, VT) by point counting using Cavalieri’s method (Gundersen & Jensen, 1987). Grid spacing for both horizontal (N = 171) and coronal (N = 171) sections was 400 μm and volume was estimated by measuring every tenth section on each of the two slides for each brain. In cases where there were missing or damaged sections, a piece-wise parabolic estimation was used (Rosen & Harry, 1990). Final volume estimates were individually corrected for histological shrinkage by determining the previously computed ratio between the brain volume at fixation (brain weight) and that after processing. For each individual, striatal volume was blindly re-estimated on 10 slides to assess intra-observer reliability. Inter-observer reliability was assessed by pairs of investigators estimating volume on 6–10 brains in common. The experimenter was blind with respect to strain and sex.
Measurement error
Intra-observer reliability was high for estimation of striatal volume. The percentage difference between the original and repeated volume estimations ranged from 0.9–8.7% and the average difference was 3.1%. These measures were highly correlated, ranging from 0.94–0.99, and a paired t-test revealed no significant differences between the two estimations (t <1, NS in all cases), indicating that technical error at this level of the analysis contributes little to case variation or strain variation. Inter-observer differences between estimations from the same brain ranged from 2.3%–9.7%, with an average difference of 5.1%. The measures were highly correlated (r = 0.87–0.94), and paired t-tests did not reveal any differences in these measures (t < 1 in all cases). Taken together, these results indicate that estimates of striatal volume were reliable.
Analysis
Data were analyzed using standard ANOVA and multiple regression techniques (JMP, SAS Institute, Cary, NC). QTL analysis was performed using the WebQTL module of GeneNetwork (GN, www.genenetwork.org). This on-line resource includes all known morphometric data for the BXD strains, several striatal transcriptome data sets, high density marker maps based on approximately 3795 full informative markers distributed on all chromosomes except Chr Y (Shifman et al., 2006), and a database containing ≈8.3 million single nucleotide polymorphisms (SNPs) taken from dbSNP (Frazer et al., 2007). WebQTL incorporates three common mapping methods: (1) simple interval mapping, (2) composite interval mapping, and (3) a scan for two-locus epistatic interactions (Wang et al., 2003). QTLs were determined by computation of a Likelihood Ratio Statistic (LRS) for each trait, and significance was assessed using 1000 permutations.
Array data
We generated expression data for the dorsal striatum for a set of 75 strains of mice, including most of the BXD strains included in the morphometric analysis, and integrated these data into www.genenetwork.org. To compare neuroanatomical and gene expression data we specifically used the GeneNetwork database named “HQF BXD Striatum ILM6.1 (Nov07) RankInv,” which can be accessed in the main search page by selecting Species = mouse, Group = BXD, Type = Striatum mRNA. Data were generated using the Illumina Sentrix Mouse-6.1 microarray. This array estimates expression for a great majority of mouse genes with confirmed protein products and consists of sets of ≈46,000 unique 50-nucleotide-long probe sequences. Like other array data in GeneNetwork (Chesler et al., 2005), the original Illumina bead array data (rank invariant transform) were logged and re-centered to a mean of 8 units and a standard deviation of 2 units—essentially a z transform of the data.
The HQF striatum data sets include information on gene expression for a total of 75 strains of mice (male and female samples) of which 47 are common to the neuroanatomical data described here. Strains for which we have morphometric data but no array data include BXD30, BXD35, BXD39, BXD63, BXD75, BXD83 and BXD92. All dissections were performed by the senior author (GDR), and we have confirmed by histology and Western Blotting of AchE that these samples are almost entirely pure dorsal striatum samples. For complete metadata on the HQF striatum data set, including quality control procedures error-checking, and normalization see www.genenetwork.org/dbdoc/UTHSC_1107_RankInv.html.
Comparing adult morphometric data with adult striatal expression data is potentially insufficient since it is possible that the transient expression of a small number of genes during development may leave a lasting imprint on differences in volume maturity (but see Zapala et al., 2005). For this reason, we generated a companion expression data set on neonatal (P1) striatal expression data for the two parental strains, C57BL/6J and DBA/2J (NIH Neuroscience Microarray Consortium <http://arrayconsortium.tgen.org>, rosen-illu-mouse-588967). Data were generated using the Illumina Sentrix Mouse-6.2 microarray. We have exploited this companion developmental data to 1) test whether genes with expression differences that covary with striatal volume at maturity are also expressed during a key stage of development in the striatum, and 2) to test whether any genes in QTL intervals have high expression only during development.
All genome coordinates in this paper are given using the mouse genome assembly of February 2006 (UCSC Genome Browser release mm8, NCBI Build 36). These position coordinated differ slightly (usually less than 1–3 Mb) from mm9 and NCBI Build 37.1.
Correlation analysis
To evaluate candidate genes and to study other covariates of variation in striatal volume we compared our data with the HQF data sets. Covariation networks were constructed using on-line tools in GN. In addition, we correlated striatal volume with the BXD Phenotype Database in GN that contains a nearly exhaustive collection of previously published and unpublished traits from the BXD RI line.
On-line data access
Phenotypes for the BXD strains generated as part of this study are all available at Genenetwork.org using the accession numbers:
Striatal volume (mm3): GN BXD Phenotypes Trait ID: 10710
Striatal volume adjusted (mm3): GN BXD Phenotypes Trait ID: 10998
Striatum expression data exploited in this study are directly accessible at www.genenetwork.org.
RESULTS
Striatal volume is highly variable
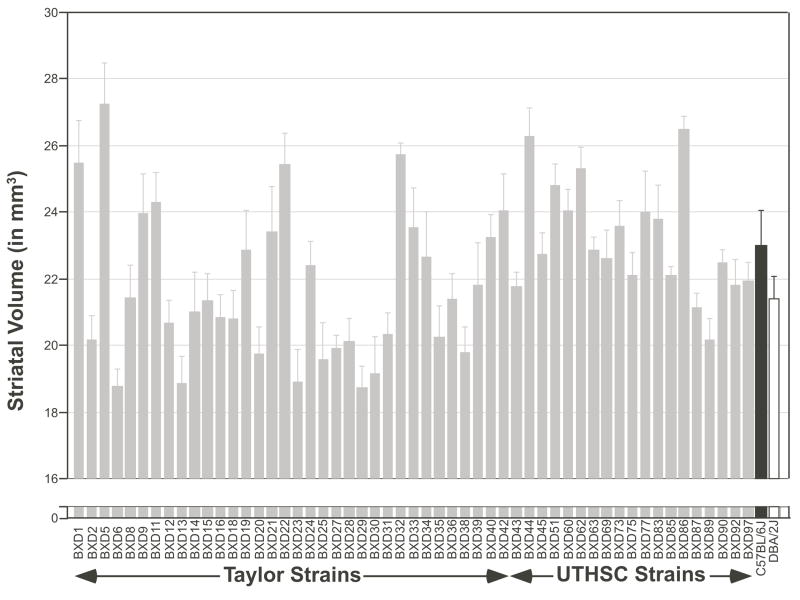
Bilateral striatal volume of all subjects ranged from 15.6 to 34.7 mm3 (mean ± SEM = 22.2 ± 0.8 mm3). In comparison, strain averages ranged from a low of 18.7 ± 0.62 mm3 in BXD29 to a high of 27.3 ± 1.2 mm3 in BXD5, with an average across all 55 strains of 22.2 ± 0.30 mm3 (see Fig 1). All estimates are fully corrected for case-by-case differences in shrinkage and should be considered close to the original size of these regions in well-fixed tissue. In vivo volumes would typically be 5–10% greater. From a genetic perspective, all of these strains can be regarded as normal wildtype but inbred mice. Right and left striatal volumes of individual mice ranged from 8.0–17.5 mm3 and 7.6–17.2 mm3. Mean striatal volume for both right and left sides were identical (11.1 ± 0.08 mm3).
Figure 1.
Mean ± SEM striatal volume in BXD RI lines (gray bars) and their parental strains, C57BL/6J (black bars) and DBA/2J (white bars).
There is no left/right asymmetry in striatal volume
We computed an asymmetry coefficient δasymm by subtracting the volume of the left from right hemisphere and dividing by one half of the total volume. To determine whether there were overall asymmetric biases irrespective of direction, we calculated an absolute asymmetry coefficient (δabs) by taking the absolute value of δasymm. We computed the distribution of both δasymm and δabs for striatal volume (mean ± SEM = −0.001 ± 0.017 and 0.031 ± 0.011 mm3, respectively), and ANOVA revealed no significant effects of strain (F52, 273 < 1, NS in both cases). These results suggest that there was no significant left/right asymmetry of striatal volume.
Striatal volume is heritable, but asymmetry is not
In order to determine the heritability of striatal volume, we computed an ANOVA with strain as the independent measure and volume as the dependent measure. We found a significant effect of strain (F54,287 = 5.2, P <.001), and this main effect accounts for 50–55% of the variance, which provides a reasonable upper bound on the fraction of variance that might be explained by additive and epistatic interactions. (Dominance effects cannot be measured using RI strains because there are no heterozygous genotypes or heterozygote phenotypes to analyze.)
There were no significant main effects for δasymm (F54,287 < 1, NS) or δabs (F54,287 <1, NS; explained variance = 14–27%). These results strongly support the notion that there are significant differences between strains on the volumetric measures, but that strain differences in asymmetry are not apparent.
We also computed heritability (h2) using inbred strain data and the adjustment method of Hegmann and Possidente (1981). Striatal volume is a moderately heritable trait (h2 = 0.33). The broad sense estimate of heritability, which takes in account the large sample sizes of genetically identical members of RI strains, yielded an h2 of 0.86. The heritability factor for δasymm and δabs in the striatum was 0.07 for both measures. These results indicate that asymmetry has a very low heritability could not be profitably mapped.
Mapping striatal volume
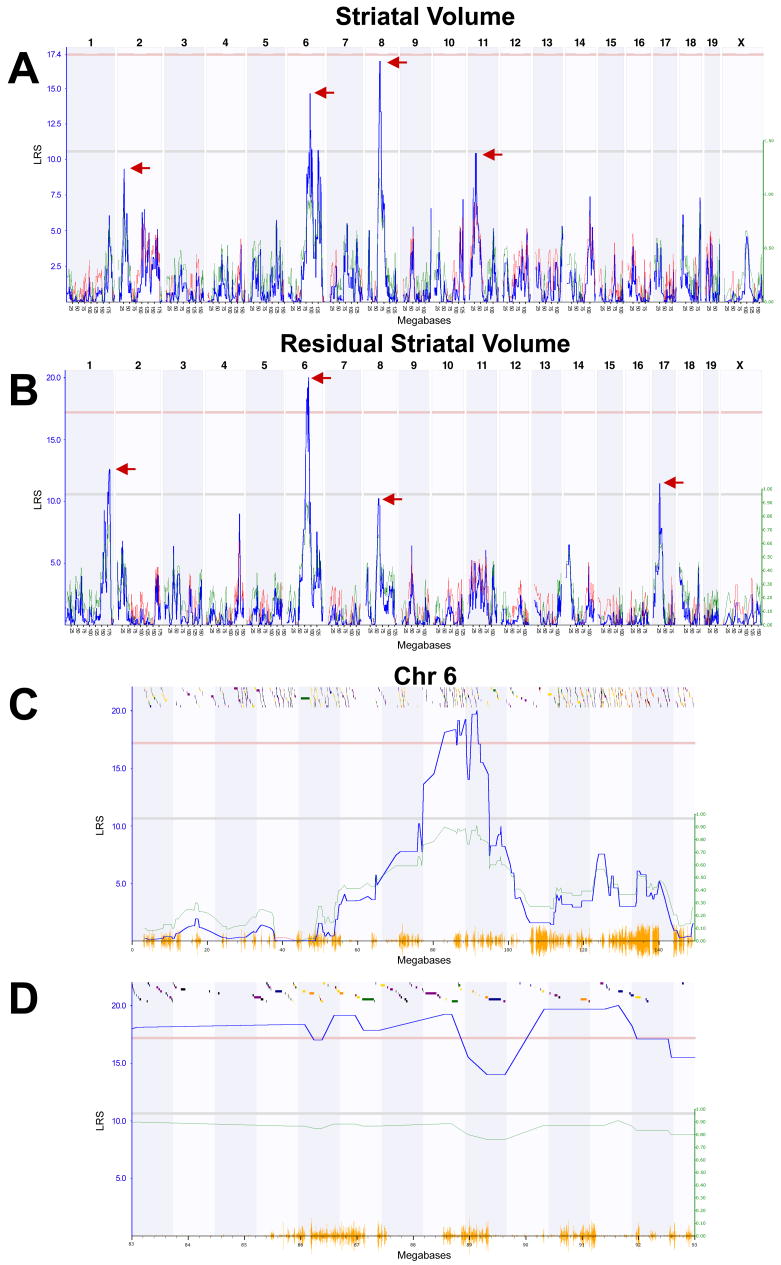
We mapped striatal volume and found a QTL on Chr 8 at 59 ± 5 Mb (LRS = 16.90, LOD = 3.7, Figure 2), which corresponds to human Chr 4 at ≈175 Mb (4q34.1). A suggestive QTL was also mapped to Chr 6 (LRS = 14.52, LOD = 3.2). This region corresponds to human Chr 3 at ≈134 Mb (3p25.1) Two other intervals contained suggestive loci with LOD scores greater than 2—one on Chr 2 at 27 ± 5 Mb (LRS = 9.31, LOD = 2.0) and another on Chr 11 at 32 ± 5 Mb (LRS = 10.41, LOD = 2.3).
Figure 2.
Mapping striatal volume. A. LRS scores for striatal volume across the entire genome. The x-axis represents the physical map of this chromosome; the y-axis and thick blue line provide the LRS of the association between the trait and the genotypes of markers. The two horizontal lines are the suggestive (blue) and significance (red) thresholds computed using 1000 permutations. There are QTLs (red arrows) on the proximal end of Chr 2, the distal end of Chr 6, the central region of Chr 8, and the proximal end of Chr 11. B. LRS for residual striatal volume (regressing out the effects of age, sex, plane of section, epoch, and non-striatal brain weight) reveals a significant QTL on the distal end of Chr 6. The LRS for the QTL on Chr 8 is now diminished (see text for explanation), and new QTLs on the distal end of Chr 1 and the proximal end of Chr 17 are revealed. C. LRS map of all of Chr 6. Orange lines on x-axis represent high density SNP map. Discontinuous track along the top are the genes on this chromosome. D. A 10 Mb interval bordering the QTL on Chr 6.
In order to map variation related to volume of striatum rather than possible confounding covariates, we performed multiple regression to remove covariance associated with differences in age, sex, plane of section, strain epoch (the original Taylor BXD set versus the newer UTHSC BXD set, (see Peirce et al., 2004, Shifman et al., 2006 for an explanation of the history of BXD strains and the subtle genetic differences between epochs), and non-striatal brain weight (brain weight–striatal weight). There were no significant effects for sex (F1,321 = 1.2, NS; females = 22.1 ± 0.2 mm3; males =22.3 ± 0.2 mm3), plane of section (F1,321 < 1, NS; horizontal = 22.2 ± 0.2 mm3; coronal = 22.2 ± 0.2 mm3), or age (F1,321 = 2.2, NS). There were significant effects of non-striatal brain weight (F1,321 = 105.2, P <.001) and epoch (F1,321 = 5.4, P <.05; Taylor strains = 21.7 ± 0.2 mm3; UTHSC strains = 23.2 ± 0.3). We regressed striatal volume for each subject against age, sex, plane of section, epoch, and non-striatal brain weight and calculated average residuals per strain. (The use of all variables, even those that do not reach an alpha level of .05, is warranted and removes that variable as a potential confound in mapping.). These residuals were used to compute adjusted strain means.
Mapping variation in adjusted striatal volume significantly improved the strength of linkage to the Chr 6 QTL (genome-wide P <.05, LRS = 19.7, LOD = 4.3) (Fig 2B–D). In contrast, the LRS at the Chr 8 QTL was substantially decreased (LRS = 10.2, LOD = 2.2). This latter finding suggests that the Chr 8 QTL is associated in part with more global differences in non-striatal brain regions and perhaps the two subgroups of BXD strains (epoch). There were two additional intervals with LOD scores greater than 2, the first on Chr 1 at 182 ± 5 Mb (LRS = 12.57, LOD = 2.7), the second on Chr 17 at 30 ± 5 Mb (LRS = 11.45, LOD = 2.5). Having a B allele on Chr 6 was associated with a 0.85 mm3 decrease in striatal volume when compared to strains having a D allele (F1,51 = 23.0, P <.001). Similarly, there was an approximately 0.7 mm3 decrease in striatal volume in strains having the B allele at the Chr 17 QTL interval (F1,51 = 12.3, P =.001), the Chr 1 QTL interval (F1,51 = 13.6, P <.001), and the Chr 8 QTL interval (F1,51 = 11.7, P <.01).
The expanded set of BXDs is now sufficiently large to search for possible two-way epistatic (non-linear) interactions among loci that contribute to variation in CNS neuroanatomical variation. This is the first study to apply this more complex genetic model in this context. A pair-scan revealed a significant interaction between the known Chr 6 and Chr 17 loci (full model LRS = 36.4, P <.05; Fig 3A, B). The genetic correlation between markers at these two loci was not significant (r = 0.16, NS), thereby excluding the possibility of non-syntenic association (see Williams et al., 2001 for an explanation of non-syntenic association). ANOVA with genotypes of markers in the Chr 6 and Chr 17 QTLs as the independent measures and the adjusted striatal volume as the dependent measure confirmed a significant main effects for both the Chr 6 QTL (F2,49 = 21.8, P <.001) and Chr 17 QTL (F2,49 = 14.7, P <.001). In addition, the interaction between these two QTLs was significant (F2,49 = 3.9, P =.05). These two QTLs interact synergistically, and those strains with D alleles at both intervals have significantly larger striata than expected from the summed effects of the two loci (Fig 3C).
Figure 3.
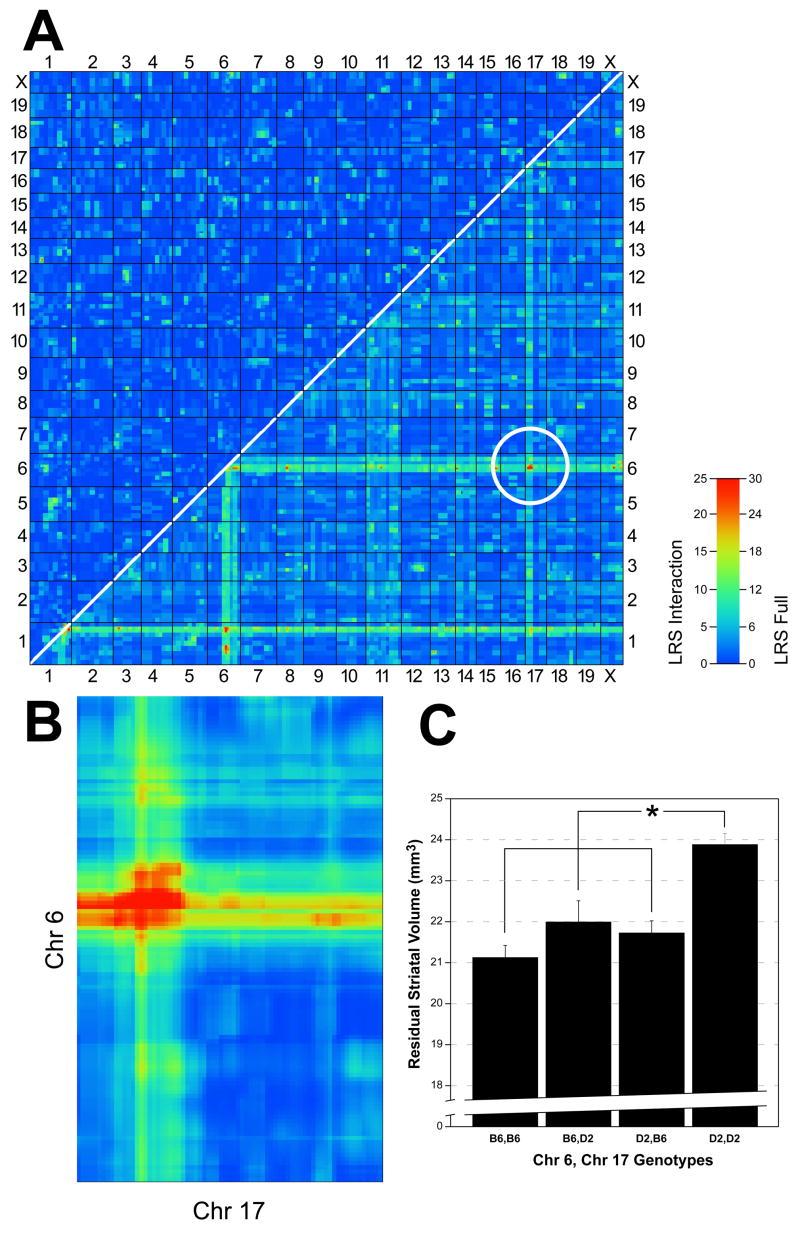
Pair-scan correlations demonstrate interactions between two QTLs for striatal volume. A. Pair-scan analysis of residual striatal volume across the genome. The upper left half of the plot highlights any epistatic interactions (corresponding to the column labeled “LRS Interact”). The lower right half provides a summary of LRS of the full model, representing cumulative effects of linear and non-linear terms (column labeled “LRS Full”). A significant interaction between QTLs on Chr 6 and 17 (white circle) is shown. B. Enlargement of intersection of Chrs 6 and 17 as seen in A, illustrating region of significance. C. Histogram illustrating the effect on adjusted striatal volume of carrying either the parental (D2) or maternal (B6) or both alleles at the Chr 6 and Chr 17 intervals. Having D2 alleles at both intervals significantly increases adjusted striatal volume when compared to all other allelic combinations.
Composite interval mapping of striatal volume data sets did not reveal any significant or consistent secondary loci.
Correlations with other BXD RI phenotypes
One advantage of employing the BXD stain is the ability to test for co-variation with other phenotypes that have also been studied in this population. We correlated striatal volume with the BXD Published Phenotypes Database of GN, which contains a large number of behavioral, anatomic, and physiologic phenotypes gathered from BXD RI sets. Alpha levels were adjusted after permuting striatal volume and adjusted striatal volume 20 times each, determining the top 50 correlations with the BXD Published Phenotypes Database for each permutation, and determining the computed alpha level of the top 5% of all correlations. From this analysis, it was determined that computed correlations by GN with P <.002 were significant at an adjusted alpha level of .05. P values represent the adjusted P values.
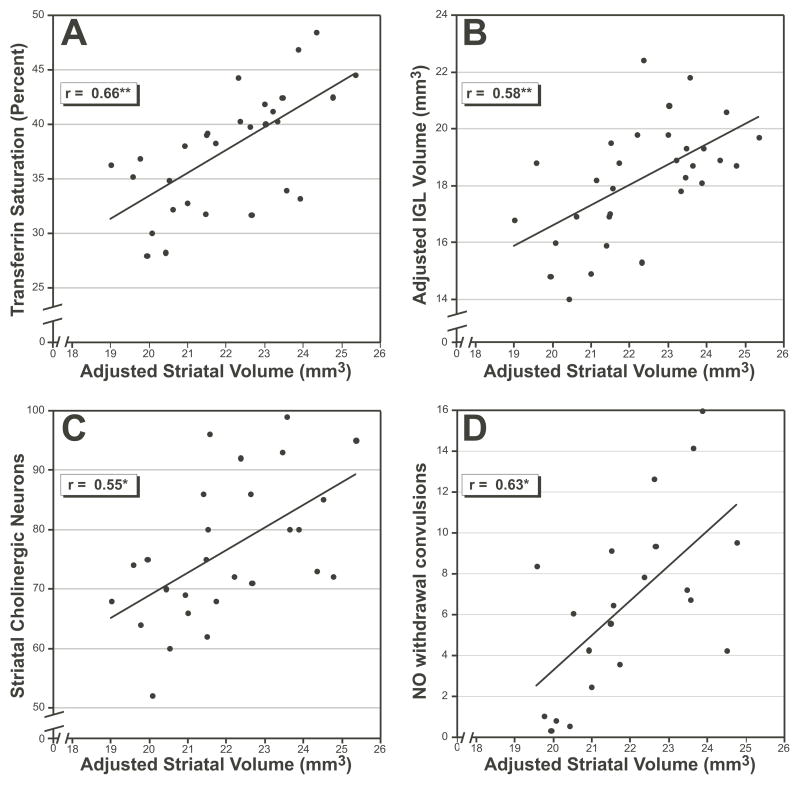
Not surprisingly, striatal volume (unadjusted) correlates primarily with other neuronanatomical measures including brain weight (r=0.72–0.77, P <.001), and volumes of the ventral hippocampus (r=0.67, P <.01), cerebellum (r=0.71, P <.01), dorsal thalamus (r=0.63, P <.01), cerebral cortex (r=0.58, P <.01) and basolateral amygdala (r=0.62, P <.01) (Airey et al., 2001, Beatty & Laughlin, 2006, Dong et al., 2007, Martin et al., 2006, Mozhui et al., 2007, Peirce et al., 2003, Seecharan et al., 2003, Williams et al., 1998). In contrast, adjusted striatal volume correlates with a variety of different phenotypes (Fig 4). The strongest correlation is with transferrin saturation percent (Jones et al., 2007), which is a measure of iron transport in the blood (r=0.66, P <.01, Fig 4A). The adjusted volume of the internal granule cell layer of the cerebellum (Airey et al., 2001) correlates with striatal volume (r=0.58, P <.01, Fig 4B). Interestingly, the number of striatal cholinergic neurons (Dains et al., 1996) correlates significantly with striatal volume (r=0.55, P <.05, Fig 4C), as did the number of convulsions following nitric oxide withdrawal (Belknap et al., 1993) (r=0.63, P <.05, Fig 4D).
Figure 4.
Scatterplots illustrating correlation of adjusted striatal volume with traits from the BXD Phenotype Database. Pearson product moment correlations (in box) of adjusted striatal volume with (A) transferrin saturation percent (Trait 10822), (B) the volume of the internal granule cell layer (IGL) of the cerebellum ITrait ID 10006), (C) the number of striatal cholinergic neurons (Trait 10106), and (D) the number of convulsions following nitrous oxide (NO) withdrawal (Trait 10027). *P <.05, **P <.01.
Candidate gene analysis
The QTLs that we identified can be subdivided into large blocks that have common haplotypes in B6 and D2 strains. These blocks are essentially identical by descent (except for any recent mutations) and have low densities of SNPs. Such regions are less likely to contain polymorphisms that modulate the volume of the striatum. In contrast, several large blocks have dissimilar haplotypes and much higher densities of SNPs (Fig 2D). Genes within these regions have a better prior probability of containing functional polymorphisms.
Chr 6 candidate analysis
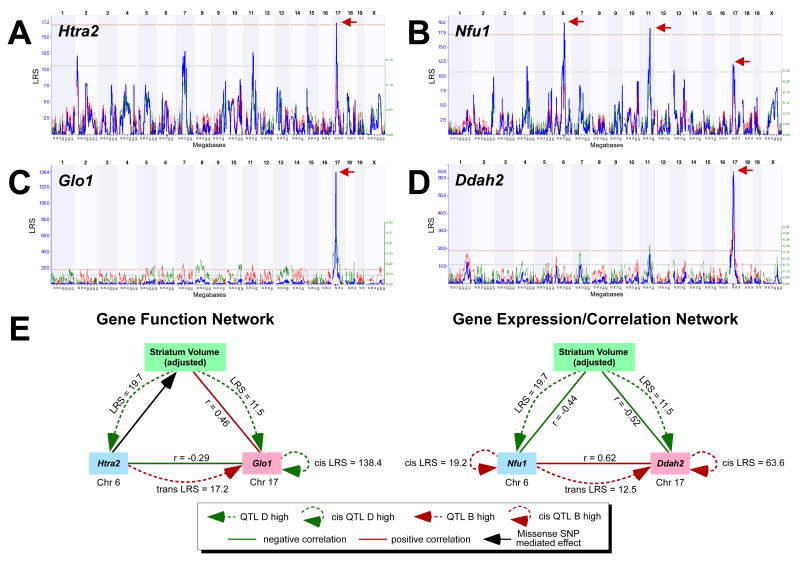
We combined data on SNP density with LRS values to rank positional candidate genes. Roughly one-quarter of ≈200 genes in the Chr 6 interval (80 and 95 Mb) were located within blocks that were highly polymorphic (1 to 6 SNPs per kilobase). Members of this subset are good positional candidates, irrespective of any role that they may have in brain development or adult forebrain structure (Supplemental Table 1). The stringency was increased by requiring that strong candidates would also have moderate to high expression in forebrain at some stage of development. To apply this filter, we extracted data on expression of all of these genes in the striatum from the HQF database and the neonatal database. A set of 34 transcripts had levels of expression that were at or above the average level (8 units) in both neonatal and adult striatum. This set of 34 candidates was ranked using data on missense SNPs in these genes and/or by evidence of local regulatory variation that controls their own expression—so-called cis-QTLs. Those genes with SNPs in Illumina microarray probes that produced high cis-QTLs were excluded from the analysis. A subset of 10 genes met these criteria: Htra2, Tia1, Mxd1, Anxa4, Aak1, Nfu1, Nup210, Hdac11, Fbln2, and Slc25a26 (Table 1). Mxd1, Aaka, Hdac11, and Slc25a26 had a greater than 2-fold increase in expression between P1 and P60, whereas the only gene with a greater than 2-fold decrease in expression was Fbln2. The remaining genes had no significant difference in expression between P1 and P60 (see Supplementary Table 1). Of all the positional candidates, Htra2 and Nfu1 were ranked at the top. Htra2 is especially noteworthy as deletion of this gene results in cell loss in the striatum and leads to neurodegenerative disorder with parkinsonian phenotype (Martins et al., 2004). Htra2 has a missense mutation in exon 8, and its expression maps as a significant trans-QTL to proximal Chr 17, close to the locus that is in epistatic interaction with the Chr 6 QTL (see Fig 5A). Nfu1 contains a missense mutation in exon 2. Functionally, its protein domain interacts with lafbrin and plays a potential role in Lafora disease, which is an autosomal recessive type of myoclonus epilepsy in humans (Ganesh et al., 2003). In addition to a highly significant cis-QTL, expression of Nfu1 maps as a suggestive trans-QTL to proximal Chr 17. (see Fig 5B).
Table 1.
Candidate genes in QTL interval on Chr 6
| Gene Symbol | Gene Description | SNP Region |
|---|---|---|
| Htra2 | HtrA serine peptidase 2 | exon 8 |
| Tia1 | T-cell restricted intracellular antigen 1 | promoter (2x), 3′ UTR (2x) |
| Mxd1 | MAX dimerization protein 1 | exon 6 |
| Anxa4 | annexin A4 | exon 13 |
| Aak1 | AP2-associated kinase 1 | exon 15 |
| Nfu1 | NFU1 iron-sulfur cluster scaffold homolog | exon 2 |
| Nup210 | nucleoporin 210 | exons 10,17,19,21 |
| Hdac11 | histone deacetylase 11 | promoter (×5) |
| Fbln2 | fibulin 2 | exons 2 (×3), 6 |
| Slc25a26 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 26 | promoter |
Figure 5.
Expression of potential candidate genes, and hypothesized gene networks. A. Interval map for Htra2 gene expression (see Fig 2 for explanation of the graph). This Chr 6 gene significantly modulates gene expression in the Chr 17 QTL interval (red arrow). B. Interval map for the Chr 6 gene Nfu1, which modulates its own gene expression, as well as gene expression on Chrs 11 and in the QTL interval on Chr 17. C and D. Interval maps for Glo1 and Ddah2, respectively, which are genes on the Chr 17 QTL interval that significantly modulate their own expression. These cis-QTLs were verified at the probe level, and are not likely due to undetected SNPs. E. Hypothetical gene networks modulating striatal volume based on gene function (left) and gene expression (right). Arrowed dotted lines indicate QTLs and circular arrowed lines indicate cis-QTLs, i.e., genes that modulate expression in the same chromosomal interval. In the case of both trans- and cis-QTLS, red lines represent contribution of the C57BL/6J and green the DBA/2J haplotype. Solid red lines represent positive correlations and solid green lines represent negative correlations, which are based on gene expression. Solid black line indicates missense SNP. See text for further explanation.
Visualization of genetic covariation
In order to assess the potential interactive effects of the Chr 6 and Chr 17 loci, we looked for correlations in expression across BXD strains in genes contained within both intervals. The Chr 17 region is gene-rich, and we selected those genes with high SNP densities, high expression in the striatum, and strong cis-QTLs. A total of 25 genes met these criteria (Supplementary Table 2). In terms of gene function, Glo1, a candidate gene in the Chr 17 QTL, may be functionally closely related to Htra2, the high priority candidate in the Chr 6 QTL. Both Htra2 and Glo1 are involved in oxidative stress response and apoptosis (Gray et al., 2000, Liu et al., 2007, Saito et al., 2004, Sakamoto et al., 2000). Glo1 is also associated with anxiety behavior in mice and mood disorder in humans (Fujimoto et al., 2008, Politi et al., 2006). Glo1 has over a 3-fold expression difference in the BXDs and is associated with a strong cis-QTL. This cis-QTL lies close to, and may overlap the trans-QTL for Htra2. Based on their functional relatedness and expression correlation, we generated a potential gene interaction network that may modulate the striatum (Fig 5E). In this model, the missense mutation in Htra2 and expression variation of Glo1 may collectively contribute to the variation in striatal volume.
Another potential gene network we generated is based mostly on strong correlations among the candidates on Chr 6 and Chr 17, and the striatum volume. Six candidate genes on Chr 17 correlate directly with adjusted striatal volume (Vps52, H2-D1, Zfp297, 0170001D07Rik, Cpne5, and Ddah2). Of these, Ddah2 also correlates strongly with Nfu1, the gene on Chr 6 previously identified as a high-ranking candidate. Expression of both Nfu1 and Ddah2 are associated with significant cis-QTLs, and both correlate strongly with volume of the striatum (Fig 5E).
To summarize these results, we constructed two models of gene variants that may modulate striatal volume. The first of these is based on known gene function and involves Htra2 and Glo1. The second involves less well-characterized genes, Nfu1 and Ddah2, and is based on both gene expression variation and correlation of expression with striatal volume (Fig 5E).
DISCUSSION
We have combined classic stereological methods, gene mapping methods, and a large-scale striatal transcriptome analysis to define and characterize sources of variation in the size of the mouse dorsal striatum. Striatal volume varies 2.2-fold among individuals and a 1.5-fold among strains. This variation is generated in part by a locus on Chr 6 and by interactions between chromosomes 6 and 17. Using whole genome array data we have extracted and ranked a small set of polymorphic candidate genes with comparatively high expression in neonatal and adult striatum.
There is a lack of anatomic asymmetry in the striatum
Because previous research has found evidence of volumetric asymmetry in the rodent brain (Caparelli-Daquer & Schmidt, 1999, Rosen et al., 1989a, Rosen et al., 1991, Rosen et al., 1993, Rosen et al., 1989b, Verstynen et al., 2001, Zilles et al., 1996), we attempted to assess this variable in the BXD RI set. We found that there was little evidence of asymmetry in the striatum. This was true both for measures that assessed directional (right vs. left) asymmetry and those that assessed magnitude (without considering directionality). Despite the fact that there are well-documented asymmetries in striatal dopamine in rats that underlie lateralized rotational behavior (Glick et al., 1976, Nielsen et al., 1997), there have been no documented anatomical asymmetries in this region (Rosen & Williams, 2001). The results reported here suggest that there are no population level anatomic asymmetries in the striatum, but do not discount the possibility that anatomic asymmetries at the individual level may correlate with functional and neurochemical asymmetries.
Correlations with BXD Phenotypes
We found a number of BXD phenotypes that significantly correlated with striatal volume. Unadjusted striatal volume correlated exclusively with other neuroanatomic measures, such as brain weight and volume of various brain regions. This result was not surprising, as we already demonstrated that a significant percent of the variance in striatal volume was accounted for by brain weight. It is therefore interesting that adjusted striatal volume—which factored out non-striatal brain weight as well as other variables—correlates strongly with behavioral, physiologic, as well as neuroanatomic phenotypes. The strongest correlation was with variables from a study of iron hemostasis in BXD lines (Jones et al., 2007), specifically the percent of transferrin saturation. Iron hemostasis is important in a number of diseases, and an increase in iron concentration in the basal ganglia has intriguingly been linked to age of onset of Parkinson’s disease (Bartzokis et al., 2004). This raises the possibility that those strains with smaller volumes (and lower transferring saturation percentage) might also be at greater risk for neurodengenerative disorders.
Adjusted striatal volume also correlated with behavioral and anatomic phenotypes. One of the first papers that used RI strains to detect non bimodal traits was that of Belknap et al., (1993), who mapped QTLs for withdrawal of nitrous oxide and ethanol withdrawal in mice. We found a significant correlation between the number of convulsions following nitric oxide withdrawal and adjusted striatal volume. This correlation is of interest given that the striatum is a target of nitrous oxide’s effects in the brain (e.g., Emmanouil et al., 2006, Henry et al., 2005). Our measure also correlated well with the number of cholinergic neurons in the striatum (Dains et al., 1996), which supports the notion that volume may be an easier to obtain first approximation of neuron number—a hypothesis that we are currently investigating. Taken together, these correlations from the BXD Phenotype Database point out the potential expansion of interesting hypotheses that can be gained through the use of the BXD genetic reference population.
It is also potentially important to note that there were a number of striatal traits in the phenotype database that were not correlated with striatal volume, including striatal copper levels (Jones et al., 2006), striatal choline uptake (Tarricone et al., 1995), and Drd1 and Drd2 levels in the striatum (Jones et al., 1999). Some of these previously published studies examined relatively small numbers of BXD strains, so the lack of a statistical significant correlation with striatal volume is perhaps not surprising. Interpreting lack of significance with correlation coefficients requires caution, but it does raise the possibility that, at least for these traits, their organization within the striatum remains relatively constant despite large changes in volume.
Previously reported QTLs for striatal volume
Using a small population of F2 mice derived from mating A/J and BXD5/TyJ mice, we had reported a QTL for striatal volume on Chr 10 (Rosen & Williams, 2001). We did not find support for this interval in the present study. There are a number of factors that would explain this difference. First, the strains used to create the cross in the previous experiment were different from those used to create the BXD RI lines in the present study. Second, and perhaps more important, is the difference in the size and types of the genetic reference populations. In our previous study, we mapped the QTL with 36 F2 mice that were genotyped at 82 markers. In the current study, by comparison, we were able to map QTLs after examining 342 mice from 55 strains that were genotyped at ~4,000 markers. Because QTL detection in RI sets improves with greater number of strains (Belknap, 1998, Crusio, 2004), it is likely that our ability to detect QTLs that modulate a smaller percent of the variance of these neuroanatomical traits was improved in this study. Moreover, the epistatic correlations that we report here could not possibly be detected with the small sample in the original study.
Positional candidate genes
The networks proposed in Fig 5 are considered at this point to be highly speculative. With this caveat in mind, the functions of some of the positional candidate genes suggest that they may play a role in various aspects important for neuronal development. Htra2 has been shown to be involved in apoptosis and neurodegeneration (Gray et al., 2000, Liu et al., 2005). Moreover, knockout mice that do not express Htra2 demonstrate specific cell loss in the striatum (Martins et al., 2004). Similarly, Glo1 has been associated with susceptibility to oxidative stress and apoptosis (Sakamoto et al., 2000, Zuin et al., 2005). Reduced expression of Glo1 has been associated with mood and panic disorders in humans (Fujimoto et al., 2008, Politi et al., 2006). The oncogene Mxd1 plays a role in cellular differentiation and is part of the MAX transcription factor network that governs many aspects of cell behavior, including cell proliferation and tumorogenesis (Hurlin & Huang, 2006, Vastrik et al., 1995). A number of the genes on Chr 6 are expressed during neural development, including Tia1, which is important for neuronal survival (Lowin et al., 1996), and Hdac11, which likely is involved in the maturation of neural cells (Liu et al., 2008). In addition, the NFU1 protein domain interacts with lafbrin and plays a potential role in Lafora disease, which is an autosomal recessive type of myoclonus epilepsy (Ganesh et al., 2003). Currently, a number of the positional candidate genes listed in Table 1 (Tia1, Aak1, Nfu1, Nup210, Hdac11, and Fbln2) are being targeted by the Knock Out Mouse Project (www.komp.org), and when these knockout mice are available, their striatal volume will be assessed.
Verification of the expression of these candidate genes in the striatum could potentially be derived from two atlases of gene expression in the mouse. The Allen Brain Atlas (ABA<http://www.brain-map.org>) uses in situ hybridization to map adult expression of most of the genes in the mouse genome. GENSAT < http://www.gensat.org/index.html> is a developing and adult mouse atlas that uses BAC-eGFP reported and BAC-Cre recombinase driver mouse lines. Of the 9 candidate genes that have been assayed by the ABA, 3 (Tia1, Hdac11, and Slc25a26) have been shown to have a high level of expression in the adult mouse striatum, which confirm our results (see Supplementary Table 1). Of the 6 genes that have moderate to low expression (Htra2, Mxd1, Anxa4, Nfu1, Nup210, and Fbln2) in the ABA, only Anxa4 and Fbln2 were moderately expressed in our dataset—the remaining 4 genes were all highly expressed in our analysis. GENSAT currently only has data for Hdac11, which is moderately expressed in the P7 striatum, and nearly absent in the adult. The P7 results are consistent with our P1 expression data, but are discrepant with both the ABA and our data. Verification of the expression of these candidate genes awaits RT-PCR of both P1 and P60 striatal tissue.
Future Directions
The QTLs that we have identified will require validation in the future. One potentially fruitful avenue would be to replicate these experiments using the Collaborative Cross (Churchill et al., 2004, Valdar et al., 2005), which is a program that is currently generating hundreds of RI lines based on an 8 strain cross. This cross—which is currently at the 13th generation—have far more recombinations than a standard RI strain, and so mapping precision will be improved. Another direction would be through the use of a Recombinant Inbred Intercross (RIX) scheme (Threadgill et al., 2002). This method involves generating all or a subset of the many pairs of intercrosses between a set of recombinant inbred strains. The derived RIX set extends the number of genomes available for phenotyping by a factor of n(n−1)/2, where n is the number of original RI strains. Each individual RIX strain has a unique but entirely predictable genome, and like RI strains, many genetically defined RIX individuals can be phenotyped to greatly improve trait reliability.
Conclusions
Our data clearly demonstrate the highly variable nature of striatal volume among inbred mouse strains, and that at least part of that variability is modulated genetically. Using bioinformatic tools and gene expression arrays, we have identified QTLs that modulate this trait. This study also represents the first use of the both the original BXD/Ty lines (Taylor, 1978) along with the newly generated BXD advanced recombinant inbred strains (Peirce et al., 2004) for the purpose of mapping neuroanatomic traits. This increase in sample size from the original 26 JAX BXD strains upon which so much work has been based (e.g., Crabbe et al., 1994, Hsu et al., 2007) to more than 50 strains has accomplished two things: 1) The power of correlation analyses and other statistical tests is now significantly greater, and 2) The power and precision of BXD mapping data are now significantly better because the new UTHSC BXD stains incorporate roughly twice as many recombinations per strain as conventional RI strains. This increased power and precision increases linkage statistics and decreases the size of confidence intervals and the number of positional candidate genes. Future research exploiting this expanded BXD genetic reference population will allow us to further dissect other important neuroanatomic phenotypes that may contribute to variation in regional volume, such as proliferation, death, number and packing density of neurons.
Supplementary Material
Acknowledgments
This work was support, in part, by NS052397 from NINDS to GDR, by DA021131 from NIDA, NIAAA, and NIMH to GDR and RWW, U01AA014425 to LL, and by U01AA13499, U01CA105417, and U24 RR021760 for support of GeneNetwork. We thank the High Q Foundation for making it possible to generate companion expression data for striatum, and the NIH Microarray Consortium and the Yale/NIH Microarray Center for performing the P1 Illumina microarray analyses. We thank Dr. Xusheng Wang for help annotating the content of the Illumina array.
References
- Airey DC, Lu L, Williams RW. Genetic control of the mouse cerebellum: Identification of quantitative trait loci modulating size and architecture. J Neurosci. 2001;21:5099–5109. doi: 10.1523/JNEUROSCI.21-14-05099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G, Tishler TA, Shin IS, Lu PH, Cummings JL. Brain ferritin iron as a risk factor for age at onset in neurodegenerative diseases. Ann N Y Acad Sci. 2004;1012:224–236. doi: 10.1196/annals.1306.019. [DOI] [PubMed] [Google Scholar]
- Beatty J, Laughlin R. Genomic regulation of natural variation in cortical and noncortical brain volume. BMC Neuroscience. 2006;7:16. doi: 10.1186/1471-2202-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H, Lauer M. The human striatum in schizophrenia. II. Increased number of striatal neurons in schizophrenics. Psychiatry Res. 1997;68:99–109. doi: 10.1016/s0925-4927(96)02947-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Metten P, Helms ML, O’Toole LA, Angeli-Gade S, Crabbe JC, Phillips TJ. Quantitative trait loci (QTL) applications to substances of abuse: physical dependence studies with nitrous oxide and ethanol in BXD mice. Behav Genet. 1993;23:213–222. doi: 10.1007/BF01067426. [DOI] [PubMed] [Google Scholar]
- Blinkov SM, Glezer II. The Human Brain in Figures and Tables. Basic Books; New York: 1968. [Google Scholar]
- Caparelli-Daquer EM, Schmidt SL. Morphological brain asymmetries in male mice with callosal defects due to prenatal gamma irradiation. Int J Dev Neurosci. 1999;17:67–77. doi: 10.1016/s0736-5748(98)00060-4. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Belknap JK, Buck KJ, Metten P. Use of recombinant inbred strains for studying genetic determinants of responses to alcohol. Alcohol Alcohol Suppl. 1994;2:67–71. [PubMed] [Google Scholar]
- Crusio WE. A note on the effect of within-strain sample sizes on QTL mapping in recombinant inbred strain studies. Genes Brain Behav. 2004;3:249–251. doi: 10.1111/j.1601-183X.2004.00079.x. [DOI] [PubMed] [Google Scholar]
- Dains K, Hitzemann B, Hitzemann R. Genetics, neuroleptic response and the organization of cholinergic neurons in the mouse striatum. J Pharmacol Exp Ther. 1996;279:1430–1438. [PubMed] [Google Scholar]
- Dong H, Martin MV, Colvin J, Ali Z, Wang L, Lu L, Williams RW, Rosen GD, Csernansky JG, Cheverud JM. Quantitative trait loci linked to thalamus and cortex gray matter volumes in BXD recombinant inbred mice. Hered. 2007;99:62–69. doi: 10.1038/sj.hdy.6800965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouil DE, Papadopoulou-Daifoti Z, Hagihara PT, Quock DG, Quock RM. A study of the role of serotonin in the anxiolytic effect of nitrous oxide in rodents. Pharmacology, biochemistry, and behavior. 2006;84:313–320. doi: 10.1016/j.pbb.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, Pethiyagoda CL, Stuve LL, Johnson FM, Daly MJ, Wade CM, Cox DR. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Uchida S, Watanuki T, Wakabayashi Y, Otsuki K, Matsubara T, Suetsugi M, Funato H, Watanabe Y. Reduced expression of glyoxalase-1 mRNA in mood disorder patients. Neurosci Lett. 2008;438:196–199. doi: 10.1016/j.neulet.2008.04.024. [DOI] [PubMed] [Google Scholar]
- Ganesh S, Tsurutani N, Suzuki T, Ueda K, Agarwala KL, Osada H, Delgado-Escueta AV, Yamakawa K. The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Genet. 2003;12:2359–2368. doi: 10.1093/hmg/ddg253. [DOI] [PubMed] [Google Scholar]
- Glick SD, Zimmerberg B, Greenstein S. Individual differences among mice in normal and amphetamine-enhanced locomotor activity: relationship to behavioral indices of striatal asymmetry. Brain Res. 1976;105:362–364. doi: 10.1016/0006-8993(76)90436-4. [DOI] [PubMed] [Google Scholar]
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, Savopoulos J, Hassan NJ, Clinkenbeard H, Hanning C, Amegadzie B, Davis JB, Dingwall C, Livi GP, Creasy CL. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. European journal of biochemistry/FEBS. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Gundersen HJG, Jensen EB. The efficiency of systematic sampling in stereology and its prediction. J Microscop. 1987;147:229–263. doi: 10.1111/j.1365-2818.1987.tb02837.x. [DOI] [PubMed] [Google Scholar]
- Harris GJ, Codori AM, Lewis RF, Schmidt E, Bedi A, Brandt J. Reduced basal ganglia blood flow and volume in pre-symptomatic, gene-tested persons at-risk for Huntington’s disease. Brain. 1999;122(Pt 9):1667–1678. doi: 10.1093/brain/122.9.1667. [DOI] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Henry ED, Ohgami Y, Li S, Chung E, Quock RM. Correlation of inbred mouse sensitivity to nitrous oxide antinociception with brain nitric oxide synthase activity following exposure to nitrous oxide. Pharmacology, biochemistry, and behavior. 2005;81:764–768. doi: 10.1016/j.pbb.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Hsu HC, Lu L, Yi N, Van Zant G, Williams RW, Mountz JD. Quantitative trait locus (QTL) mapping in aging systems. Methods Mol Biol. 2007;371:321–348. doi: 10.1007/978-1-59745-361-5_23. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Seminars in cancer biology. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Jones BC, Beard JL, Gibson JN, Unger EL, Allen RP, McCarthy KA, Earley CJ. Systems genetic analysis of peripheral iron parameters in the mouse. Am J Physiol Regul Integr Comp Physiol. 2007;293:R116–124. doi: 10.1152/ajpregu.00608.2006. [DOI] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, Rodriguez LA, Reed CL, McClearn GE, Plomin R, Erwin VG. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9:607–617. [PubMed] [Google Scholar]
- Jones LC, McCarthy KA, Beard JL, Keen CL, Jones BC. Quantitative genetic analysis of brain copper and zinc in BXD recombinant inbred mice. Nutritional neuroscience. 2006;9:81–92. doi: 10.1080/00268970600691365. [DOI] [PubMed] [Google Scholar]
- Kreczmanski P, Heinsen H, Mantua V, Woltersdorf F, Masson T, Ulfig N, Schmidt-Kastner R, Korr H, Steinbusch HW, Hof PR, Schmitz C. Volume, neuron density and total neuron number in five subcortical regions in schizophrenia. Brain. 2007;130:678–692. doi: 10.1093/brain/awl386. [DOI] [PubMed] [Google Scholar]
- Lauer M, Beckmann H. The human striatum in schizophrenia. I. Increase in overall relative striatal volume in schizophrenics. Psychiatry Res. 1997;68:87–98. doi: 10.1016/s0925-4927(96)02946-0. [DOI] [PubMed] [Google Scholar]
- Liu H, Hu Q, Kaufman A, D’Ercole AJ, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J Neurosci Res. 2008;86:537–543. doi: 10.1002/jnr.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Liu ML, Shen YF, Kim JM, Lee BH, Lee YS, Hong ST. Transgenic mice with neuron-specific overexpression of HtrA2/Omi suggest a neuroprotective role for HtrA2/Omi. Biochem Biophys Res Commun. 2007;362:295–300. doi: 10.1016/j.bbrc.2007.07.118. [DOI] [PubMed] [Google Scholar]
- Liu ML, Liu MJ, Kim JM, Kim HJ, Kim JH, Hong ST. HtrA2 interacts with A beta peptide but does not directly alter its production or degradation. Molecules and cells. 2005;20:83–89. [PubMed] [Google Scholar]
- Lowin B, French L, Martinou JC, Tschopp J. Expression of the CTL-associated protein TIA-1 during murine embryogenesis. J Immunol. 1996;157:1448–1454. [PubMed] [Google Scholar]
- Martin MV, Dong H, Vallera D, Lee D, Lu L, Williams RW, Rosen GD, Cheverud JM, Csernansky JG. Independent quantitative trait loci influence ventral and dorsal hippocampal volume in recombinant inbred strains of mice. Genes Brain Behav. 2006;5:614–623. doi: 10.1111/j.1601-183X.2006.00215.x. [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K, Hamre KM, Holmes A, Lu L, Williams RW. Genetic and structural analysis of the basolateral amygdala complex in BXD recombinant inbred mice. Behav Genet. 2007;37:223–243. doi: 10.1007/s10519-006-9122-3. [DOI] [PubMed] [Google Scholar]
- Nielsen DM, Visker KE, Cunningham MJ, Keller RW, Glick SD, Carlson JN. Paw preference, rotation, and dopamine function in Collins HI and LO mouse strains. Physiol Behav. 1997;61:525–535. doi: 10.1016/s0031-9384(96)00496-9. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Chesler EJ, Williams RW, Lu L. Genetic architecture of the mouse hippocampus: identification of gene loci with selective regional effects. Genes Brain Behav. 2003;2:238–252. doi: 10.1034/j.1601-183x.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi P, Minoretti P, Falcone C, Martinelli V, Emanuele E. Association analysis of the functional Ala111Glu polymorphism of the glyoxalase I gene in panic disorder. Neurosci Lett. 2006;396:163–166. doi: 10.1016/j.neulet.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Reiss AL, Faruque F, Naidu S, Abrams M, Beaty T, Bryan RN, Moser H. Neuroanatomy of Rett syndrome: a volumetric imaging study. Ann Neurol. 1993;34:227–234. doi: 10.1002/ana.410340220. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Goodman J, Chen YI, Jenkins BG, Kennedy DN, Makris N, Patti M, Seidman LJ, Beal MF, Koroshetz WJ. Striatal volume loss in HD as measured by MRI and the influence of CAG repeat. Neurology. 2001;57:1025–1028. doi: 10.1212/wnl.57.6.1025. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Harry JD. Brain volume estimation from serial section measurements: A comparison of methodologies. J Neurosci Meth. 1990;35:115–124. doi: 10.1016/0165-0270(90)90101-k. [DOI] [PubMed] [Google Scholar]
- Rosen GD, La Porte NT, Diechtiareff B, Pung CJ, Nissanov J, Gustafson C, Bertrand L, Gefen S, Fan Y, Tretiak O, Manly KF, Park MR, Williams AG, Connolly MT, Capra JA, Williams RW. Informatics center for mouse genomics: The dissection of complex traits of the nervous system. Neuroinformatics. 2003;1:327–342. doi: 10.1385/NI:1:4:327. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Sherman GF, Galaburda AM. Interhemispheric connections differ between symmetrical and asymmetrical brain regions. Neuroscience. 1989a;33:525–533. doi: 10.1016/0306-4522(89)90404-1. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Sherman GF, Galaburda AM. Ontogenesis of neocortical asymmetry: A [3H]thymidine study. Neuroscience. 1991;41:779–790. doi: 10.1016/0306-4522(91)90368-x. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Sherman GF, Galaburda AM. Neuronal subtypes and anatomic asymmetry: Changes in neuronal number and cell-packing density. Neuroscience. 1993;56:833–839. doi: 10.1016/0306-4522(93)90131-x. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Sherman GF, Mehler C, Emsbo K, Galaburda AM. The effect of developmental neuropathology on neocortical asymmetry in New Zealand Black mice. Int J Neurosci. 1989b;45:247–254. doi: 10.3109/00207458908986237. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Williams RW. Complex trait analysis of the mouse striatum: Independent QTLs modulate volume and neuron number. BMC Neuroscience. 2001;2:5. doi: 10.1186/1471-2202-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Modulation of the Omi/HtrA2 signaling pathway after transient focal cerebral ischemia in mouse brains that overexpress SOD1. Brain Res Mol Brain Res. 2004;127:89–95. doi: 10.1016/j.molbrainres.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Mashima T, Kizaki A, Dan S, Hashimoto Y, Naito M, Tsuruo T. Glyoxalase I is involved in resistance of human leukemia cells to antitumor agent-induced apoptosis. Blood. 2000;95:3214–3218. [PubMed] [Google Scholar]
- Seecharan DJ, Kulkarni AL, Lu L, Rosen GD, Williams RW. Genetic control of interconnected neuronal populations in the mouse primary visual system. J Neurosci. 2003;23:11178–11188. doi: 10.1523/JNEUROSCI.23-35-11178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS biology. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone BJ, Hwang WG, Hingtgen JN, Mitchell SR, Belknap JK, Nurnberger JI., Jr Identification of a locus on mouse chromosome 17 associated with high-affinity choline uptake using BXD recombinant inbred mice and quantitative trait loci analysis. Genomics. 1995;27:161–164. doi: 10.1006/geno.1995.1019. [DOI] [PubMed] [Google Scholar]
- Taylor BA. Recombinant inbred strains. Use in gene mapping. In: Morse H, editor. Origins of inbred mice. Academic; New York: 1978. pp. 423–438. [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R. Simulating the collaborative cross: power of QTL detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2005 doi: 10.1534/genetics.104.039313. genetics. 104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, Kaipainen A, Penttila TL, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstynen T, Tierney R, Urbanski T, Tang A. Neonatal novelty exposure modulates hippocampal volumetric asymmetry in the rat. NeuroReport. 2001;12:3019–3022. doi: 10.1097/00001756-200110080-00008. [DOI] [PubMed] [Google Scholar]
- Voelbel GT, Bates ME, Buckman JF, Pandina G, Hendren RL. Caudate nucleus volume and cognitive performance: Are they related in childhood psychopathology? Biol Psychiatry. 2006;60:942–950. doi: 10.1016/j.biopsych.2006.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Williams RW, Manly KF. WebQTL: Web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:0046. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Strom RC, Goldowitz D. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci. 1998;18:138–146. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RJ, Mozhui K, Karlsson RM, Cameron HA, Williams RW, Holmes A. Variation in Mouse Basolateral Amygdala Volume is Associated With Differences in Stress Reactivity and Fear Learning. Neuropsychopharmacology. 2008 doi: 10.1038/sj.npp.1301665. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Lockhart DJ, Barlow C. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Dabringhaus A, Geyer S, Amunts K, Qü M, Schleicher A, Gilissen E, Schlaug G, Steinmetz H. Structural asymmetries in the human forebrain and the forebrain of non-human primates and rats. Neurosci Biobehav Rev. 1996;20:593–605. doi: 10.1016/0149-7634(95)00072-0. [DOI] [PubMed] [Google Scholar]
- Zuin A, Vivancos AP, Sanso M, Takatsume Y, Ayte J, Inoue Y, Hidalgo E. The glycolytic metabolite methylglyoxal activates Pap1 and Sty1 stress responses in Schizosaccharomyces pombe. J Biol Chem. 2005;280:36708–36713. doi: 10.1074/jbc.M508400200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.