Abstract
The study aimed to reveal alterations in expression and methylation levels of the growth-related imprinted genes H19 and Igf2 in fetuses of diabetic mice. Diabetes was induced in female mice by intraperitoneal injection of streptozotocin. DNA and total RNA were extracted from fetuses obtained from diabetic and control dams on embryonic day (E) 14. Real-time RT-PCR analysis revealed that the mRNA expression of Igf2 in fetuses from diabetic mice was 0.65-fold of the control counterparts. Bisulfite genomic sequencing demonstrated that the methylation level of the H19–Igf2 imprint control region was 19.1% higher in diabetic fetuses than in those of control dams. In addition, the body weight of pups born to diabetic dams was 26.5% lower than that of the control group. The results indicate that maternal diabetes can affect fetal development by means of altered expression of imprinted genes. The modified genomic DNA methylation status of imprinting genes may account for the change in gene expression.
Abbreviations: E, embryonic day
Human epidemiologic and experimental animal studies strongly suggest that maternal diabetes influences adult susceptibility to obesity, glucose intolerance, and type 2 diabetes in the offspring. Gestational diabetes is characterized by an increased placental transport of glucose and other nutrients, resulting in fetal and neonatal macrosomia or microsomia.1,2 The level of nutrition available during gestation has been proposed to influence developmental programming through predictive adaptive responses that set appropriate postnatal growth and metabolic criteria.15 Diabetes during pregnancy induces marked abnormalities in glucose homeostasis and insulin secretion in the fetus that result in aberrant fetal growth and have long-term consequences for the offspring,16 including increased risk for obesity, glucose intolerance, and type 2 diabetes in later childhood and adulthood.3,26,30 A genetic contribution to the development of diabetic fetuses is commonly assumed. Several studies have shown that the effects of maternal diabetes on embryopathy are associated with abnormal expression of genes involved in developmental control, metabolism, and signal transduction.29,44 Growth of early mammalian embryos and fetal growth and development can be modulated by genomic imprinting.21,35 Because of the unique epigenetic requirements associated with allele-specific expression, genomically imprinted genes may be especially sensitive to environmental influences during development.28,36,40 Indeed, recent results from animal models and human epidemiologic studies support this hypothesis. For example, subtle differences in the medium used to culture preimplantation mouse embryos and pregnant mice fed a methyl-supplemented diet affected methylation and expression of imprinted genes.21,41 These data indicate that the environment of the early embryo can affect the establishment and maintenance of epigenetic mechanisms regulating the expression of genomically imprinted genes.
H19 and Igf2 are neighboring imprinted genes on mouse distal chromosome 7. Igf2 is expressed from the paternal allele, whereas H19 is from the maternal allele.5,10 These 2 genes are expressed widely during fetal development in the same tissues and are downregulated shortly after birth.8 Alterations in the expression of Igf2 severely affect fetal growth in the mouse.9 Deletion of H19 leads to the birth of pups that are 27% heavier than their wild-type littermates and increases Igf2 expression.23
Genome-wide methylation can change extensively during early embryonic development, and several imprinted genes undergo allele-specific changes in methylation during both gametogenesis and early embryogenesis.13 Any change in methylation status could result in deregulation of development at later stages; therefore imprinted genes are perhaps most vulnerable to alterations induced by exogenous factors and other nutritional changes. The allelic methylation status of the imprint control region upstream of the H19 gene is critical to imprinted expression of both H19 and Igf2.37,38 The imprint control region on the maternal chromosome is unmethylated, which enables the zinc-finger protein CTCF to bind to the ICR and blocks access of the enhancers located downstream of H19 to the Igf2 promoter (Figure 1). However, the enhancers can interact with the H19 promoter, and H19 is active. When the imprint control region at the CTCF binding site is methylated, Igf2 promoters can interact with enhancers, thereby triggering the Igf2 genes.11,45
Figure 1.
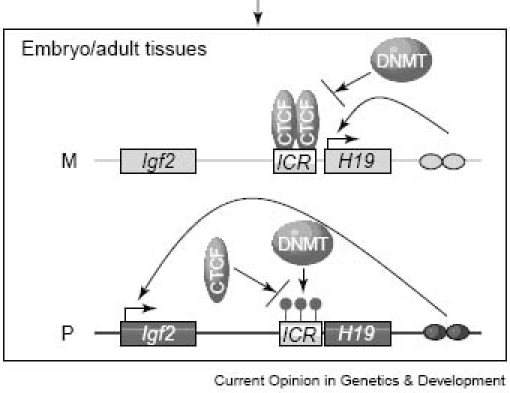
Establishment and maintenance of DNA methylation at the imprint control region (ICR) of the H19 –Igf2 locus. M, maternal allele; P, paternal allele; ovals, enhancers; CTCF, CCCTC-binding factor; DNMT, DNA methyltransferase. Adapted from reference 11.
Maternal dietary restriction, the culture medium, environmental contamination, and radiation all might alter H19 and Igf2 gene expression and methylation level of imprinted control region in animal fetuses.22,39,45,47 Such alterations would be maintained somatically and might affect gene expression at later stages of development.43 However, whether embryo environment in vivo, such as maternal diabetes, may similarly alter the pattern of embryonic imprinted gene expression with lasting consequences is unknown. To evaluate this possibility, the present study assessed expression levels of imprinted genes with the use of real-time PCR and analyzed the methylation levels of the imprinted control region of H19–Igf2 by using bisulfite genomic sequencing and restriction endonuclease digests.
Materials and Methods
Animals.
ICR strain mice (specific pathogen-free; age, 6 to 8 wk) were purchased from Shanghai Laboratory Animal Center (Chinese Academy of Science, Shanghai, PR China).
Female mice were induced to develop diabetes by means of intraperitoneal injection of 150 mg/kg streptozotocin (Sigma-Aldrich, Steinheim, Germany). One week after injection, the glucose concentration of a blood sample from the cut tip of the tail was measured in a glucometer, Free Style Mini Glucose Meter, Accu-Chek (Roche Diagnostics, Shanghai, PR China). Mice with a glucose concentration exceeding 20 mM were considered to be diabetic. Sodium citrate injected female mice were used as controls. After the diabetic status was verified, diabetic and control mice were mated with normal male mice. The presence of a vaginal plug the morning after mating indicated gestational day 0.5.
Mating ability and fetal mortality were monitored. Pregnant mice were killed by cervical dislocation after light ether anesthesia, and fetuses (n = 8 per group) were obtained on E14. Total RNA and DNA were purified from each fetus; 4 pregnancies per group were used for the experiment. Some dams were maintained until the pups were born (E21), when they were weighed.
The animals had free access to food and water and were maintained at an ambient temperature of 22 °C on a 12:12-h light:dark cycle. All animal protocols were approved by the Shanghai Laboratory Animal Care and Ethics Committee.
Preparation of total RNA.
Total RNA was extracted from whole fetuses by using RNeasy Mini Kits (Qiagen, Hilden, Germany). RNA concentration was quantified by measuring the absorbance at 260 nm in a photometer (Biophotometer, Eppendorf, Hamburg, Germany); ratios of absorption (260:280 nm) of all preparations were between 1.9 and 2.0.
Reverse transcription.
Aliquots (1 μg) of total RNA were reverse-transcribed by incubation at 37 °C for 1 h in a 30-μl reaction volume that consisted of 10 U AMV reverse transcriptase (Promega, Madison, WI), 40 U RNase inhibitor, 0.17 μmol/l random primers (9 bp), 250 mmol/l Tris-HCl (pH 8.3), 50 mmol/l MgCl2, 250 mmol/l KCl, 2.5 mmol/l spermidine, 50 mmol/l dithiothreitol, and 1.0 mmol/l each dNTP. The reaction was terminated by heating at 95 °C for 5 min and quickly cooling on ice.
Fluorescent real-time quantitative PCR.
mRNA transcription of Igf2 and H19 was quantified relative to that of 18S rRNA by using the Quantum RNA 18S Internal Standards Kit (Ambion, Austin, TX). Quantitative real-time PCR analysis was performed using a thermocycler (Opticon 2, MJ Research, Miami, FL) and the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA); 1 μl of each reverse transcription reaction mix was amplified in a 20-μl volume that contained 2 μl iQ SYBR Green Supermix and 0.2 μmol/l of a primer pair specific for Igf2, H19, or 18S rRNA. After completion of the final cycle, a melting curve analysis was performed to monitor the purity of the PCR products. We used this information to calculate the differential expression of the genes of interest. Gene expression levels were calculated and presented as 2 –ΔΔc(t) values (n = 8).25
The primer pairs for H19 and Igf2 were designed as described45 and synthesized (Shengneng Bicolor Biotech, Shanghai, PR China). The nucleotide sequences of these primers and the PCR conditions set for the genes of interest are shown in Table 1.
Table 1.
Nucleotide sequences of specific primers and PCR conditions
| Target gene | GenBank accession no. | PCR product (bp) | Primer sequences | PCR conditions |
| Igf2 | U71085 | 253 | forward: gtg tgt gtc agc caa gca tg | 95 °C ×15 min; |
| reverse: caa atg tgg gga cac aga gg | 95 °C ×15 s, | |||
| 56°C × 20 s, | ||||
| 72 °C × 20 s; | ||||
| 28 cycles | ||||
| H19 | Af049091 | 185 | forward: tac ccc ggg atg act tca tc | 95°C×15 min; |
| reverse: tat ctc cgg gac tcc aaa cc | 95°C×15 s, | |||
| 56°C×20 s, | ||||
| 72°C×20 s; | ||||
| 28 cycles | ||||
| Imprint control region of H19–Igf2 | U19619 | 498 | forward: gta taa gaa ttt tgt aag gag att atg ttt | 95 °C × 5 min; |
| reverse: ata aat caa ata cct aaa ata act ctt aaa | 95 °C × 1 min, | |||
| 55°C × 1 min, | ||||
| 72 °C × 1 min; | ||||
| 40 cycles | ||||
| Imprint control region of H19–Igf2 | U19619 | 430 | forward: ttt gta agg aga tta tgt ttt att ttt gga | 95 °C × 5 min; |
| reverse: ccc taa cct cat aaa acc cat aac tat aaa | 95 °C × 1 min, | |||
| 55 °C × 1 min, | ||||
| 72 °C × 1 min; | ||||
| 35 cycles | ||||
| Imprint control region of H19–Igf2 | U19619 | 408 | forward: gga acc gcc aac aag aaa gt | 95 °C × 10 min |
| reverse: ggt cttt cca ctc aca acg g | 95 °C × 30 s, | |||
| 53 °C × 1 min, | ||||
| 72 °C × 1 min; | ||||
| 32 cycles |
DNA isolation and bisulfite treatment.
Genomic DNA of E14 fetuses from both diabetes and control groups was isolated by using DNeasy Mini Kits (Qiagen) according to the manufacturer's protocol. The DNA concentration and purity were evaluated spectrophotometrically (Eppendorf). DNA samples were digested overnight with NotI and then subjected to sodium bisulfite treatment by using the CpGenome DNA Modification Kit (Chemicon, Billerica, MA) according to the manufacturer's protocol.
PCR amplification, cloning, and sequencing.
The 430-bp imprint control region of the H19–Igf2 locus was PCR-amplified with AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). Two rounds of PCR were performed with the fully nested primer pairs shown in Table 1. Each reaction mixture for the first-round PCR (product, 498 bp) contained 2 µl sodium bisulfite-treated DNA, 2 µl 10× PCR buffer II (provided with the polymerase), 2 mmol/l MgCl2, 200 µmol/l dNTPs, 0.2 µmol/l of each primer, and 0.4 µl AmpliTaq Gold polymerase (5 U/µl) in a total volume of 20 µl.
For the second-round PCR, 1 µl of the first-round products was used as template. The PCR products (430 bp) were separated on 1% agarose gels, and the bands were purified with Agarose Gel DNA Purification Kit (Qiagen). The purified DNA was subcloned into pGEM-T vector (Promega) by using T4 DNA ligase and transformed into JM109 cells. Colonies were picked and their DNA amplified, positive colonies were sequenced (BigDye Terminator Cycle Sequencing Kit, version 3.1, Applied Biosystems) with standard primers (M13 forward and reverse) on an automated sequencer (Prism 3700, Applied Biosystems). A total of 16 CpG sites in the 5′ end of the imprint control region of the H19–Igf2 region were examined. On E14, 2 or 3 clones corresponding to each of 8 fetuses in the diabetes and control groups were selected; in total, 21 clones from the diabetes group and 17 clones from control group were sequenced.
Restriction endonuclease digests and quantitative PCR.
Quantitative analysis of DNA methylation was performed at the imprint control region of H19–Igf2.20 The locus contains 3 methylation sites that can be detected by differential restriction endonuclease digestion. Fetus genomic DNA (<1 µg) was digested with methylation-sensitive (HpaII) or methylation-insensitive endonucleases (MspI) and used as templates for PCRs (408 bp). It was quantified relative to undigested genomic DNA. PCR primer pairs were designed to amplify regions flanking 3 restriction sites in the imprinted control region of H19–Igf2 gene locus (Table 1). Quantitative real-time PCR analysis was performed by using a thermocycler (Opticon 2, MJ Research) and the iQ SYBR Green Supermix (Bio-Rad). Aliquots (1 μl) of digested and undigested DNA were used for PCR in 20-μl reaction volumes containing 2 μl iQ SYBR Green Supermix and 0.2 μmol/l of the appropriate primer pair. After completion of the final cycle, a melting curve analysis was performed to monitor the purity of the PCR products. The DNA undigested levels were calculated and presented with 2 –ΔΔc(t) values (n = 8).25
Statistical analysis.
SPSS13.0 for Windows (StatSoft, Tulsa, OK) was used for the statistical analysis. All results are expressed as mean ± SEM. Differences in methylation levels were analyzed by χ2 test, and differences in birth weight and gene expression were analyzed by 1-way ANOVA. Statistical significance was indicated by a P value of less than 0.05.
Results
Embryonic development.
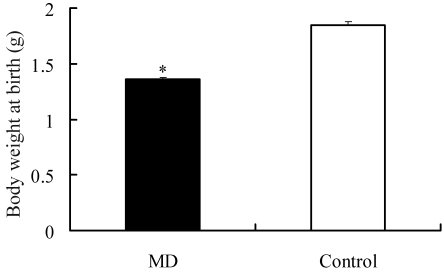
Fetal mortality among the progeny of diabetic dams was 20.5%. The birth weight of pups born to diabetic dams was 26.5% (P < 0.01) lower than that of controls (Figure 2). These data were obtained from 35 fetuses of diabetic dams (5 pregnancies) and 32 control fetuses (3 pregnancies).
Figure 2.
Effect of maternal diabetes on fetal birth body weight. Body weight of E21 fetuses in diabetic and control groups. Data for 35 diabetic (MD) and 32 control fetuses from 8 recipients in each group were compared. Results are expressed as mean ± SEM. **, P < 0.01.
Expression of Igf2 and H19 mRNAs.
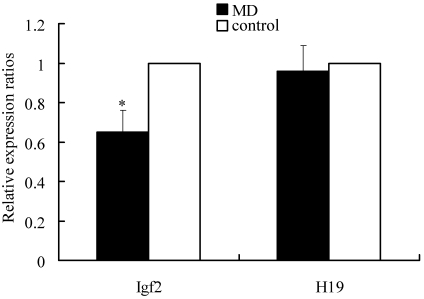
Real-time quantitative PCR analysis revealed that the ratio of mRNA expression in fetuses from diabetic dams to that in control fetuses was 0.65 for Igf2 (P < 0.05) and 0.98 for H19 (Figure 3). These data were obtained from 8 control fetuses and 8 insulin-exposed fetuses of 4 recipients for each treatment.
Figure 3.
Analysis of relative gene expression fluorescent quantitative RT-PCR. The 2−ΔΔCt method was used to analyze gene expression in E14 mouse fetuses; 18S rRNA was used as a reference gene. Data for diabetes-exposed (MD) and control fetuses (n = 8 fetuses each) from 4 recipients were compared. *, P < 0.05.
Methylation patterns of CpG islands in the imprint control region of H19–Igf2 in E14 mouse fetuses.
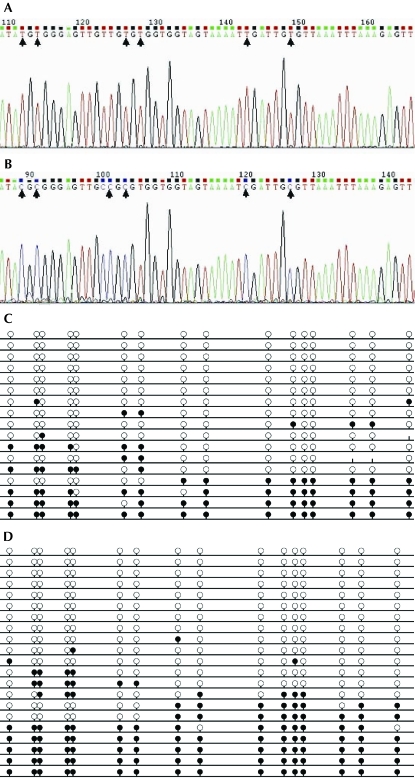
Seventeen clones from control E14 fetuses and 21 clones from E14 fetuses from diabetic dams were sequenced. The nucleotide sequences of representative clones are shown in Figure 4 A, B. Among the progeny of diabetic dams, 4 (19%) of the 21 clones prepared from 8 individual PCR products exhibited methylation at all CpG islands. In comparison, in the control group, 1 (5.9%) of the 17 clones assayed from 8 individual PCR products was fully methylated. On average, 34.5% of CpG sites were methylated in the diabetes group compared with 27.9% of CpGs in the control group (P < 0.05). Therefore the CpG methylation ratio in the targeted region of genomic DNA was increased by 19.1% in the diabetes group, compared with the control group.
Figure 4.
Summary of clone data for the imprint control region of H19–Igf2 in E14 mouse fetuses. Nucleotide sequences of representative clones from the (A) control group and (B) insulin-exposed group. Methylation patterns of CpG islands in (C) 17 clones of the control group and (D) 21 clones of the diabetes group. Each line represents a separate clone; a black circle indicates a methylated CpG, whereas an open circle indicates an unmethylated CpG. Differences in methylation between the diabetes group and control group were significant (P < 0.05).
Methylation levels of the imprint control region of H19–Igf2 in E14 fetuses.
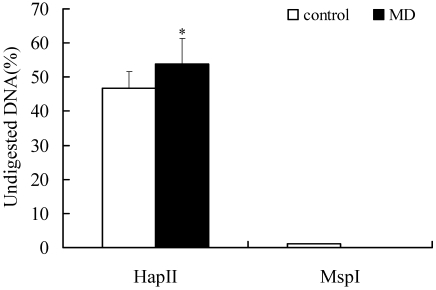
In the control group, 46.7% of DNA was undigested by the methylation-sensitive endonuclease (HpaII), whereas 53.9% was undigested in the diabetes group (Figure 5). Therefore the methylation ratio of the targeted region of genomic DNA was increased by 13.4% in the diabetes group, compared with the control group (P < 0.05).
Figure 5.
Quantitative analysis of DNA methylation in the imprint control region of H19–Igf2 in E14 fetuses. Genomic DNA from E14 fetuses of MD and control (n = 8 per group) was digested with methylation-sensitive (HapII) or –insensitive (MspI) endonucleases. The proportion of DNA that remained undigested was measured by fluorescent quantitative RT-PCR. The 2−ΔΔCt method was used to analyze gene expression. *, P < 0.05.
Discussion
These data confirm that maternal diabetes increases fetal mortality and decreases birth weight, as has been previously found.6,18,27 Streptozotocin-induced diabetes is dose-dependent: high doses induce insulin-deficient diabetes related to fetal growth restriction.17 The present study found that streptozotocin-induced diabetes is associated with intrauterine growth restriction.
The mRNA expression level of Igf2 in E14 fetuses from diabetic dams was significantly lower than in controls. Alterations in the expression of Igf2 severely affect the growth of mouse fetuses.9 Several authors have proposed that nutrition and other environmental stimuli during development may induce persistent changes in the epigenetic regulation of genomically imprinted genes.24,33,42 For example, pregnant mice fed a methyl-supplemented diet containing methionine, betaine, folic acid, and vitamin B12 at conception showed increased DNA methylation in Avy agouti mice offspring.41 Further, feeding pregnant rats a low-protein diet that contained excess methionine relative to other amino acids led to hypermethylation of DNA in fetal liver.32 Our results indicate that maternal diabetes altered DNA methylation in the imprint control region of H19–Igf2. Therefore, the embryonic environment can affect the establishment or maintenance of epigenetic mechanisms that persist in cell-specific patterns of gene activity throughout life. Moreover, these data suggest that imprinted genes are susceptible to environmental perturbation that increases the possibility of their involvement in metabolic imprinting.4
The regulation of imprinted genes correlates well with their DNA methylation.14 DNA methylation is involved in transcriptional silencing of genes, leading to regulation of expression of imprinted genes.34 Our data show that maternal diabetes affected fetal development and suggest a link between alterations in H19–Igf2 mRNA expression and genomic methylation status in the imprint control region of H19–Igf2. Expression of both H19 and Igf2 is dependent on an H19-Igf2 imprint control region, which is located 2 kb upstream of the H19 gene. Typically, when enhancers interact with promoters, transcription is turned on.45 In the present study, we found a higher methylation level in the H19-Igf2 imprint control region of the maternal diabetes group. Whereas increased Igf2 and decreased H19 expression levels would be expected in this situation, expression levels of Igf2 were decreased on E14 of fetuses exposed to maternal diabetes. This finding is agreement with a previous report of a higher methylation level in the H19-Igf2 imprint control region of fetuses exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin.45 The mechanisms of the decreased expression Igf2 are unknown, but differential histone modification has been reported at many imprinted domains. The presence of histone modifications may be a cause or a consequence of monoallelic gene expression.12
Alteration of the DNA methylation status is associated with methyltransferase.7 Three functional DNA methyltransferases—Dnmtl, Dnmt3a, and Dnmt3b—are involved in the genomic methylation pattern in mammals. Dnmt3a and Dnmt3b have been shown to be important for de novo methylation and are essential for embryonic development.12 Dnmt3a is essential for maintenance and establishment of maternal and paternal imprints.19
To our knowledge, the present study is the first to provide evidence that maternal diabetes can alter the expression level of H19–Igf2 and the genomic DNA methylation status of imprinted genes. The present findings may provide insights into early fetus endocrine disorders in other mammalian species, including human. Any endocrine disruption induced by environmental or nutritional factors, clinical treatments, or disease condition during ‘critical windows’ of early embryonic development may cause epigenetic alterations, which may be transmitted to the subsequent generations to induce phenotypic changes.38
The development of embryos exposed to maternal diabetes is complex, involving aberrant glucose and lipid metabolism, disturbed cell signaling, and altered gene expression.46 Hyperglycemia can induce altered gene expression, resulting in aberrant cell signaling, morphogenesis, and embryopathy.31 In future experiments, we will systematically examine differential gene expression profiles in the fetuses of diabetic and normal mice.
In conclusion, maternal diabetes can affect fetal development by increasing fetal mortality and decreasing birth weight. Our data also suggest a link between H19–Igf2 mRNA expression and genomic methylation status in the imprint control region of H19–Igf2.
Acknowledgments
This work was supported by National Basic Research Program of China (2004CB117505) and Shanghai Science and Technology Committee (054909001).
References
- 1.Aerts L, Holemans K, Van Assche FA. 1990.. Impaired insulin response and action in offspring of severely diabetic rats. In: Frontiers in diabetes research. Lessons from animal diabetes, III. p 561–566 [Google Scholar]
- 2.Aerts L, Holemans K, Van Assche FA. 1990. Maternal diabetes during pregnancy: consequences for the offspring. Diabetes Metab Rev 6:147–167 [DOI] [PubMed] [Google Scholar]
- 3.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. 1993. Type 2 (noninsulin-dependent) diabetes mellitus, hypertension, and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia 36:62–67 [DOI] [PubMed] [Google Scholar]
- 4.Barlow DP. 1995. Gametic imprinting in mammals. Science 270:1610–1613 [DOI] [PubMed] [Google Scholar]
- 5.Bartolomei MS, Zemel S, Tilghman SM. 1991. Parental imprinting of the mouse H19 gene. Nature 351:153–155 [DOI] [PubMed] [Google Scholar]
- 6.Beebe LF, Kaye PL. 1991. Maternal diabetes and retarded preimplantation development of mice. Diabetes 40:457–461 [DOI] [PubMed] [Google Scholar]
- 7.Bestor TH. 2000. The DNA methyltransferases of mammals. Hum Mol Genet 9:2395–2402 [DOI] [PubMed] [Google Scholar]
- 8.Cooney CA, Dave AA, Wolff GL. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 132 Suppl:2393S–2400S [DOI] [PubMed] [Google Scholar]
- 9.DeChiara TM, Efstratiadis A, Robertson EJ. 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345:78–80 [DOI] [PubMed] [Google Scholar]
- 10.DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859 [DOI] [PubMed] [Google Scholar]
- 11.Delaval K, Feil R. 2004. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 14:188–195 [DOI] [PubMed] [Google Scholar]
- 12.Edwards CA, Ferguson-Smith AC. 2007. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19:281–289 [DOI] [PubMed] [Google Scholar]
- 13.Feil R. 2006. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res 600:46–57 [DOI] [PubMed] [Google Scholar]
- 14.Gebert C, Wrenzycki C, Herrmann D, Groger D, Reinhardt R, Hajkova P, Lucas-Hahn A, Carnwath J, Lehrach H, Niemann H. 2006. The bovine IGF2 gene is differentially methylated in oocyte and sperm DNA. Genomics 88:222–229 [DOI] [PubMed] [Google Scholar]
- 15.Gluckman PD, Hanson MA. 2004. Maternal constraint of fetal growth and its consequences. Semin Fetal Neonatal Med 9:419–425 [DOI] [PubMed] [Google Scholar]
- 16.Gluckman PD, Harding J. 1997. Fetal growth retardation: underlying endocrine mechanisms and postnatal consequences. Acta Paediatr Suppl 422:69–72 [DOI] [PubMed] [Google Scholar]
- 17.Holemans K, Aerts L, Van Assche FA. 2003. Fetal growth restriction an consequences for the offspring in animal models. J Soc Gynecol Investig 10:392–399 [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Kondo Y, Nakatani A, Hayashi K, Naruse A. 2001. Characterization of low dose streptozotocin-induced progressive diabetes in mice. Environ Toxicol Pharmacol 9:71–78 [DOI] [PubMed] [Google Scholar]
- 19.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429:900–903 [DOI] [PubMed] [Google Scholar]
- 20.Katoh M, Curk T, Xu QK, Zupan B, Kuspa A, Shaulsky G. 2006. Developmentally regulated DNA methylation in dictyostelium discoideum. Eukaryot Cell 5:18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khosla S, Dean W, Brown D, Reik W, Feil R. 2001. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod 64:918–926 [DOI] [PubMed] [Google Scholar]
- 22.Kwong WY, Miller DJ, Ursell E, Wild AE, Wilkins AP, Osmond C. 2006. Imprinted gene expression in the rat embryo-fetal axis is altered in response to periconceptional maternal low protein diet. Reproduction 132:265–277 [DOI] [PubMed] [Google Scholar]
- 23.Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 9:2079–2089 [DOI] [PubMed] [Google Scholar]
- 24.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. 2005. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135:1382–1386 [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–ΔΔC(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 26.Martyn CN, Hales CN, Barker DJ, Jespersen S. 1998. Fetal growth and hyperinsulinaemia in adult life. Diabet Med 15:688–694 [DOI] [PubMed] [Google Scholar]
- 27.Palomar-Morales M, Baiza LA, Verdín-Terán L, Román-Ramos R, Altamirano-Lozano M, Méndez JD. 1998. Fetal development in alloxan-treated rats. Reprod Toxicol 12:659–665 [DOI] [PubMed] [Google Scholar]
- 28.Pembrey M. 1996. Imprinting and transgenerational modulation of gene expression: human growth as a model. Acta Genet Med Gemellol (Roma) 45:111–125 [DOI] [PubMed] [Google Scholar]
- 29.Phelan SA, Ito M, Loeken MR. 1997. Neural tube defects in embryos of diabetic mice: role of the Pax3 gene and apoptosis. Diabetes 46:1189–1197 [DOI] [PubMed] [Google Scholar]
- 30.Plagemann A, Harder T, Franke K, Kohlhoff R. 2002. Long-term impact of neonatal breast-feeding on body weight and glucose tolerance in children of diabetic mothers. Diabetes Care 25:16–22 [DOI] [PubMed] [Google Scholar]
- 31.Reece EA, Ji I, Wu YK, Zhao Z. 2006. Characterization of differential gene expression profiles in diabetic embryopathy using DNA microarray analysis. Am J Obstet Gynecol 195:1075–1080 [DOI] [PubMed] [Google Scholar]
- 32.Rees WD, Hay SM, Brown DS, Antipatis C, Palmer RM. 2000. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J Nutr 130:1821–1826 [DOI] [PubMed] [Google Scholar]
- 33.Ross MG, Desai M, Khorram O, McKnight RA, Lane RH, Torday J. 2007. Gestational programming of offspring obesity: a potential contributor to Alzheimer's disease. Curr Alzheimer Res 4:213–217 [DOI] [PubMed] [Google Scholar]
- 34.Serman A, Vlahovic M, Serman L, Bulic-Jakus F. 2006. DNA methylation as a regulatory mechanism for gene expression in mammals. Coll Antropol 30:665–671 [PubMed] [Google Scholar]
- 35.Smith FM, Garfield AS, Ward A. 2006. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res 113:279–291 [DOI] [PubMed] [Google Scholar]
- 36.Thompson SL, Konfortova G, Gregory RI, Reik W, Dean W, Feil R. 2001. Environmental effects on genomic imprinting in mammals. Toxicol Lett 120:143–150 [DOI] [PubMed] [Google Scholar]
- 37.Thorvaldsen JL, Duran KL, Bartolomei MS. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12:3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tremblay KD, Duran KL, Bartolomei MS. 1997. A 5′ 2-kb region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 17:4322–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterland RA, Jirtle RL. 2003. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterland RA, Jirtle RL. 2004. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20:63–68 [DOI] [PubMed] [Google Scholar]
- 41.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. 2006a. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis 44:401–406 [DOI] [PubMed] [Google Scholar]
- 42.Waterland RA, Lin JR, Smith CA, Jirtle RL. 2006b. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet 15:705–716 [DOI] [PubMed] [Google Scholar]
- 43.Weber M, Milligan L, Delalbre A, Antoine E, Brunel C, Cathala G, Forne T. 2001. Extensive tissue-specifc variation of allelic methylation in the Igf2 gene during mouse fetal development: relation to expression and imprinting. Mech Dev 101:133–141 [DOI] [PubMed] [Google Scholar]
- 44.Wentzel P, Wentzel CR, Gareskog MB, Eriksson UJ. 2001. Induction of embryonic dysmorphogenesis by high glucose concentration, disturbed inositol metabolism, and inhibited protein kinase C activity. Teratology 63:193–201 [DOI] [PubMed] [Google Scholar]
- 45.Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. 2004. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and Igf2. Biol Reprod 70:1790–1797 [DOI] [PubMed] [Google Scholar]
- 46.Zhao Z, Reece EA. 2005. Experimental mechanisms of diabetic embryopathyand strategies for developing therapeutic interventions. J Soc Gynecol Investig 12:549–557 [DOI] [PubMed] [Google Scholar]
- 47.Zhu B, Huang XH, Chen JD, Lu YC, Chen Y, Zhao JY. 2006. Methylation changes of H19 gene in sperms of X-irradiated mouse and maintenance in offspring. Biochem Biophys Res Commun 340:83–89 [DOI] [PubMed] [Google Scholar]