Abstract
Matrix Attachment Therapy (MAT) is an enzyme prodrug strategy that targets hyaluronan in the tumor extracellular matrix to deliver a prodrug converting enzyme near the tumor cells. A recombinant fusion protein containing the hyaluronan binding domain of TSG-6 (Link) and yeast cytosine deaminase (CD) with an N-terminal His(×6) tag was constructed to test MAT on the C26 colon adenocarcinoma in Balb/c mice that were given 5-fluorocytosine (5-FC) in the drinking water. LinkCD was expressed in E.coli and purified by metal-chelation affinity chromatography. The purified LinkCD fusion protein exhibits a Km of 0.33 mM and Vmax of 15 μM/min/μg for the conversion of 5-FC to 5-fluorouracil (5-FU). The duration of the enzyme activity for LinkCD was longer than that of CD enzyme at 37 °C: the fusion protein retained 20% of its initial enzyme activity after 24 hr, and 12% after 48 hr. The LinkCD fusion protein can bind to a hyaluronan oligomer (12-mer) at a KD of 55 μM at pH 7.4 and a KD of 5.32 μM at pH 6.0 measured using surface plasmon resonance (SPR). To evaluate the anti-tumor effect of LinkCD/5-FC combination therapy in vivo, mice received intratumoral injections of LinkCD on days 11 and 14 after C26 tumor implantation and the drinking water containing 10 mg/mL of 5-FC starting on day 11. To examine if the Link domain by itself was able to reduce tumor growth, we included treatment groups that received LinkCD without 5-FC and Link-mtCD (a functional mutant that lacks cytosine deaminase activity) with 5-FC. Animals that received LinkCD/5-FC treatment showed significant tumor size reduction and increased survival compared to the CD/5-FC treatment group. Treatment groups that were unable to produce 5-FU had no effect on the tumor growth despite receiving the fusion protein that contained the Link domain. The results indicate that a treatment regime consisting of a fusion protein containing the Link domain, the active CD enzyme, and the prodrug 5-FC are sufficient to produce an anti-tumor effect. Thus, the LinkCD fusion protein is an alternative to antibody-directed prodrug enzyme therapy (ADEPT) approaches for cancer treatment.
Keywords: cancer, drug delivery, enzyme prodrug therapy, protein engineering, tumor matrix
INTRODUCTION
A variety of strategies have been proposed to improve the selectivity of chemotherapeutic drugs. Antibody-Directed Enzyme Prodrug Therapy (ADEPT) is a strategy devised to target cancer cells with an antibody-enzyme conjugate (1). The antibody targets the enzyme to the cancer cell and the enzyme converts a relatively non-toxic prodrug to a cytotoxic drug. In theory, targeting the enzyme to the cancer results in local production of the cytotoxic drug; the high drug concentration could eradicate the cancer cells in the vicinity of the enzyme. Gene-Directed Enzyme Prodrug Therapy (GDEPT, or VDEPT for Viral-Directed Enzyme Prodrug Therapy) is a variant of ADEPT where the enzyme is expressed in the tumor cells. ADEPT and GDEPT systems exploit the Enhanced Permeability and Retention (EPR) effect of the leaky tumor vasculature to access the cancer while reducing accumulation of the system in normal tissue (2). ADEPT and GDEPT approaches to treat cancer have been investigated since the late 1980's (3-11) without clinical success. A major disadvantage for both ADEPT and GDEPT is the lack of a robust target. Tumor specific antigens, which are usually growth factor receptors, are heterogeneous. Moreover, not all patients over-express the same cancer cell marker; for instance, only 25% of breast cancer patients over-express Her-2/neu (12). Thus, a single antibody is unable to target all variants. An unresolved challenge to GDEPT is the lack of a safe and effective vector. Inefficient transfer of genetic materials into cancer cells, and the short survival of the genetically modified cells are additional barriers for the GDEPT approach (13).
To overcome several of the obstacles of current enzyme prodrug therapies, we devised the Matrix Attachment Therapy (MAT) strategy (14) to target hyaluronan in the extracellular matrix of cancer cells to localize a prodrug converting enzyme in the tumor. Hyaluronan is a linear polysaccharide (Mw > 106 Da) consisting of repeating disaccharides of N-glucosamine and D-glucuronate (15). Hyaluronan is implicated in the proliferation, migration, and metastasis of cancer (16, 17). Although present in normal tissues, hyaluronan is up-regulated in many cancers including: breast, ovarian, colon, lung, and prostate (18). In particular, breast and ovarian cancers have high levels of hyaluronan where its up-regulation is associated with poor patient survival (19, 20).
Using the MAT strategy directed towards hyaluronan has several distinct advantages for targeting cancer: 1) there is no known heterogeneity of hyaluronan; 2) hyaluronan in the tumor matrix can be targeted by the EPR effect. Extravasation into normal tissue is greatly diminished because of their intact endothelial cell barrier. Thus, the hyaluronan content in the tumor does not have to be elevated for MAT to achieve selective tumor targeting; 3) there are many binding sites on hyaluronan; 4) the cytotoxic drug generated in the tumor matrix can also target stromal cancer cell-associated fibroblasts that support tumor metastasis (21,); and 5) disrupting cell-to-hyaluronan association inhibits tumor progression in some experimental models (22-25).
CD44, TSG-6 (Tumor necrosis factor alpha-stimulated Gene-6), and RHAMM (Receptor for hyaluronan-mediated mobility) are major hyaluronan binding proteins found in human and other mammals (26). The function and structure of hyaluronan binding domains of CD44 and TSG-6 (also known as the Link domain) are well characterized (27, 28). The TSG-6 Link domain binds to hyaluronan tighter than does the CD44 Link domain (29), so we used the Link domain from TSG-6 as the hyaluronan binding portion of the fusion protein. We selected cytosine deaminase (CD) from yeast to create a chimeric fusion protein, LinkCD, for a proof of concept of the MAT strategy. Cytosine deaminase is only found in bacteria and fungi, and has been extensively employed for ADEPT and GDEPT to generate cytotoxic 5-fluorouracil (5-FU) from the prodrug 5-fluorocytosine (5-FC). 5-FC is a widely used orally available antimicrobial prodrug with high bioavailability (30). In this manuscript, we describe the creation and functional characterization of a LinkCD fusion protein. We then demonstrate an anti-tumor effect of the LinkCD fusion protein in combination with 5-FC delivered in the drinking water, in Balb/c mice bearing C26 murine colon adenocarcinoma flank tumors.
EXPERIMENTAL SECTION
All restriction enzymes and T4 ligase used for cloning were purchased from New England Biolabs (Beverly, MA). Primers used for cloning and performing mutagenesis were synthesized by IDT (Coralville, IA) and Invitrogen (Carlsbad, CA). The bacterial expression vectors pET41a and pET15b were purchased from Novagen (San Diego, CA). All chemicals used in our experiments were purchased from Sigma-Aldrich (St. Louis, MO).
Construction of bacterial expression vector containing the ORF of LinkCD
To create a bacterial expression construct for producing the LinkCD fusion protein in E.coli, the cloning was performed in two steps. First, we created GST-tag fusion in pET41a vector. Using cDNA of yeast CD (Invivogen; San Diego, CA), the ORF was amplified by the following primers: 5’-primer containing SacII restriction site (GCTCGTCCGCGGGTCTGGTGCCACGCGGTAGTGTCACAGGAGGCATGGC) and 3’-primer with XhoI restriction site (GCTCGTCTCGAGTTACTCCCCAATGTCCTCAAAC). The ORF was ligated into SacII and XhoI restriction sites of pET41a vector. Then the ORF of the Link domain of TSG-6 (Link) was amplified from cDNA of TSG-6 (American Type Culture Collection; Manassas, VA) using a 5’-primer containing SacII followed by a thrombin cleavage site (GCTCGTCCGCGGGTCTGGTGCCACGCGGTAGTGGTGTGTACCACAGAGAAGCAC) and a 3’-primer containing a SacII site (GCTCGTCCGCGGCTTTGCGTGTGGGTTGTAG). The Link ORF was inserted into the SacII site of pET41-CD to create pET41-LinkCD.
We then created a LinkCD with an N-terminal His(×6) tag and a flexible linker between the Link and CD domains. Using pET41-LinkCD as a template, the CD gene was amplified using 5’-primer containing the sequence for a (Gly4Ser)3 flexible linker. The primer sequences are: 5’-primer GCTCGTCATATGGGTGGTGGTGGTAGCGGTGGTGGTGGTAGCGGTGGTGGTGGTAGCATGGTCACAGGAGGCAT, and 3’-primer GCTCGTGGATCCTTACTCCCCAATGTCCTCAAACCAGT, and the amplicon was inserted into pET15b vector between NdeI and BamHI restriction sites to produce pET-CD plasmid. The ORF of the Link domain was amplified using 5’-primer containing NdeI restriction site (GCTCGTCATATGGGTGTGTACCACAGAGAAGCA) and 3’-primer with AseI restriction site (GCTCGTATTAATCTTTGCGTGTGGGTTGTAGC), and it was inserted into the NdeI site of pET-CD to produce pET-LinkCD (compatible-end ligation was used between ORFs of Link and (Gly4Ser)3-CD). The final ligation product was transformed into DH-5α E.coli cells (Invitrogen; San Diego, CA) for screening, and the nucleotide sequence of the fusion protein expression construct was verified by DNA sequencing reactions (SeqWrite DNA Technology Services; Houston, TX).
A functional mutant of LinkCD that lacks the CD activity was created by a single point mutation, resulting in a Glu to Ala change at the residue 64 in CD using the QuikChange® II Site Directed Mutagenesis Kit (Stratagene; La Jolla, CA) and designed primers for the point mutation (forward primer: CCCTGCATGGGGCGATCAGCACCCTG, and reverse primer: CAGGGTGCTGATCGCCCCATGCAGGG). Using pET-LinkCD as a template, the mutagenesis was done with 12 cycles of denaturing (95 °C), annealing (55 °C), and extension (68 °C). After DpnI treatment on the reaction mixture to digest methylated template plasmid, 2 μl of PCR reaction mixture was transformed into DH-5α E.coli cells for screening to confirm the mutation sequence.
Protein expression and purification in E.coli
For protein expression, pET-LinkCD (the N-terminal His(×6)-tag version in pET15b vector) was transformed into BL21-Codon Plus (DE3)-RIPL E.coli cells (Stratagene; La Jolla, CA), which contains tRNAs for rare codons to express mammalian proteins. E.coli cells containing pET-LinkCD plasmid were grown in 1 L LB containing 50 μg/mL of carbenicillin at 35 °C until OD600nm reached between 0.6 and 0.9. The protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.5 mM final concentration), and the expression was done at 23 °C overnight with constant shaking at 200 rpm. Bacterial cells containing the expressed protein were collected by centrifugation at 6,000×g for 30 min at 4 °C, and resuspended in 20 mL of Lysis Buffer containing 300 mM NaCl, 20 mM Tris-Cl, 40 mM imidazole, 0.2% Triton X-100, and 0.5 mM PMSF. After one cycle of freeze-thawing, the cells were lysed by combination of lysozyme and sonication treatments in an ice-water bath. The cell lysate was treated with DNase I and RNase A to digest nucleic acid, and centrifuged at 10,000×g for 45 min at 4 °C. The supernatant containing soluble proteins was collected, and 10 mM β-mercaptoethanol was added to the supernatant fraction and filtered through a 0.4 μm membrane. The filtered supernatant fraction was applied on to a HisTrap™ FF affinity column (Amersham Biosciences; Piscataway, NJ) that was pre-equilibrated with Binding Buffer containing 300 mM NaCl, 20 mM Tris-Cl, and 40 mM imidazole, pH 8. After the entire soluble fraction was passed through the column, the column was washed with 20 column volumes of the Binding Buffer, followed by 10 column volumes of 2 M NaCl, 20 mM Tris-Cl, and 40 mM imidazole, pH 8. The bound protein was eluted using Elution Buffer containing 300 mM NaCl, 20 mM Tris-Cl, and 500 mM imidazole, pH 8. The eluates were pooled and applied onto an Econo-Pac® 10DG desalting column (Bio-Rad; Hercules, CA) to remove imidazole and to exchange buffer to PBS. The purified protein was concentrated using Amicon® Ultra Centrifugal Filter Unit (Millipore; Billerica, MA). The Link-mtCD fusion protein was expressed and purified using the same procedure described above. The N-terminal His(×6)-tagged CD enzyme was purified as described in Huang et al (2006). All proteins used for animal studies were chromatographed on Detoxi™ Gel polymixin B column (Pierce; Rockford, IL) to remove endotoxin prior to injections. The removal of endotoxin was confirmed by measurement of endotoxin level using Limulus Amebocyte Lysate Kit (Cambrex Bio Science; Walkersville, MD). All proteins chromatographed on the polymixin B column contained less than 1 endotoxin unit per mg protein.
Size exclusion column
To determine the native molecular weights of LinkCD and CD, a Sephacryl™ S-100HR (Amersham Biosciences; Piscataway, NJ) size exclusion column was used. The column was calibrated using a Low Molecular Weight Gel Filtration Calibration Kit (Amersham Biosciences; Piscataway, NJ) containing blue dextran 2000 (for the void volume measurement), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (13.7 kDa). The column was operated on Dionex HPLC, with a constant flow-rate of 0.5 mL/min using PBS. The chromatograms containing the retention time of the protein standards, LinkCD, and CD were recorded by the UV absorbance at 280 nm. The gel-phase distribution coefficient (Kav) for protein standards, LinkCD, and CD were calculated and a calibration standard curve plotted. The standard curve of Kav versus MW was used to estimate the molecular weights of LinkCD and CD.
Enzyme assay and kinetics measurements
Enzyme assays for measuring cytosine deaminase activity were performed at 37 °C in PBS. A 1-mL reaction mixture containing 5 mM of 5-FC and 5 μg of LinkCD fusion protein was incubated in a 37 °C water bath, and 20 μL of aliquots were taken out at various time points (10−30 min) and quenched in 980 μL PBS containing 0.1 N HCl. The concentrations of 5-FU in quenched solutions were calculated by measuring absorbance at 255 nm and 290 nm, as described by Senter and coworkers (4):
For enzyme kinetics measurement, 5 μg of LinkCD was added to a pre-warmed 1-mL reaction volume with various concentrations of 5-FC. A 20 μL of aliquot was removed from the reaction mixture every 90 sec for 12 min and quenched in 980 μL PBS containing 0.1 N HCl. The Km and Vmax parameters were calculated from double-reciprocal plots.
To determine the stability of the enzyme activity at 37 °C, 50 μg of LinkCD or CD was stored in 1-mL volume of PBS at 37°C. After various periods of incubation at 37 °C (1−48 hr), the enzyme activity was measured using the method described above, and activity was normalized as the percent of enzyme activity at time zero.
Circular dichroism
Jasco J-715 spectropolarimeter with a Peltier temperature controller was used to measure the circular dichroism spectra on CD and LinkCD. All protein samples were prepared in PBS for circular dichroism studies. Circular dichroism spectrum measurements were done using wavelength scans from 270 nm to 200 nm using a 0.1-cm path length cuvette at 25 °C. To determine thermal denaturation of LinkCD, circular dichroism at a fixed wavelength (222 nm or 223 nm) was monitored in a 1-cm path length cuvette as a function of increasing temperature starting at 20 °C. EXAM software (31) was used to fit the melting temperature curves and calculate Tm for each protein sample.
Hyaluronan binding affinity measurement
Biacore T100 surface plasmon resonance (SPR) machine was used to measure the binding affinity of hyaluronan oligomer toward LinkCD. The surface of CM5 chip was modified with tri-nitrilotriacetic acid (tri-NTA) by the following protocol: 1) Activate the CM5 chip by flowing the mixture of EDC (0.4 M)/NHS (0.1 M) at 10 μL/min for 7 minutes. 2) Flow the solution of amino tri-NTA (10 mM) in 50 mM pH 8.5 borate buffer at 10 μL/min in 2 minutes pulses for 15 cycles. 3) Block the remaining activated dextran with 1 M pH 8 ethanolamine (two 4 minutes cycles). 4) Wash the chip with 0.25% SDS and 0.35 M EDTA alternatively until a stable baseline is reached. The tri-NTA chip was then loaded with 5 mM NiCl2, and LinkCD was immobilized on the surface through His-tag. The kinetics data were measured by flowing the hyaluronan oligomers (12-mer or 8-mer) through the LinkCD loaded chip at pH 7.4 and pH 6.0 respectively. Data were processed and analyzed with the Biacore T100 Evaluation software. Hyaluronan oligomers were digested from crude hyaluronan and purified by the published method (32). The molecular weights of the hyaluronan oligomers used are short enough so that they cannot bind to more than one binding site.
In vivo anti-tumor experiment using C26 tumor model
The C26 murine adenocarcinoma model in female Balb/c mice was used for anti-tumor studies. 6−8 week old female Balb/c mice were purchased from Charles River Laboratories (Wilmington, MA). All animals were housed in UCSF Animal Facility under strict protocols recommended by the National Institute of Health Guide for the Care and Use of Laboratory Animals, and approved by the UCSF Institutional Animal Care and Use Committee. C26 cells were obtained from UCSF Cell Culture Facility, and were cultured in RPMI-1640 medium containing 10% FBS. To implant tumors in mice, 3×105 cells in 50 μl volume were injected subcutaneously on shaved right flank of each mouse. On day 11 after tumor implantation, each mouse received 0.2 units of either purified LinkCD or CD via intratumoral injection (1 unit is defined as the amount of protein that can generate 1 μmole of 5-FU per min using 5 mM 5-FC). On the same day, drinking water was replaced with water containing 10 mg/mL 5-FC until day 40. 5-FC water was replaced every two days to reduce microbial contaminations in the bottles. Also, the amount of 5-FC intake was monitored by weighing the water bottles every two days. Three days after the first dose, the animals were given a second dose of the protein, 0.6 units per mouse (day 14). The tumor sizes and body weights were measured every 2−3 days. Tumor volume (cm3) was calculated by 0.5 × height (cm) × width (cm) × length (cm).
Statistics
Statistical analysis was performed using MedCalc Software version 9.1.0.1 (Belgium). To find statistically significant tumor size reduction, one-way ANOVA and Student-Newman-Keuls pairwise comparisons were performed using tumor volume data from all treatment groups. Survival data was analyzed by the log rank test and a p value < 0.05 was considered significant.
RESULTS
Expression and Purification of LinkCD
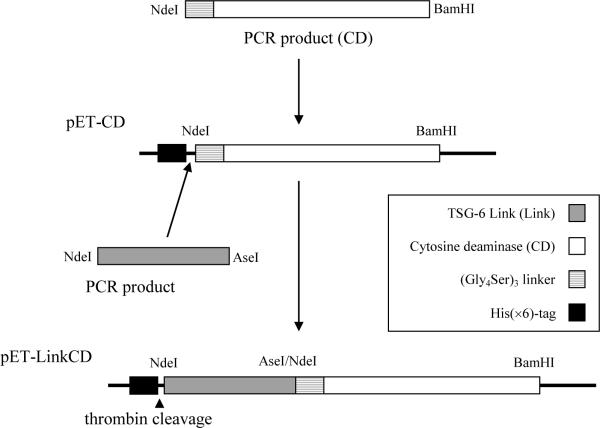
The pET-LinkCD construct was created as shown in Figure 1 (see SI Figure 1 for restriction enzyme digest). BL21-Codon Plus® (DE3) RIPL E.coli cells were used as a host to express the fusion protein. We first attempted protein expression with an N-terminal GST-tag using the pET41 vector, but were unable to obtain the fusion protein with reasonable purity (data not shown). The ORF was moved into pET15b, which contained only an N-terminal His(×6) tag followed by a thrombin cleavage site. Also, a (Gly4Ser)3 linker was added to provide a 15-residue long space between the two functional groups (33). Most of the expressed proteins aggregated in the inclusion bodies (Figure 2A), in spite of efforts to increase accumulation of the soluble protein by lowering post-induction temperature while increasing the incubation time. The LinkCD fusion protein in the soluble fraction was purified using a HisTrap™ FF, a Ni2+-NTA column. On average, about 0.5 mg of the purified protein was recovered from the soluble fraction per liter culture medium. All proteins used in this study contain N-terminal His(×6)-tags.
Figure 1. Construction of pET-LinkCD for protein expression in E.coli.
As described in Materials and Methods, the ORF for LinkCD was inserted into pET15 vector containing the nucleotide sequence for an N-terminal His(×6) residue tag followed by a thrombin cleavage site (LVPRGS) for the expression of the LinkCD fusion protein in E.coli. The length of the fusion gene ORF is 885 bp.
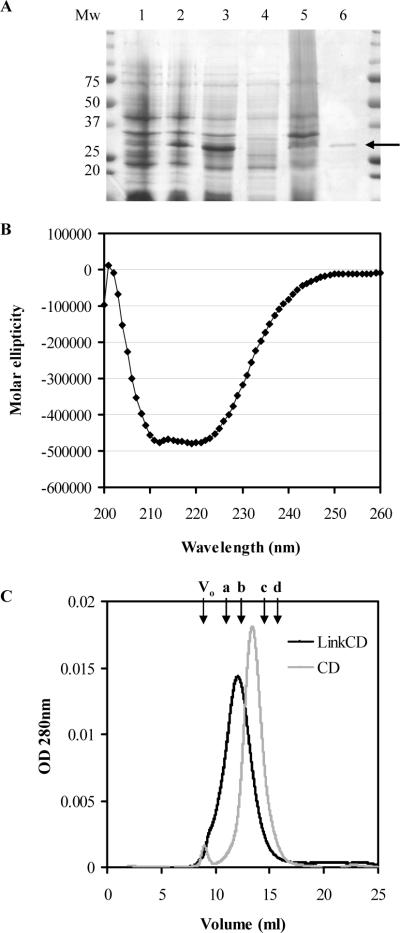
Figure 2. Purified LinkCD fusion protein.
A. SDS-PAGE showing purified LinkCD from the soluble fraction (the expected mass of LinkCD under denatured condition is at 32 kDa). Cell lysate before (lane 1) and after (lane 2) IPTG induction. Lanes 3−6 were collected during protein purification steps: lane 3) cell lysate, 4) soluble fraction, 5) insoluble fraction, and 6) the arrow indicates purified LinkCD. B. Circular dichroism spectrum of purified LinkCD showing the secondary structure of the protein (Molar elipticity: 103×mdeg cm2/dmol). C. The chromatogram of LinkCD (black) and CD (grey) on Sephacryl S-100HR size exclusion column (100kD cut-off). The left shift of the retention volume suggests that LinkCD forms a dimer. The arrows indicate the void volume of the column (Vo) and retention volumes of protein standards: a) albumin (67 kDa), b) ovalbumin (43 kDa), (c) chymotrypsinogen A (25 kDa), and d) ribonuclease A (13.7 kDa).
Secondary structure of LinkCD
Circular dichroism scan on LinkCD was performed to examine if the purified fusion protein from the soluble fraction contains secondary structure. The spectrum of LinkCD in the far-UV range showed a significant alpha-helix signal between 210 nm and 222 nm, which confirmed that the purified protein is folded, and contains secondary structure (Figure 2B).
Measurement of the native Mw by size exclusion chromatography
To examine if the soluble LinkCD is a monomer, dimer, or aggregate, the purified protein was applied on to a Sephacryl™ S-100HR size exclusion column. The theoretical monomer sizes of LinkCD and CD are 32 kDa and 19 kDa, respectively. A calibration standard curve was used to calculate the apparent molecular weights of LinkCD and CD. As shown in Figure 2C, LinkCD eluted at the retention volume corresponding to 45 kDa, which is between the sizes of monomer and dimer (32 and 64 kDa, respectively). The CD enzyme, which has been shown to form a dimer (34, 35), eluted at the retention volume corresponding to 29 kDa. The apparent molecular weight of the CD enzyme is also between the sizes of its monomer and dimer (19 and 38 kDa, respectively), and it is close to the reported mass of native CD (31.4 kDa, with a His-tag) determined by size exclusion chromatography using a Superose™ column (36).
Enzymatic activity of LinkCD
Using 5-FC as a substrate, the enzyme kinetics parameters for the conversion of 5-FC to 5-FU by LinkCD were measured at 37 °C. As shown in Table 1, Km and Vmax values were 0.33 mM and 15 μM/min/μg, respectively, for the LinkCD fusion protein. These values were significantly different from the reported kinetics parameters for CD enzyme (6). The ratio of Vmax/Km , which is a good indicator for the efficiency of catalytic activity, was 53% of that ratio observed in CD. We also created Link-mtCD, a functional mutant that lacks cytosine deaminase activity. In the proposed catalytic scheme of CD enzyme described by Ko et al (35), the negatively charged Glu 64 is a key residue in the active site of the enzyme to initiate the conversion of cytosine or 5-FC to uracil or 5-FU, respectively. Therefore, we performed point mutagenesis to replace the Glu 64 residue with an Ala. The Link-mtCD fusion protein lacked enzyme activity as shown on SI Figure 2B. The purified Link-mtCD fusion protein was used as a negative control in the in vivo anti-tumor experiment (Figure 5).
Table 1.
Functional characteristics of LinkCD fusion protein
| Enzyme kinetics (5-FC) |
Ligand binding (HA12) |
||
|---|---|---|---|
| Km (mM) | Vmax (μM/min/μg) | KD (μM) | |
| LinkCD | 0.33 ± 0.04 | 15 ± 1 | 5.3 at pH 6.0 |
| 55 at pH 7.4 | |||
| CD | 0.51 (*0.8 ± 0.2) | 120 (*68 ± 10) | - |
| Link** | - | - | 0.3 at pH 6.0 |
Km and Vmax for the LinkCD fusion protein were obtained from three repeated experiments using 5-FC as a substrate (mean ± sem). The kinetics parameters of recombinant CD enzyme obtained from one experiment were 0.51 mM for Km and 120 μM/min/μg for Vmax. The ligand binding activity of LinkCD to hyaluronan oligomer (12-mer HA) was measured at pH 6.0 and pH 7.4 using SPR.
Literature values from *Kievit et al. 1999 and **Kahmann et al. 2000.
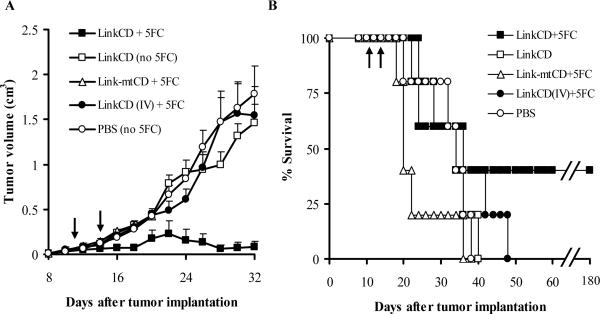
Figure 5. Anti-tumor effect of the LinkCD requires the Link domain and conversion of 5-FC to 5-FU.
A. The tumor growth curves (n ≥ 3) show that the anti-tumor effect is only observed with enzymatically active LinkCD in the presence of 5-FC. All data points represent mean + sem. Tumor volumes in the LinkCD/5FC treatment group were significantly smaller than the other groups (p < 0.05 for days 16 − 20, and 24 − 32). B. Survival curve for the all treatment groups in the study. LinkCD/5-FC treatment group has 40% long-term survivors. The arrows indicate the protein injection on days 11 and 14 after tumor implantation..
Stability of LinkCD enzymatic activity
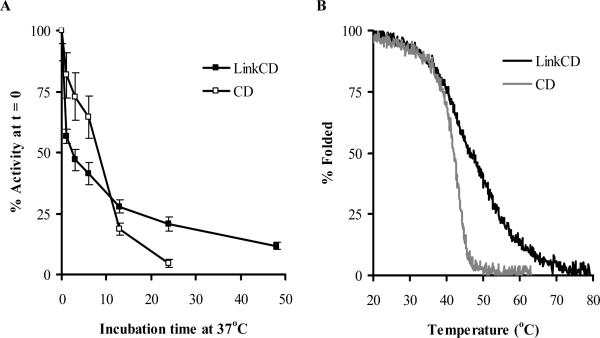
As the stability of CD is known to be limited at 37 °C (6, 36, 37), we were interested to see if the Link moiety could increase the stability of the enzymatic activity of LinkCD. The LinkCD fusion protein lost almost 45% of its enzymatic activity within the first hour of incubation (Figure 3A). However, the rate of activity loss after the first hour decreased, and after 24 hr incubation at 37 °C it LinkCD retained more than 20% of the initial activity. At 48 hr and 124 hr time points, the enzymatic activity of LinkCD decreased to about 12% and 6% respectively of its time zero activity (124 hr time point data is not shown on Figure 3A). In contrast, the activity of CD decreased rapidly at 37 °C, and retained only about 5% of initial activity after 24 hr. In the presence of hyaluronan (0.1 mg/mL) in PBS containing Ca2+ and Mg2+ after 24 hr incubation at 37 °C, LinkCD activity remained slightly higher than in the absence of hyaluronan (SI Figure 3).
Figure 3. Stability of LinkCD fusion protein.
A. Enzyme activity as a function of time at 37 °C. An aliquot of 50 μg of purified protein in 1mL of PBS was incubated at 37 °C for each time point. The enzymatic activities for all time points were normalized by the enzyme activity at the zero hr time point (mean ± propagated error). B. Thermostabilities of LinkCD and CD were measured by circular dichroism at 222nm with increasing temperature from 20 °C to 80 °C. Tm for LinkCD fusion protein was about 6 °C higher than that of CD enzyme.
The thermal denaturation of the protein was examined by measuring circular dichroism at 222 nm as a function of increasing temperature (Figure 3B). The melting curve for LinkCD was shifted to the right of the CD enzyme melting curve: using the EXAM curve fitting software (31), Tm for LinkCD fusion protein was calculated to be at 48 °C, whereas the Tm for the CD enzyme was 42 °C. Therefore, the LinkCD fusion protein is both functionally and structurally more thermal stable than the CD enzyme.
Hyaluronan binding activity
The binding affinity of hyaluronan oligomers toward CD44 and TSG6 have been measured by either isothermal titration calorimetry (ITC) or surface plasmon resonance (SPR) (28, 38). In order to determine the affinity between hyaluronan and LinkCD by the SPR method, we need to immobilize either hyaluronan or LinkCD on the surface of the SPR chip. A common strategy is to immobilize a biotinylated ligand or protein on a streptavidin chip. Streptavidin showed strong nonspecific interaction with both hyaluronan and LinkCD which interfered with the measurement of binding between hyaluronan and LinkCD (data not shown). To circumvent this problem, we developed a tri-NTA chip by modifying the commercially available CM5 chip with our recently reported tri-NTA reagent that has a much higher affinity toward His-tag than mono NTA (37). LinkCD can be reversibly immobilized on the tri-NTA chip through the His-tag without obvious dissociation when stabilized. The binding kinetics of 12-mer hyaluronan toward LinkCD at both pH 7.4 and 6.0 are summarized in Table 1. The equilibrium affinity KD at pH 6.0 is 5.32 μM which is about 10 times stronger than the KD at pH 7.4 (55 μM). There was no detectable interaction between 8-mer hyaluronan and LinkCD at pH 7.4 though others have reported that a 6-mer hyaluronan can bind to TSG6 Link (38). The summary of the binding kinetics between LinkCD and 12-mer hyaluronan is shown on SI Table 1.
Anti-tumor effect of LinkCD/5FC on C26 tumor bearing mice
We hypothesized that the LinkCD fusion protein should be retained in the extracellular matrix of tumor because it can bind to hyaluronan, and in the presence of 5-FC, generate 5-FU to kill surrounding tumor cells. To test this hypothesis we examined if the LinkCD induced a better therapeutic effect than CD when injected into the C26 murine colon adenocarcinoma in female Balb/c mice. Two intratumoral injections of LinkCD, CD, or PBS were given: the first injection on day 11 when the tumor was large enough to accept an intratumoral injection, and the second on day 14. 5-FC was administered in the drinking water starting on day 11 for 30 days. We selected this dosing schedule for the LinkCD based upon the thermal stability of the fusion protein so that active enzyme would still be at the site if it was retained and because we wanted to avoid any immune responses to the LinkCD which would have taken more than 1 week to arise. We dosed 5-FC in the water because of the reduced stress on the animals compared to oral gavage or intraperitoneal administration. The oral delivery of 5-FC is the route of choice for investigators studying the antifungal effects of 5-FC (48, 49). We showed that animals housed in individual cages consumed similar amounts of 5-FC (SI, Figure 5).
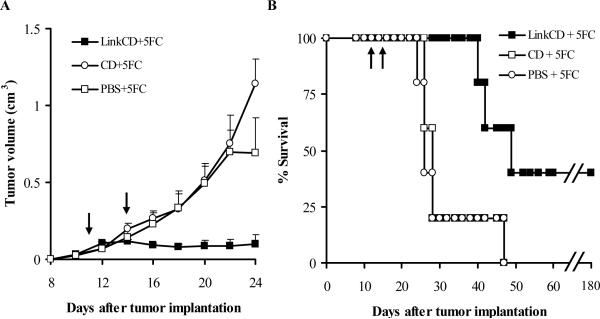
The mean tumor volume of the mice treated with LinkCD/5-FC was significantly lower than CD/5-FC and PBS/5-FC controls (p < 0.05 on days 20−24, Figure 4A). In the CD/5-FC and PBS/5-FC treatment groups there was no noticeable effect on tumor progression. The absence of an effect of CD enzyme is most likely due to a rapid elimination of CD from the tumor site; CD lacks a matrix binding domain and was probably not retained in the vicinity of the tumor. As shown on Figure 4B, the survival of LinkCD/5-FC treatment group animals was significantly better than the groups that received either CD/5-FC or PBS/5-FC (p < 0.025 for both comparisons by log rank test). The median survival time for LinkCD/5-FC group was 45 days with 40% long-term survivors, whereas those of CD and PBS control groups were 28 and 26 days, respectively.
Figure 4. Anti-tumor effect of LinkCD/5-FC treatment on C26 tumor bearing mice.
A. Tumor growth curves (n ≥ 3) show that LinkCD was able to reduce tumor growth in the presence of 5-FC, whereas CD with 5-FC did not show anti-tumor effect. All data points represent mean + sem. Tumor volumes were significantly reduced by LinkCD/5-FC treatment (p < 0.05, days 20 − 24). B. Survival curve for LinkCD, CD, and PBS groups that received 5-FC in drinking water. The survival of the animals in the LinkCD + 5FC treatment was statistically significant over that of the other treatment groups (p < 0.025) with 40% long-term survivors. The arrows indicate the protein injection on days 11 and 14 after tumor implantation.
Several groups have published that the disruption of the extracellular matrix-tumor cell interaction through competition between cell surface CD44 and soluble CD44 or hyaluronan binding peptides for the hyaluronan in the matrix, inhibits tumor growth (22-25). These observations raised the possibility that the Link domain could be responsible for the anti-tumor effect. To examine whether the Link domain contributed to the anti-tumor effect, we repeated the in vivo experiment with two treatment groups that contained the Link domain but could not generate 5-FU: 1) LinkCD with no 5-FC in drinking water and 2) Link-mtCD with 5-FC in the drinking water. Link-mtCD fusion protein is a functional mutant that lacks cytosine deaminase activity (see SI Figure 3). There were no significant tumor reductions in any groups that were unable to generate 5-FU, whereas animals treated with LinkCD/5-FC treatment had a reduced tumor progression (p < 0.05 for days 16−20, and 24−32) and 40% long-term survivors (Figure 5). The median survival time for the LinkCD/5-FC group in the repeat experiment was 36 days, which did not reach the p < 0.05 significance level for survival when compared to all other treatment groups. However the survival data, when combined from the two experiments for the LinkCD/5-FC treatment groups (n=10), is highly significant when compared to the survival data combined from the LinkCD and Link-mtCD/5-FC treatment groups that could not generate 5-FU (p < 0.002, with n=10). The consistency of the results in the two experiments strongly suggests that the tumor regression is only observed when three conditions are present: 1) the Link, a hyaluronan binding protein, 2) active CD enzyme that converts prodrug 5-FC to cytotoxic 5-FU, and 3) the prodrug 5-FC. Absence of any one of these conditions did not result in a tumor reduction or lead to long-term survival for C26 tumor bearing mice.
DISCUSSION
MAT for cancer is based on the hypothesis that the generation of cytotoxic drug in the extracellular matrix of a tumor can effectively eradicate nearby cancerous cells. This strategy is fundamentally different from ADEPT: 1) there is no heterogeneity in the target site; 2) the level of hyaluronan in the tumor does not need to be significantly higher than hyaluronan in normal tissue because the fusion protein is either administered into the tumor site or reaches the site via the enhanced permeability and retention phenomenon, an effect that is not present in normal tissues; 3) hyaluronan, the target for MAT is more accessible than the tumor-specific antigen on the cell surfaces of tumors; 4) there are more binding sites available on the hyaluronan matrix than for most, if not all, tumor antigens; 5) the tumor is unlikely to down regulate or shed the matrix. MAT for cancer has the potential to overcome some of the limitations of ADEPT, which are the heterogeneity of the cancer specific antigen that may compromise the binding of the designed antibody, the limited number of available binding sites on the cancer cell surface, and the internalization of the antibody-enzyme conjugate into the tumor cells.
To demonstrate a proof-of-concept for MAT, we constructed a recombinant fusion protein, LinkCD. LinkCD was designed to target hyaluronan in the extracellular matrix and convert 5-FC to 5-FU surrounding tumor cells. The rate of catalysis of 5-FC to 5-FU by LinkCD was significantly slower than the recombinant CD. Although we do not know the reason for the decrease in Vmax, it is possible that the Link domain has a subtle influence on the mobility of the catalytic domains in CD; since the active site of CD is completely occluded by the C-terminal helix, the enzyme must relax to open the active site for a substrate to bind and its product to be released after enzymatic reaction (34, 35). The Km of 5-FC for LinkCD was lower than the Km of 5-FC for CD: this provides an advantage for LinkCD to generate 5-FU at lower concentrations of 5-FC. The enzymatic activity is retained at 37 °C for the LinkCD fusion protein longer than for CD, which is consistent with the observation that the secondary structure of LinkCD is more thermostable than CD.
The LinkCD protein retained the ability to bind hyaluronan. The affinity for a hyaluronan oligomer (12 mer) at pH 6.0 was 5.32 μM. The affinity of the LinkCD for 12 mer hyaluronan at pH 7.4 was an order of magnitude weaker (55 μM). This is an advantage for delivery of the LinkCD via the circulation since hyaluronan affinity at pH 7.4 will be low but will increase at the lower pH values found in tumors (39). This low binding affinity of LinkCD for hyaluronan at pH 7.4 will reduce the likelihood that plasma hyaluronan will occupy the Link domain during the delivery phase to obstruct LinkCD binding to the tumor matrix.
The size exclusion chromatography results show that the LinkCD elutes between where the monomer and dimer would elute. This has previously been found to be the case for the CD enzyme, which also elutes from a sizing column between the expected monomer and dimer masses (36). It may be that the dimeric protein is more compact than the proteins used to calibrate the column so would elute in a larger volume than expected.
One of the factors impeding the development of ADEPT is the low production levels of therapeutic proteins containing antibody fragments originated from mammalian cells using bacterial expression system (9, 33). The low yield of the soluble LinkCD fusion protein from E.coli was also a significant challenge for this study. Yeast CD enzyme is readily expressed as soluble protein in E.coli with a yield with at least 30 mg per liter of culture (34, 35, 40). However, the Link domain of human TSG-6 was found to be toxic to the host E.coli cells and could only be expressed as an inclusion body protein (41). The Link domains from both TSG-6 and CD44 were recovered from the inclusion bodies after denaturation and refolding steps with yields of 10−20 mg per liter culture medium (41-43). A TSG-6 Link-luciferase fusion protein was expressed and purified from the inclusion bodies with yield of 75 mg per liter culture medium (44). Although the majority of LinkCD was found in the inclusion bodies, we were able to recover a sufficient quantity of the functional LinkCD from the soluble fraction to avoid a refolding step in purification. Recovery of the LinkCD might have been aided by the soluble nature of the CD enzyme. Attempts to prepare a fusion protein between TSG-6 Link with one, two, or three mutations in the hyaluronan binding site (45) were frustrated by very low expression and low recovery of the fusion protein in the soluble fraction. So we were unable to prepare a fusion protein that could not bind to hyaluronan, but otherwise was the same as the LinkCD used in these studies. A comparison between a non-binding LinkCD and LinkCD is required to confirm that it is the ability of the LinkCD to bind to the hyaluronan matrix that is responsible for the improved therapeutic effect observed here.
In this study, two intratumoral injections of the LinkCD fusion protein were able to inhibit the tumor growth significantly with the prodrug 5-FC, and 4 out of the 10 treated mice from the in vivo experiments are tumor-free and remain healthy beyond 180 days post treatment. There was no anti-tumor effect by LinkCD/5-FC treatment when the same amount of the LinkCD fusion protein was injected intravenous (i.v.). For this treatment group, the dosing amount of LinkCD was probably too small for i.v. administrations, and the injected protein was possibly rapidly eliminated from the systemic circulation. In the study design used in in vivo experiments reported here, we did not include a 5-FU treatment as a control group. The reason for this is that 5-FU treatment (from 25 mg/kg to 100 mg/kg) in the C26 tumor models have delayed tumor growth, but no animals in the 5-FU treatment groups survived longer than 50 days (46, 47). This is presumably due to the toxicity of 5-FU. The results herein demonstrate superior efficacy of LinkCD/5-FC treatment over published protocols using 5-FU drug treatment alone.
In most ADEPT and GDEPT experiments that used CD enzyme, the major route of 5-FC administration was daily i.p. injections (400−500 mg/kg) for up to 2 weeks (5-7, 10, 11). We took advantage of the relatively high oral bioavailability of 5-FC, which is between 76% and 89% in humans (30), and administered the drug in the drinking water at a concentration of 10 mg/mL. Oral administration of 5-FC from the drinking water (10 mg/mL) was previously done for targeting CD-expressing tumor cells in rats (48) and in antifungal combination therapy experiments in mice (49). 5-FC administration via drinking water is less likely to stress the animals compared to multiple day regimes of intraperitoneal (i.p.) administration. The daily ingestion of 5-FC per mouse ranged from 20 mg to 75 mg (SI Figure 4). We also compared the amount of the prodrug consumed by individual animals and observed that all animals consumed similar amounts of 5-FC (SI Figure 5). Animals that were sacrificed in the LinkCD/5-FC treatment group had relatively small tumor volumes at the times of deaths compared to animals sacrificed in the other treatment groups. The deaths in the LinkCD/ 5-FU treatment group might have been due to systemic toxicity of 5-FU generated from 5-FC in these animals. The blood and tumor levels of 5-FC and 5-FU are a subject for future studies that will enable a rationale dosing strategy (50).
The disruption of the hyaluronan-cell interaction by over-expression of soluble CD44 demonstrated tumor growth inhibition in mammary carcinoma and melanoma models, although long-term survival of animals in these studies was not reported (22, 23). Synthetic hyaluronan-binding peptides also reduce tumor progression (24, 25). These studies were done using stably transfected tumor cell lines that secreted soluble CD44 or hyaluronan-binding peptide (22, 23, 25), or by co-injection of a hyaluronan-binding peptide with tumor cells in vivo at the time of tumor implantation (24). A direct effect of the Link domain on tumor progression does not appear to be operative in established tumors since neither the LinkCD without 5-FC treatment group nor the Link-mtCD with 5-FC treatment group reduced C26 tumor growth (Figure 5A) or resulted in long-term survivors (Figure 5B). It is possible that the dose of the protein injected into the tumor in our studies was insufficient to disrupt the cell-to-matrix interaction, or the growth of C26 tumor cells may be independent of their interaction with hyaluronan. Nonetheless, in the absence of long-term survival data from the earlier studies, the therapeutic benefit of using just a hyaluronan-binding protein, or peptide, for anti-tumor treatment does not appear promising.
An antibody-CD enzyme conjugate was created and employed for ADEPT approach for cancer in the early 1990's, and was successful in targeting tumor and generating 5-FU both in vitro and in vivo in H3719 human colon carcinoma model in nude mice (4, 5). However, because of the extended circulation time of antibody-CD conjugate, which potentially could cause systemic toxicity by 5-FU generation in the blood, a secondary antibody specific for the CD enzyme was used to clear the antibody-CD conjugate in vivo (51). To reduce the circulation time of antibody-enzyme conjugate, a recombinant fusion protein containing single chain variable fragment of antibody (scFv) and the CD enzyme was characterized in vitro (9, 52). However, the therapeutic efficacy of antibody-CD conjugates in vivo remains to be tested. Recently, Schellenberger and coworkers have characterized and shown an in vivo anti-tumor effect of a recombinant fusion protein containing scFv and β-lactamase conjugate in combination with a prodrug GM-Mel up to day 36 (11). Significant drug toxicity was reported with their treatment regime; thus, unwanted toxicity from the conversion of active drug remains a challenge for ADEPT.
The MAT strategy would also appear to be better than the GDEPT approach to treat cancer, with the ability to deliver higher amount of prodrug converting enzyme at the tumor site with local delivery. Based on the enzyme stability test at 37 °C in PBS (Figure 3A), 0.2 units of LinkCD can generate 9.4 μmole of 5-FU after 60 min, with assumptions that the enzyme activity does not change in vivo and 5-FC concentration is at steady state. Since the expression level of CD enzyme in stably transduced cells that express CD has not been quantified in the GDEPT approach, the amount of the expressed CD enzyme can only be estimated from the intratumoral 5-FU concentration after 5-FC administration. Using stably transfected HT29 colon carcinoma model in mice, Stegman et al (1999) reported that 60 min after 1 g/kg of 5-FC administration, the peak level of 5-FU in the ∼250 mm3 tumor was measured to be 3.4 mM or 0.85 μmole. Assuming saturation kinetics from time zero, about 1 μg of CD enzyme is needed to produce 3.4 mM 5-FU in the tumor after 60 min. The 0.2 units of LinkCD injected in our studies is greater than the amount of CD enzyme expressed in the subpopulation of transfected tumor cells.
A limited amount of 5-FU is generated in GDEPT. As a consequence, there are few reports of long-term survivors (> 100 days) in GDEPT studies where the CD enzyme/5-FC combination was employed for therapy. One GDEPT study where the U-87 glioma was transduced by replication-competent retrovirus vector in athymic mice 7 days after tumor implantation, resulted in 90% long-term survivors up to 110 days (10). A combined therapy using the 5-FC, CD-uracil phosphoribosyltransferase genes delivered in an adenoviral vector at 4 days post tumor implantation and radiation treatment in 9L brain tumor in rats produced 80% long-term survivors at 90 days (8). In both of these studies, the viral vector was inoculated into the tumor site soon after tumor implantation; the tumor volume at the time of treatment (7 or 4 days after tumor implantation) is expected to be smaller than the tumor volume in the studies reported here. It will be of interest to learn if a local delivery of LinkCD into the brain via convection enhanced delivery (CED) (53-56) can provide a similar therapeutic effect to that observed in these GDEPT studies (8, 10).
The MAT strategy is not limited to the use of recombinant proteins—matrix targeting ligands and prodrug converting enzymes can be conjugated to other widely used drug delivery vehicles, such as liposomes and biocompatible polymers. There is an additional advantage for the combined formulation of recombinant proteins and vehicles for drug delivery: the properties of the delivery vehicles can be modified to optimize the blood circulation time for systemic delivery or CED for local administrations for brain tumor therapy (56, 57). In this regard CD has been PEGylated to increase the circulation time (36). Although CD enzyme activity seemed to be relatively stable with 2 PEG chains, attachment of 3 or more PEG chains on CD reduced the thermal stability of the enzyme at 37 °C with a half-life to less than 2 hr (36). In spite of the reduced stability, this general approach of PEGylating CD enzymes seems promising, and could be applied to LinkCD developed here.
The anti-tumor activity of the LinkCD fusion protein supports the MAT approach to cancer therapy. Using a recombinant fusion protein containing the hyaluronan-binding domain of TSG-6 and the CD enzyme from yeast, we show that the minimal elements for successful MAT against C26 tumor are 1) a matrix binding domain, 2) a prodrug-converting enzyme, and 3) the prodrug. However, due to our inability to yet produce a mutated LinkCD fusion protein that lacks hyaluronan binding capability to compare to the LinkCD in the tumor model, we cannot know if binding to hyaluronan is essential for the therapeutic effect. It is important to undertake such experiments to learn if MAT is a versatile prodrug activation strategy that complements ADEPT and GDEPT, and if it has the potential to target neoplastic and inflammatory diseases where components of the matrix such as hyaluronan are an accessible target.
Supplementary Material
Supporting Information Figure 1
Constructed pET-LinkCD plasmid for the expression of the LinkCD fusion protein. The insert size is 885 bp. 1) Double digestion of pET-LinkCD with NdeI and BamHI restriction enzymes. 2) double digestion of empty plasmid, 3) uncut pET-LinkCD, 4) uncut pET15, 5) PCR amplified LinkCD fragment.
Supporting Information Figure 2
Link-mtCD: a functional mutant that lacks the enzyme activity. A. A sequence alignment between the pET-Link-mtCD (mutant) and pET-LinkCD (reference) showing the point mutation of A to C, indicated by the arrow. B. Enzyme assay shows that Link-mtCD does not convert 5-FC to 5-FU. C) SDS-PAGE gel showing purified LinkCD (lane 1) and Link-mtCD (lane 2).
Supporting Information Figure 3
Cytosine deaminase activity of the LinkCD fusion protein after 24 hr incubation at 37°C in PBS. Enzyme activity of LinkCD in the presence of 0.1 mg/ml hyaluronan (HA). PBS+ contained Ca2+ and Mg2+ whereas PBS did not contain these divalent cations.
Supporting Information Figure 4
5-FC consumption by animals in therapeutic experiments. The amount of 5-FC consumption was estimated by weighing water bottles containing 5-FC (10mg/mL). A. Anti-tumor experiment with LinkCD + 5FC, CD + 5FC, and PBS + 5FC groups. B. Second experiment with LinkCD + 5FC, LinkCD (IV) + 5FC, and Link-mtCD + 5FC groups.
Supporting Information Figure 5
5-FC consumption by individual animal that received LinkCD treatment. Each animal was housed separately and the daily intake of 5-FC by individual animal was monitored. The survival of animals showed similar results as shown in Figure 4 and 5.
Supporting Information Table 1: Summary of SPR binding kinetics data
ACKNOWLEDGEMENTS
We thank Allen Graves, Neema Salimi, and Peter Hwang for their technical assistance with circular dichroism and Biacore instruments, and Nichole Macaraeg for careful animal experimentation. Supported by NIH R01-CA107268.
REFERENCES
- 1.Senter PD, Springer CJ. Selective activation of anticancer prodrugs by monoclonal antibody-enzyme conjugates. Adv. Drug Deliv. Rev. 2001;53:247–264. doi: 10.1016/s0169-409x(01)00206-x. [DOI] [PubMed] [Google Scholar]
- 2.O'Connor SW, Bale WF. Accessibility of circulating immunoglobulin G to the extravascular compartment of solid rat tumors. Cancer Res. 1984;44:3719–3723. [PubMed] [Google Scholar]
- 3.Bagshawe KD. Antibody directed enzymes revive anti cancer prodrugs concept. Br. J. Cancer. 1987;56:531–532. doi: 10.1038/bjc.1987.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senter PD, Su PC, Katsuragi T, Sakai T, Cosand WL, Hellstrom I, Hellstrom KE. Generation of 5-fluorouracil from 5-fluorocytosine by monoclonal antibody-cytosine deaminase conjugates. Bioconjug. Chem. 1991;2:447–451. doi: 10.1021/bc00012a012. [DOI] [PubMed] [Google Scholar]
- 5.Wallace PM, MacMaster JF, Smith VF, Kerr DE, Senter PD, Cosand WL. Intratumoral generation of 5-fluorouracil mediated by an antibody-cytosine deaminase conjugate in combination with 5-fluorocytosine. Cancer Res. 1994;54:2719–2723. [PubMed] [Google Scholar]
- 6.Kievit E, Bershad E, Ng E, Sethna P, Dev I, Lawrence TS, Rehemtulla A. Superiority of yeast over bacterial cytosine deaminase for enzyme/prodrug gene therapy in colon cancer xenografts. Cancer Res. 1999;59:1417–1421. [PubMed] [Google Scholar]
- 7.Miller CR, Williams CR, Buchsbaum DJ, Gillespie GY. Intratumoral 5-fluorouracil produced by cytosine deaminase/5-fluorocytosine gene therapy is effective for experimental human glioblastomas. Cancer Res. 2002;62:773–780. [PubMed] [Google Scholar]
- 8.Kambara H, Tamiya T, Ono Y, Ohtsuka S, Terada K, Adachi Y, Ichikawa T, Hamada H, Ohmoto T. Combined radiation and gene therapy for brain tumors with adenovirus mediated transfer of cytosine deaminase and uracil phosphoribosyltransferase genes. Cancer Gene Ther. 2002;9:840–845. doi: 10.1038/sj.cgt.7700506. [DOI] [PubMed] [Google Scholar]
- 9.Deckert PM, Renner C, Cohen LS, Jungbluth A, Ritter G, Bertino JR, Old LJ, Welt S. A33scFv-cytosine deaminase: a recombinant protein construct for antibody-directed enzyme-prodrug therapy. Br. J. Cancer. 2003;88:937–939. doi: 10.1038/sj.bjc.6600751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai CK, Wang WJ, Chen TC, Kasahara N. Single-shot, multicycle suicide gene therapy by replication-competent retrovirus vectors achieves long-term survival benefit in experimental glioma. Mol. Ther. 2005;12:842–851. doi: 10.1016/j.ymthe.2005.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alderson RF, Toki BE, Roberge M, Geng W, Basler J, Chin R, Liu A, Ueda R, Hodges D, Escandon E, Chen T, Kanavarioti T, Babe L, Senter PD, Fox JA, Schellenberger V. Characterization of a CC49-based single-chain fragment-beta-lactamase fusion protein for antibody-directed enzyme prodrug therapy (ADEPT). Bioconjug. Chem. 2006;17:410–418. doi: 10.1021/bc0503521. [DOI] [PubMed] [Google Scholar]
- 12.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 13.Greco O, Dachs GU. Gene directed enzyme/prodrug therapy of cancer: historical appraisal and future prospectives. J. Cell. Physiol. 2001;187:22–36. doi: 10.1002/1097-4652(2001)9999:9999<::AID-JCP1060>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Platt VM, Szoka FC., Jr. Anticancer therapeutics: targeting molecules and nanocarriers to hyaluronan or CD44, a hyaluronan receptor. Mol. Pharmaceutics. 2008;5:474–486. doi: 10.1021/mp800024g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J. Intern. Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 16.Itano N, Sawai T, Miyaishi O, Kimata K. Relationship between hyaluronan production and metastatic potential of mouse mammary carcinoma cells. Cancer Res. 1999;59:2499–2504. [PubMed] [Google Scholar]
- 17.Kosaki R, Watanabe K, Yamaguchi Y. Overproduction of hyaluronan by expression of the hyaluronan synthase Has2 enhances anchorage-independent growth and tumorigenicity. Cancer Res. 1999;59:1141–1145. [PubMed] [Google Scholar]
- 18.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 19.Anttila MA, Tammi RH, Tammi MI, Syrjanen KJ, Saarikoski SV, Kosma VM. High levels of stromal hyaluronan predict poor disease outcome in epithelial ovarian cancer. Cancer Res. 2000;60:150–155. [PubMed] [Google Scholar]
- 20.Auvinen P, Tammi R, Parkkinen J, Tammi M, Agren U, Johansson R, Hirvikoski P, Eskelinen M, Kosma VM. Hyaluronan in peritumoral stroma and malignant cells associates with breast cancer spreading and predicts survival. Am. J. Pathol. 2000;156:529–536. doi: 10.1016/S0002-9440(10)64757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahrens T, Sleeman JP, Schempp CM, Howells N, Hofmann M, Ponta H, Herrlich P, Simon JC. Soluble CD44 inhibits melanoma tumor growth by blocking cell surface CD44 binding to hyaluronic acid. Oncogene. 2001;20:3399–3408. doi: 10.1038/sj.onc.1204435. [DOI] [PubMed] [Google Scholar]
- 23.Peterson RM, Yu Q, Stamenkovic I, Toole BP. Perturbation of hyaluronan interactions by soluble CD44 inhibits growth of murine mammary carcinoma cells in ascites. Am. J. Pathol. 2000;156:2159–2167. doi: 10.1016/S0002-9440(10)65086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mummert ME, Mummert DI, Ellinger L, Takashima A. Functional roles of hyaluronan in B16-F10 melanoma growth and experimental metastasis in mice. Mol. Cancer. Ther. 2003;2:295–300. [PubMed] [Google Scholar]
- 25.Xu XM, Chen Y, Chen J, Yang S, Gao F, Underhill CB, Creswell K, Zhang L. A peptide with three hyaluronan binding motifs inhibits tumor growth and induces apoptosis. Cancer Res. 2003;63:5685–5690. [PubMed] [Google Scholar]
- 26.Day AJ, Prestwich GD. Hyaluronan-binding proteins: tying up the giant. J. Biol. Chem. 2002;277:4585–4588. doi: 10.1074/jbc.R100036200. [DOI] [PubMed] [Google Scholar]
- 27.Blundell CD, Almond A, Mahoney DJ, DeAngelis PL, Campbell ID, Day AJ. Towards a structure for a TSG-6 hyaluronan complex by modeling and NMR spectroscopy: insights into other members of the link module superfamily. J. Biol. Chem. 2005;280:18189–18201. doi: 10.1074/jbc.M414343200. [DOI] [PubMed] [Google Scholar]
- 28.Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, Jackson DG. Structures of the Cd44-hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007;14:234–239. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 29.Lesley J, English NM, Gal I, Mikecz K, Day AJ, Hyman R. Hyaluronan binding properties of a CD44 chimera containing the link module of TSG-6. J. Biol. Chem. 2002;277:26600–26608. doi: 10.1074/jbc.M201068200. [DOI] [PubMed] [Google Scholar]
- 30.Cutler RE, Blair AD, Kelly MR. Flucytosine kinetics in subjects with normal and impaired renal function. Clin. Pharmacol. Ther. 1978;24:333–342. doi: 10.1002/cpt1978243333. [DOI] [PubMed] [Google Scholar]
- 31.Kirchhoff WH. NIST Tech. Note 1401. U.S. Government Printing Office; Washington, DC.: 1993. Exam: A Two-State Thermodynamic Analysis Program. pp. 1–103. [Google Scholar]
- 32.Ruhela D, Riviere K, Szoka FC., Jr Efficient synthesis of an aldehyde functionalized hyaluronic acid and its application in the preparation of hyaluronan lipid conjugates. Bioconjug. Chem. 2006;17:1360–1363. doi: 10.1021/bc0600721. [DOI] [PubMed] [Google Scholar]
- 33.Asai T, Trinh R, Ng PP, Penichet ML, Wims LA, Morrison SL. A human biotin acceptor domain allows site-specific conjugation of an enzyme to an antibody-avidin fusion protein for targeted drug delivery. Biomol. Eng. 2005;21:145–155. doi: 10.1016/j.bioeng.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Ireton GC, Black ME, Stoddard BL. The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003;11:961–972. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 35.Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 36.Xiong MP, Kwon GS. PEGylation of yeast cytosine deaminase for pretargeting. J. Pharm. Sci. 2005;94:1249–1258. doi: 10.1002/jps.20354. [DOI] [PubMed] [Google Scholar]
- 37.Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahmann JD, O'Brien R, Werner JM, Heinegard D, Ladbury JE, Campbell ID, Day AJ. Localization and characterization of the hyaluronan-binding site on the link module from human TSG-6. Structure. 2000;8:763–774. doi: 10.1016/s0969-2126(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 39.Gerweck LE. Tumor pH: implications for treatment and novel drug design. Semin. Radiat. Oncol. 1998;8:176–182. doi: 10.1016/s1053-4296(98)80043-x. [DOI] [PubMed] [Google Scholar]
- 40.Huang Z, Park JI, Watson DS, Hwang P, Szoka FC., Jr Facile synthesis of multivalent nitrilotriacetic acid (NTA) and NTA conjugates for analytical and drug delivery applications. Bioconjug. Chem. 2006;17:1592–1600. doi: 10.1021/bc0602228. [DOI] [PubMed] [Google Scholar]
- 41.Day AJ, Aplin RT, Willis AC. Overexpression, purification, and refolding of link module from human TSG-6 in Escherichia coli: effect of temperature, media, and mutagenesis on lysine misincorporation at arginine AGA codons. Protein Expr. Purif. 1996;8:1–16. doi: 10.1006/prep.1996.0068. [DOI] [PubMed] [Google Scholar]
- 42.Kahmann JD, Koruth R, Day AJ. Method for quantitative refolding of the link module from human TSG-6. Protein Expr. Purif. 1997;9:315–318. doi: 10.1006/prep.1996.0694. [DOI] [PubMed] [Google Scholar]
- 43.Banerji S, Day AJ, Kahmann JD, Jackson DG. Characterization of a functional hyaluronan-binding domain from the human CD44 molecule expressed in Escherichia coli. Protein Expr. Purif. 1998;14:371–381. doi: 10.1006/prep.1998.0971. [DOI] [PubMed] [Google Scholar]
- 44.Chang TS, Wan HM, Chen CC, Giridhar R, Wu WT. Fusion protein of the hyaluronan binding domain from human TSG-6 with luciferase for assay of hyaluronan. Biotechnol. Lett. 2003;25:1037–1040. doi: 10.1023/a:1024178110310. [DOI] [PubMed] [Google Scholar]
- 45.Mahoney DJ, Blundell CD, Day AJ. Mapping the hyaluronan-binding site on the Link module from human tumor necrosis factor stimulated gene-6 by site-directed mutagenesis. J. Biol. Chem. 2001;276:22764–22771. doi: 10.1074/jbc.M100666200. [DOI] [PubMed] [Google Scholar]
- 46.Evrard A, Cuq P, Robert B, Vian L, Pelegrin A, Cano JP. Enhancement of 5-fluorouracil cytotoxicity by human thymidine-phosphorylase expression in cancer cells: in vitro and in vivo study. Int. J. Cancer. 1999;80:465–470. doi: 10.1002/(sici)1097-0215(19990129)80:3<465::aid-ijc21>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Bourquin C, Schreiber S, Beck S, Hartmann G, Endres S. Immunotherapy with dendritic cells and CpG oligonucleotides can be combined with chemotherapy without loss of efficacy in a mouse model of colon cancer. Int. J. Cancer. 2006;118:2790–2795. doi: 10.1002/ijc.21681. [DOI] [PubMed] [Google Scholar]
- 48.Gavelli A, Baque P, Brossettej N, Bourgeon A, Staccini P, Rossi B, Pierrefite-Carle V. Per os administration of 5-fluorocytosine is effective in the regression of CD-expressing liver metastases in rats. Int. J. Mol. Med. 2004;14:323–325. [PubMed] [Google Scholar]
- 49.Larsen RA, Bauer M, Weiner JM, Diamond DM, Leal ME, Ding JC, Rinaldi MG, Graybill JR. Effect of fluconazole on fungicidal activity of flucytosine in murine cryptococcal meningitis. Antimicrob. Agents Chemother. 1996;40:2178–2182. doi: 10.1128/aac.40.9.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stegman LD, Rehemtulla A, Beattie B, Kievit E, Lawrence TS, Blasberg RG, Tjuvajev JG, Ross BD. Noninvasive quantitation of cytosine deaminase transgene expression in human tumor xenografts with in vivo magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9821–9826. doi: 10.1073/pnas.96.17.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerr DE, Garrigues US, Wallace PM, Hellstrom KE, Hellstrom I, Senter PD. Application of monoclonal antibodies against cytosine deaminase for the in vivo clearance of a cytosine deaminase immunoconjugate. Bioconjug. Chem. 1993;4:353–357. doi: 10.1021/bc00023a008. [DOI] [PubMed] [Google Scholar]
- 52.Coelho V, Dernedde J, Petrausch U, Panjideh H, Fuchs H, Menzel C, Dubel S, Keilholz U, Thiel E, Deckert PM. Design, construction, and in vitro analysis of A33scFv::CDy, a recombinant fusion protein for antibody-directed enzyme prodrug therapy in colon cancer. Int. J. Oncol. 2007;31:951–957. [PubMed] [Google Scholar]
- 53.Bobo RH, Laske DW, Akbasak A, Morrison PF, Dedrick RL, Oldfield EH. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2076–2080. doi: 10.1073/pnas.91.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voges J, Reszka R, Gossmann A, Dittmar C, Richter R, Garlip G, Kracht L, Coenen HH, Sturm V, Wienhard K, Heiss WD, Jacobs AH. Imaging guided convection enhanced delivery and gene therapy of glioblastoma. Ann. Neurol. 2003;54:479–487. doi: 10.1002/ana.10688. [DOI] [PubMed] [Google Scholar]
- 55.Voges J, Weber F, Reszka R, Sturm V, Jacobs A, Heiss WD, Wiestler O, Kapp JF. Clinical protocol. Liposomal gene therapy with the herpes simplex thymidine kinase gene/ganciclovir system for the treatment of glioblastoma multiforme. Hum. Gene Ther. 2002;13:675–685. doi: 10.1089/10430340252837260. [DOI] [PubMed] [Google Scholar]
- 56.MacKay JA, Deen DF, Szoka FC., Jr Distribution in brain of liposomes after convection enhanced delivery; modulation by particle charge, particle diameter, and presence of steric coating. Brain Res. 2005;1035:139–153. doi: 10.1016/j.brainres.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Gillies ER, Dy E, Frechet JM, Szoka FC. Biological evaluation of polyester dendrimer: poly(ethylene oxide) “bow-tie” hybrids with tunable molecular weight and architecture. Mol. Pharm. 2005;2:129–138. doi: 10.1021/mp049886u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1
Constructed pET-LinkCD plasmid for the expression of the LinkCD fusion protein. The insert size is 885 bp. 1) Double digestion of pET-LinkCD with NdeI and BamHI restriction enzymes. 2) double digestion of empty plasmid, 3) uncut pET-LinkCD, 4) uncut pET15, 5) PCR amplified LinkCD fragment.
Supporting Information Figure 2
Link-mtCD: a functional mutant that lacks the enzyme activity. A. A sequence alignment between the pET-Link-mtCD (mutant) and pET-LinkCD (reference) showing the point mutation of A to C, indicated by the arrow. B. Enzyme assay shows that Link-mtCD does not convert 5-FC to 5-FU. C) SDS-PAGE gel showing purified LinkCD (lane 1) and Link-mtCD (lane 2).
Supporting Information Figure 3
Cytosine deaminase activity of the LinkCD fusion protein after 24 hr incubation at 37°C in PBS. Enzyme activity of LinkCD in the presence of 0.1 mg/ml hyaluronan (HA). PBS+ contained Ca2+ and Mg2+ whereas PBS did not contain these divalent cations.
Supporting Information Figure 4
5-FC consumption by animals in therapeutic experiments. The amount of 5-FC consumption was estimated by weighing water bottles containing 5-FC (10mg/mL). A. Anti-tumor experiment with LinkCD + 5FC, CD + 5FC, and PBS + 5FC groups. B. Second experiment with LinkCD + 5FC, LinkCD (IV) + 5FC, and Link-mtCD + 5FC groups.
Supporting Information Figure 5
5-FC consumption by individual animal that received LinkCD treatment. Each animal was housed separately and the daily intake of 5-FC by individual animal was monitored. The survival of animals showed similar results as shown in Figure 4 and 5.
Supporting Information Table 1: Summary of SPR binding kinetics data