Abstract
Molecular simulation is increasingly demonstrating its practical value in the investigation of biological systems. Computational modelling of biomolecular systems is an exciting and rapidly developing area, which is expanding significantly in scope. A range of simulation methods has been developed that can be applied to study a wide variety of problems in structural biology and at the interfaces between physics, chemistry and biology. Here, we give an overview of methods and some recent developments in atomistic biomolecular simulation. Some recent applications and theoretical developments are highlighted.
Keywords: biomolecular simulation, molecular modelling, molecular dynamics, force fields, quantum mechanics/molecular mechanics, quantum chemical modelling
1. Introduction
‘Can I believe modelling?’ is a question often asked by biologists and biochemists. Answering it requires informed understanding of the strengths and limitations of current computational biomolecular modelling and simulation methods, and their ranges of application. Knee-jerk scepticism of all biomolecular modelling is sometimes encountered among experimentalists even today; equally misguided is a blind acceptance of modelling results without critical analysis. However, demonstrations of the practical contribution made by biomolecular modelling have led to a growing recognition of its worth. This is a fertile and growing area, with exciting opportunities and an enormous range of potential applications. It is crucial for the biomolecular modeller to understand the issues of interest to biologists, the complexity of biological systems and how to tackle them effectively by modelling. Vast amounts of data are being provided by large-scale research efforts in genomics, proteomics, glycomics and structural biology. Also, sophisticated physical techniques are increasingly being applied to the study of biomolecular systems. The challenge for biomolecular modelling is to help in efforts to use these diverse data to develop new drugs, therapies, catalysts and biologically based nanotechnology.
Molecular modelling and simulation methods are increasingly making important and indeed often uniquely detailed contributions to the study of the structure and function of biological macromolecules. Applications include studies of protein folding and conformational changes (Daggett 2006; Elcock 2006), association of proteins with small molecules (Moitessier et al. 2008) or other proteins (McGuffee & Elcock 2006; Ritchie 2008), structure-based drug design (Taft et al. 2008), computation of binding free energies for ligands (Gilson & Zhou 2007), modelling the dynamics of ion channels and transport across membranes (Beckstein et al. 2003) and modelling and analysis of enzyme mechanisms (Warshel et al. 2006b; Mulholland 2008). This last area (biological catalysis), for example, is fascinating from a chemical point of view, and is an important interface between chemistry and biology.
Improvements in computer hardware continue to deliver more computational power, which, when combined with theoretical and algorithmic developments, have led to an increasing range and depth of applications of molecular modelling in biology. Indeed, the whole field of biomolecular modelling is now too large to be reasonably covered in a single review. Here, the focus is on atomistic molecular simulation, in particular of proteins. This review aims to highlight some exciting recent applications of molecular modelling and simulation methods to biological systems, and outlines important current methods as well as notable current theoretical developments in the field.
Perhaps the most obvious challenge in simulating biological macromolecules is their large size, exacerbated by the need to include at least a representative part of their environment (i.e. the surrounding solvent, perhaps membrane or other proteins, or cofactors or DNA which may be bound to a protein). A key decision in beginning a simulation of a biomolecular system is the choice of an appropriate method for that particular system and for the questions of interest. A modelling method should be capable of delivering a reliable result in a reasonable time. Some key strengths and weaknesses of various current methods are outlined here. The field of biomolecular simulation is still evolving, and it is not yet at the stage where quantitative, exact predictions, of (for example) relative binding free energies, reaction rates or the effects of mutation, can routinely be made (Van Gunsteren et al. 2006). For this reason, it is important to try to link with experiment to validate predictions from modelling: prediction of pKa values of functional groups in proteins provides a useful and demanding example of this type of test (Nielsen & McCammon 2003; Jensen et al. 2005; Warshel et al. 2006a; Stanton & Houk 2008; Bas et al. 2008). Similarly, it can be useful to compare activation barriers for a series of alternative substrates with the activation energies derived from experimental rates: demonstration of a correlation can validate mechanistic calculations as being truly predictive (Ridder et al. 2003).
2. Biomolecular structure and modelling
The number of structures of biological macromolecules found by experiments is large and ever increasing. Making use of this wealth of data is a challenge to biomolecular modellers. One important current source is the Research Collaboratory for Structural Bioinformatics (RCSB, www.rcsb.org), which makes available (via the web) three-dimensional biological macromolecular structural data from all experimental techniques. The RCSB Protein Data Bank (PDB) is the single worldwide repository for processing and distribution of three-dimensional structure data of large molecules, such as proteins and nucleic acids (Berman et al. 2000), and is a vital resource for biomolecular simulation.
A wide variety of experimental methods have provided insight into the structure of biological macromolecules, and these structures are the starting points for many simulations. The most important experimental technique for studying protein structure to date has been X-ray crystallography. A well-ordered crystal is needed, and finding appropriate crystallization conditions can be difficult, particularly for membrane proteins. One indication of the precision of a crystallographic protein structure determined by X-ray crystallography is the resolution, ranging from very low resolution where perhaps just the overall shape of the protein may be revealed, to higher resolution (1–2 Å), where most atomic positions can be determined, at least for heavy atoms. However, modellers should remember that the quoted resolution is a measure of global model quality (e.g. dependent on the nature of the crystal and experimental conditions) and, even in high-resolution structures, there can be considerable uncertainty due to the dynamic nature of proteins, which can cause conformational variability. The molecular models of protein structure provided by crystallography are the product of considerable subjective human intervention (e.g. in model building and refinement) and, more importantly, these structures represent an average over all the molecules in the crystal and over the whole time course of the experiment. One obvious result of this averaging is the presence of alternative conformations for some groups in many protein crystal structures—two or more well-ordered conformations are often observed, for example, for some amino acid side chains. Similarly, some parts of the structure may not be resolved by crystallography, in particular surface loops or terminal regions of the protein; these may be very mobile and have no well-defined conformation and position in the very large numbers of molecules in the crystal. It is important to bear in mind that protein crystal structures are not the equivalent of small molecule crystal structures. Crystallographic structures of biological macromolecules should not be thought of as the structure of a single molecule—they are the best fit to the available experimental data, which, as well as sources of experimental errors, contain the effects of both static and dynamic disorders. It may be surprising to find that a molecular mechanics (MM) energy minimization (including the effects of solvation) of a protein crystal structure will typically reduce the energy of a protein crystal structure by a large amount (e.g. by relaxing large numbers of close interatomic contacts), changing the structure in subtle but important ways. This does not mean, however, that the MM method cannot be trusted, but nor does it indicate that the crystal structure is ‘wrong’. MM methods aim to give a good structure of a single protein molecule, whereas a crystallographic structure is an average, as described previously, and the best fit to experimental diffraction data. Proteins undergo a wide range of complex internal motions. A crystal structure contains the effects of averaging many different protein conformations produced by these motions and the effects of the motions themselves during the experiment. It is revealing that, for high-resolution structures, combinations of two or more different structural models may give a better fit to the experimental data than a single structure. Protein crystallography gives only very limited information about dynamics; typically, only isotropic temperature factors (Debye or B factors) can be found, although at very high resolutions (e.g. less than 1 Å), more detailed information (e.g. anisotropic temperature factors) can sometimes be extracted.
Nuclear magnetic resonance (NMR) methods are increasingly important for protein structure determination. In NMR, the magnetic spin properties of atomic nuclei are used to build up a list of distance constraints between atoms in an enzyme, from which the three-dimensional structure of the protein can be determined. This method does not require the growth of crystals as it can be used on concentrated protein solutions. Direct determination of structure by NMR is generally restricted to smaller proteins. High-resolution X-ray powder diffraction has also been used to solve and refine protein structures (Von Dreele et al. 2000). This method shares the advantage of not requiring a protein crystal.
3. Molecular dynamics simulations of biological macromolecules
The impact of biomolecular simulation on biology has probably been greatest in the study of the dynamics of biological macromolecules (Karplus & Kuriyan 2005). A close integration of experiment and modelling has built up in this area. Molecular dynamics simulations of proteins have been instrumental in demonstrating that proteins flex and undergo complex internal motions, which in some cases are directly related to function (Karplus et al. 2005). Molecular dynamics simulations of protein folding and unfolding (Daggett & Fersht 2003) have shown their value in the interpretation of experimental data and complementing experiments (Mayor et al. 2003). Molecular dynamics simulations also assist in the refinement of biomolecular structures in structural investigations by X-ray crystallography (Brunger & Adams 2002) and NMR (Chen et al. 2005; Fossi et al. 2005) and also in the analysis of NMR data on spin relaxation (Case 2002) and dynamics, e.g. dynamics of protein side chains (Best et al. 2005). Other important areas of application of biomolecular dynamics simulation include studies of protein conformational changes (Elber 2005; Woods et al. 2005; Pentikäinen et al. 2008; figure 1), simulations of ion channels and other membrane proteins (Roux 2002; Warshel 2002; Gumbart et al. 2005; Sansom et al. 2005), studies of the role of biomolecular dynamics in enzyme catalysis, photosynthesis and vision (Warshel 2002) and studies of functional macromolecular assemblies such as F1-ATPase (Strajbl et al. 2003b; Dittrich et al. 2004). The now wide application of biomolecular dynamics methods is a testament to the growing maturity of the field. Studies involving multinanosecond dynamics simulations are now common. However, expert knowledge is still required, and care needs to be taken to ensure that the application of a biomolecular simulation method to a particular problem is meaningful and useful.
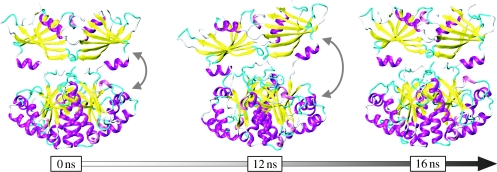
Figure 1.
Molecular dynamics simulations of the human scavenger decapping enzyme, DcpS (Pentikäinen et al. 2008), identify large-scale conformational changes that are probably important in its function. DcpS is a dimeric enzyme with two active sites, which catalyses the hydrolysis of the cap of mRNA. During the simulation, the conformation of the enzyme changes from (approx.) symmetric to asymmetric (where one active site is closed and the other open) and back to symmetric, indicating the cooperative behaviour of the binding sites of the protein.
In addition to their use to study structure, dynamics and kinetics, biomolecular dynamics simulations can also be employed to calculate thermodynamic properties. A molecular dynamics simulation provides a means to sample configurational phase space, thereby generating an ensemble of structures from which thermodynamic averages may be accumulated. For example, molecular dynamics simulations can be used to generate the ensembles necessary to calculate protein–ligand relative binding free energies (Charlier et al. 2007; Deng & Roux 2008), solvation free energies (Johansson & Lindahl 2008) and activation free energies for enzyme-catalysed reactions (Garcia-Viloca et al. 2004; Pang et al. 2006; Bowman et al. 2007; Liu & Warshel 2007). A problem in using molecular dynamics as a sampling algorithm is that the potential energy surfaces of biomolecular systems can be highly frustrated, and so the trajectory can easily become trapped sampling within a high-energy local minimum (Friesner & Gunn 1996). Several methods have been developed to enhance sampling, for example, applying a biasing ‘umbrella’ potential to force sampling along a particular reaction coordinate (Torrie & Valleau 1977; Bowman et al. 2007; Piccinini et al. 2008), coupling multiple trajectories together and allowing them to exchange parameters (Sugita & Okamoto 1999; Kamberaj & van der Vaart 2007; Mu et al. 2007; Nymeyer 2008; Sindhikara et al. 2008), such as temperature (Hansmann 1997; Hofinger et al. 2007; Huang et al. 2007; Patriksson & van der Spoel 2008) and employing generalized ensembles (Kinnear et al. 2004; Okamoto 2004), such as the multicanonical ensemble (Kamiya et al. 2008). The field of enhanced sampling methods for molecular dynamics is large and is covered by several excellent reviews (Kinnear et al. 2004; Okamoto 2004; Tai 2004; Christen & Van Gunsteren 2008; Gao et al. 2008; Piccinini et al. 2008).
Work on the protein α-synuclein (which is intrinsically disordered and involved in the pathogenesis of Parkinson's disease; Dedmon et al. 2005) provides an example of the synergy between experiments and biomolecular dynamics simulations. This involved mapping long-range interactions using a combination of ensemble molecular dynamics simulations and spin-label NMR. Distance restraints derived from paramagnetic relaxation enhancement NMR spectroscopy were applied to ensemble molecular dynamics simulations containing 20 protein replicas, with the CHARMM19 force field (see §4). The results showed that the native state of α-synuclein is made up of a broad distribution of conformers. The ensemble-averaged hydrodynamic radius found was significantly smaller than expected for a simple random coil structure. The structural studies showed that this contraction is driven by interactions between the highly charged C-terminus and a large hydrophobic central region of the protein sequence. Based on these findings, these workers suggested that this type of structure may be responsible for inhibiting the formation of α-synuclein aggregates (thought to be the cytotoxic species responsible for neurodegeneration in Parkinson's disease). Molecular dynamics simulations have similarly been used to generate conformations in the experimental determination of an ensemble of structures representing the denatured state of the bovine acyl-coenzyme A-binding protein (Lindorff-Larsen et al. 2004).
Recent work by Leontiadou et al. (2007) demonstrates some of the strengths, and limitations, of current state-of-the-art atomistic biomolecular dynamics simulations. This study investigated the effects of ionic concentration on the transport of ionic species across a pore in a lipid membrane. This work involved a number of large simulations, involving 128 dipalmitoylphosphatidylcholine lipids (each containing 130 atoms) and approximately 6000 water molecules. This work pushed the limits of what is achievable with current atomistic molecular dynamics. Using approximations, such as modelling long-range electrostatics using a reaction field, and using bond constraints so that a 5 fs integration time step could be used, it was possible to run several simulations of between 50 and 100 ns each in length. Despite the impressive size of these simulations, they are still limited to biologically small length- and time scales. One hundred and twenty-eight lipids are only an eight-by-eight membrane bilayer, which is too small to model effects such as membrane curvature or membrane waves (Ayton & Voth 2004; Chu et al. 2007). One hundred nanoseconds are also not enough time to capture events such as membrane protein aggregation or lipid raft formation within a membrane (Ayton & Voth 2004).
Coarse-grained models provide a route to longer time and length scales in biomolecular simulations. They are a class of mesoscale model, in which the groups of atoms are treated by grouping them together and modelling them as a single interaction site. In effect, groups of atoms are collected together into ‘beads’. Coarse-grained models were introduced by Levitt and Warshel for proteins in their pioneering 1970s papers (Levitt & Warshel 1975; Levitt 1976). Levitt & Warshel's (1975) paper introduced the first coarse-grained model of a globular protein. It used two coarse-grain particles per residue: one that was centred on the α-carbon of an amino acid (Cα) and the other that represented the side chain atoms. A torsion potential acted about the Cα particles, while a Lennard-Jones-type potential acted between pairs of side chain particles. In this way, a residue is represented as a pair of beads, and a protein as a string of beads. This coarse-grained model was built to represent bovine pancreatic trypsin inhibitor (BPTI), and was found to be successful, being able to correctly refold the protein starting from a completely denatured configuration. Coarse-grained model simulations are less computationally expensive than their atomistic counterparts, because coarse graining reduces the number of interaction sites. In addition, coarse-grained models contain fewer degrees of freedom and use force fields that lead to smoother potential energy surfaces (see §4). The smoother potential energy surface reduces the problems associated with frustration or non-ergodic trapping, thereby leading to more efficient sampling and a lower correlation time. Also, coarse graining typically removes the stiffest degrees of freedom from the model (e.g. the carbon–hydrogen bond vibrational modes), thereby allowing coarse-grained models to use longer time steps. Altogether, this means that coarse-grained simulations can access length- and time scales far beyond those that are practically achievable by atomistic molecular dynamics. Coarse-grained modelling methods are the subject of considerable current interest, and significant effort is being put into the development and application of coarse-grained methods for simulations of biological systems. It is not possible here to discuss these recent developments in detail; several excellent recent reviews give good descriptions of the development and the application of coarse-grained methods (Nielsen et al. 2004; Tozzini 2005; Venturoli et al. 2006; Sansom et al. 2008).
Coarse-grained models allow biomolecular simulations to investigate time- and length scales that are not feasible with atomistic modelling methods. However, because they do not represent molecules in full atomistic detail, coarse-grained models may not represent some important effects. There is now significant interest in developing frameworks for multiscale modelling, for example, interfacing atomistic and coarse-grained models, to overcome these problems (Woods & Mulholland 2008).
4. Empirical ‘MM’ force fields for biomolecules
Empirical force fields for biomolecular simulation were first developed nearly 40 years ago (Warshel et al. 1970; Warshel & Karplus 1972; Hagler et al. 1974; Warme et al. 1974), with the first simulations of biomolecular dynamics carried out soon after, e.g. investigating the photoisomerization dynamics of retinal (Warshel 1976) and, seminally, modelling the dynamics of a small protein, BPTI, in the gas phase (McCammon et al. 1977). Biomolecular simulation has come a long way in the intervening years. A number of empirical ‘MM’ force fields have been developed for simulations of proteins, nucleic acids, lipids and other biological molecules. It is important to make the distinction between programs used for biomolecular simulation and the MM parameter sets that have been developed for them, in particular as the names may be the same, or similar, and sometimes are used interchangeably. A number of good-quality parameter sets have been developed, and may be applied with several different programs, because the functional forms used are often the same or very similar. The quality of the particular parameter set is something to consider independently of the quality of the computer program itself. Of course, it is essential that a particular force field parameter set should be implemented in any program exactly as it was designed to be, and this should be carefully checked. Different protocols may apply in different programs, perhaps with different hidden assumptions. Tests on model systems are important to ensure that interactions are treated consistently and correctly by a given force field.
Among the most widely used computer programs used for biological molecular dynamics simulations, particularly in academic research, are Amber (Case et al. 2005), CHARMM (Brooks et al. 1983), Gromos (Scott et al. 1999), NAMD (Phillips et al. 2005) and Tinker (Ponder & Richards 1987). Several other molecular dynamics simulation packages are available, including commercial and academic programs.
Programs for molecular simulation should not be confused with the force fields used, as mentioned previously. A force field consists of an energy function together with the parameters. A simple energy function has to be used to allow large systems to be studied for long (multinanosecond) time scales. Current protein force fields use similar, simple potential energy functions (Mackerell 2004), in which, for example, bonds and bond angles are represented by harmonic terms, electrostatic interactions are included through atomic point partial charges, and dispersion and exchange repulsion are included by a simple Lennard-Jones function (usually of the 12–6 variety).
There are important limitations to this simple MM approach. For example, electrostatic interactions are represented by including a point charge on each atom (and only on atoms). This simple model cannot capture the full electrostatic properties (e.g. multipole moments) of a molecule, a particular problem for less polar species. For example, it is not possible to represent the quadrupole moment of benzene using atom-centred charges. Models including point charges off atomic centres, representing the π-electron clouds in benzene, give a better description of the condensed-phase behaviour of benzene and other aromatic molecules (Baker & Grant 2006, 2007). Models of aromatic amino acid residues have been developed that use off-centre charges (Xu et al. 2007), which have been shown to better reproduce condensed-phase properties. Modelling electronic polarization is also a challenge for MM approaches. While MM force fields that explicitly include polarization via a range of approaches are being developed and applied to biomolecular systems (e.g. the Amber polarizable force field, FF04, has recently been optimized for simulations of proteins and peptides; Wang et al. 2006), the majority of biomolecular force fields are non-polarizable. In these force fields, electronic polarization is not included (except in an implicit, indirect sense): that is, the atomic charges are invariant, they do not change in response to changes in the molecular environment or conformation. The attractive (r−6) component of the Lennard-Jones potential has some physical justification for modelling dispersion interactions. The repulsive (r−12) term is chosen simply for computational convenience to represent exchange repulsion at short distances. It is known that an exponential description is more physically realistic, but in the context of the overall MM description this is typically a small error for ‘organic’-type molecules. Simple harmonic terms represent the energy of bond stretching and valence angle bending, with simple periodic terms for torsion angles, and terms for other intramolecular interactions where necessary. Energy functions of this type cannot model the changes in bonding involved in a chemical reaction: the bond terms do not allow for bond making or breaking, and electronic redistribution is not allowed for. Also, the MM force field parameters are developed based on the properties of stable molecules, and so will usually not be applicable to transition states and intermediates. It is possible to develop MM functions and parameters specifically for reactions, and this has been highly successful in application to organic reactions in solution (Lim et al. 1999). This is a laborious process, however, and the parameters are typically applicable only to a particular reaction type, meaning that reparametrization may be required for each new application. Also, the form of the potential function imposes important limitations, such as the neglect of electronic polarization.
Force fields for biological macromolecules fall into two classes: all atom and united atom. All-atom force fields, as the name suggests, explicitly represent all atoms in a molecular system. United-atom force fields, by contrast, include only the heavy (non-hydrogen) atoms and polar hydrogen atoms explicitly, while non-polar hydrogen atoms are not included explicitly, but instead represented as part of the carbon atom to which they are bonded (which will have an enlarged van der Waals radius, i.e. Lennard-Jones collision diameter).
Currently, the most widely used all-atom force fields for proteins are OPLS/AA (Jorgensen et al. 1996; Kaminski et al. 2001), CHARMM22 (MacKerell et al. 1998) and Amber (PARM99; Cornell et al. 1995; Case et al. 2005). A number of good reviews of the performance of protein MM force fields have been published (Okur et al. 2003; Ponder & Case 2003; MacKerell 2005; Hornak et al. 2006). Parametrization of these force fields is increasingly based on fitting to experimental condensed-phase data (such as free energies of solvation for amino acid side chains), particularly in the optimization of Lennard-Jones parameters. This is in contrast to the previously dominant role of gas-phase data (e.g. ab initio calculations of heterodimers) in parametrization. Force fields for other types of biological macromolecules (e.g. lipids, nucleic acids (Cheatham 2004, 2005) and saccharides, as well as many small molecules and ligands) consistent with these protein force fields have also been developed, which, for example, allow simulations of proteins interacting with DNA and embedded in membranes. It is important to ensure that force fields are consistent and well balanced: different force fields should not be mixed together.) Examples include the CHARMM27 force field for nucleic acids (Foloppe & MacKerell 2000; MacKerell & Banavali 2000), Amber nucleic acid parameters (Cornell et al. 1995; Cheatham et al. 1999), CHARMM parameters for lipids (Feller et al. 1997) and several different MM parameter sets for common carbohydrates. For example, Kuttel et al. (2002) have developed carbohydrate parameters for use with the CHARMM force field, suitable for nanosecond molecular dynamics simulations in aqueous solution. Free energy profiles for rotation of the hydroxymethyl group for two monosaccharides (β-d-glucose and β-d-galactose) with this parameter set showed equilibrium rotamer populations in very good agreement with NMR data; the primary alcohol rotational frequency in solution and the gas-phase vibrational frequencies were also found to be in excellent agreement with experiment. Similarly, Woods et al. (1995) have developed carbohydrate parameters, called GLYCAM_93, for use with the Amber force field. These were shown to reproduce structural features and conformational preferences of a series of tetrahydropyran derivatives, based on ab initio calculations. Its newest incarnation, GLYCAM06 (Kirschner et al. 2008), is no longer specific to carbohydrates nor reliant on the Amber force field and work is underway on a polarizable version. Hemmingsen et al. (2004) have tested the performance of 20 different MM carbohydrate force fields, by comparison with (gas phase) ab initio and hybrid density functional calculations on monosaccharides. Geometry-optimized structures (B3LYP/6-31G(d)) and relative energies in the gas phase for monosaccharide carbohydrate benchmark systems were used. It was found that most carbohydrate force fields give an incorrect value for the interaction energy of the α-d-glucopyranose–H2O complex, compared with the ab initio result (at the coupled cluster CCSD(T) level); no single force field performed consistently better than the others for a variety of test cases (e.g. for conformational energies of methyl 5-deoxy-β-d-xylofuranoside, methyl α-d-glucopyranoside and methyl α-d-galactopyranoside). A statistical assessment of the performance of the force fields suggested that CHEAT95 (a united-atom model; Kouwijzer & Grootenhuis 1995) and some MM parametrizations developed based on the Amber (Senderowitz et al. 1996), CFF(consistent force field; Siebert et al. 2000) and MM3 (Allinger et al. 1990; Stortz & Cerezo 2003) force fields have the best overall performance for the gas-phase monosaccharide systems studied. It is important to point out that several of these force fields employ more complicated and sophisticated potential energy functions than those typically used for protein simulations. Developing MM parameters for (poly)saccharides is notoriously difficult (Imberty & Perez 2000), owing to their conformational complexity, large range of possible substitution patterns and the particular difficulty of balancing inter- and intramolecular interactions (because sugars contain very large numbers of hydrogen-bonding groups). There are clear limitations of the invariant atomic point charge model for carbohydrates. A quantum mechanics/molecular mechanics (QM/MM) approach (see §7), treating the sugar by a quantum mechanical (QM electronic structure) method, may be an improvement in many cases (French et al. 2001). Standard semi-empirical molecular orbital methods have significant shortcomings for carbohydrates, but reparametrized variants have been developed, which give better descriptions of carbohydrate conformation (e.g. PM3CARB-1; McNamara et al. 2004).
Examples of united-atom protein force fields for proteins are Gromos87 and Gromos96 (Scott et al. 1999; Schuler et al. 2001), CHARMM19 (Neria et al. 1996), OPLS/UA (united atom; Jorgensen & Tirado-Rives 1988) and the original force fields developed for the Amber program (Weiner et al. 1984). United-atom force fields were developed to reduce the computer time required for molecular dynamics simulations by reducing the number of atoms. They are still important today, in studies using either explicit or implicit solvation models. They are particularly widely used in studies of protein folding, often employing a continuum solvation description. Implicit solvent models avoid an explicit representation of water molecules and so reduce computational demands, significantly helping in accessing longer time scales. Many continuum solvation models have been developed, including the Poisson–Boltzmann (Baker 2005a,b) and generalized Born (Chen et al. 2006; Yu et al. 2006) models. These represent the solvent as a dielectric continuum, and calculate the polarization of that continuum caused by the charge distribution of the solute. This polarization leads to an electrostatic reaction field with which the solute then interacts. Continuum solvent models have been used for many years in combination with QM (Klamt & Schuurmann 1993; Park et al. 2000; Jang et al. 2003; Tomasi 2004), MM (Brown & Muchmore 2007; English 2007) and coarse-grain (Brannigan et al. 2006; Lotan & Head-Gordon 2006) solute models. These represent a large class of methods and have been reviewed in detail by many workers (Tomasi 2004; Brannigan et al. 2006; Carlsson et al. 2006; Im et al. 2006; Koehl 2006; Warshel et al. 2006a). Assessment of the performance (both accuracy and efficiency) of implicit solvent models (e.g. by comparison with explicit solvent simulations) is a highly active area of research.
Most biomolecular MM force fields have been developed to be consistent with simple point charge models of water, in particular the TIP3P water model (Jorgensen & Tirado-Rives 2005) and variants of it. In models such as TIP3P, the dipole moment is higher than that observed in the gas phase, so that electronic polarization is included in an approximate, invariant way, similar to protein MM force fields. Polarizable force fields for biological molecules are the subject of much current research and development effort (Kaminski et al. 2002; Ren & Ponder 2003; Kaminski et al. 2004; Patel et al. 2004; Anisimov et al. 2005; Gresh et al. 2005; Harder et al. 2005; Vorobyov et al. 2005; Wang et al. 2006; Warshel et al. 2007). It is likely that the next generation of protein MM force fields will treat electronic polarization explicitly. Other improvements to protein MM force fields include the use of results of high-level ab initio calculations to correct for the two-dimensional potential energy surface for peptide backbone dihedral angle rotation. MacKerell et al. (2004a,b) have developed the grid-based CMAP correction for the CHARMM22 force field that modifies the potential of the backbone φ and ψ torsion angles. It has been shown to improve overall agreement between order parameters derived from molecular dynamics simulations and experimental NMR order parameters of hen egg white lysozyme (Buck et al. 2006). Similar modifications in Amber have led to the derivation of the ff99SB (Hornak et al. 2006) and parmbsc0 (Perez et al. 2007) corrections. A more extensive modification led to ff03 (Duan et al. 2003), which can be considered as a distinct force field model rather than an extension of previous Amber force fields.
An illustration of how far biomolecular dynamics simulations have advanced is provided by that ‘guinea-pig’ of biological simulation, BPTI. Simulations of this protein have examined the effects of solvent and protein polarizability on its structure, solvation and dynamics (Kim et al. 2005). Molecular dynamics simulations of BPTI were performed in explicit water, using MM force fields that include polarization for both the water and the protein. Three model potentials for water and two model potentials for the protein were used, of which two of the water models and one of the protein models were polarizable. Six systems were simulated, covering all combinations of these polarizable and non-polarizable protein and water force fields. It was found that all six systems behave similarly in less polar parts of the protein (either hydrophobic or weakly hydrophilic). However, close to parts of the protein in which relatively strong electrostatic fields occur (i.e. near positively or negatively charged residues), it was found that the structure and the dynamics of water were clearly dependent on the model of both the protein and the water used.
5. Empirical valence bond methods
For investigating some important questions relating to enzyme action (e.g. to analyse the causes of catalysis, i.e. why an enzymic reaction proceeds faster than the equivalent, uncatalysed reaction in solution), it is necessary to use a method that not only captures the essential details of the chemical reaction but also includes the explicit effects of the enzyme and solvent environment. One notable method in this area is the empirical valence bond (EVB) model (Warshel 2003; Hong et al. 2006; Truhlar 2007). In the EVB approach, resonance structures (e.g. ionic and covalent resonance forms) are chosen to represent the reaction. The energy of each resonance form is given by a simple empirical force field (e.g. with realistic treatment of the stretching of important bonds, e.g. by a Morse function). The potential energy is given by solving the related secular equation. The EVB Hamiltonian is calibrated to reproduce experimental data for a known and relevant solution reaction, or alternatively ab initio results can be used (Bentzien et al. 1998). The surrounding protein and solution are modelled by an empirical force field with appropriate treatment of long-range electrostatics. The free energy of activation for the reaction in solution, and in the enzyme, can be calculated using free energy perturbation simulations (Warshel 1997).
One of the main advantages of the EVB method is that the free energy surfaces can be calibrated by comparison with experimental data for reference reactions in solution. However, as in any valence bond representation, it is essential that the valence bond forms should represent all the resonance forms that are important in the reaction. An appealing feature of the EVB method is that it makes it straightforward to use non-geometrical reaction coordinates in modelling a reaction, which may be significantly more accurate for some condensed-phase reactions. Energy can be used as a reaction coordinate by following a path between valence bond optima. A mapping procedure is followed, which moves gradually from the reactant to the product. In this mapping, the change in both the solute structure and charge is taken into account. This EVB umbrella sampling method locates the correct transition state in the combined solute–solvent reaction coordinate. This allows the evaluation of non-equilibrium solvation effects (e.g. Warshel 2003). Other strengths of the method have been discussed elsewhere (Villa & Warshel 2001). The EVB method is a powerful and useful approach, which has now become a widely adopted tool for studying reactions in condensed phases. Illustrative simulations with EVB methods have included a study of alternative nucleotide insertion mechanisms for T7 DNA polymerase (Florian et al. 2003), the investigation of proton transfer in 1-(trifluoroacetylamino)-naphthaquinone (Cembran & Gao 2007), modelling proton transport in an ion channel (Chen et al. 2007), a study of the reaction mechanism of human aldose reductase (Varnai & Warshel 2000) and investigation of the nature of the proton bottleneck in redox-coupled proton transfer in cytochrome c oxidase (Olsson et al. 2005).
In another study applying EVB techniques, Bjelic & Åqvist (2004) used a well-validated homology model to examine the substrate-binding mode and reaction mechanism of a malaria protease with a novel active site. This enzyme (histo-aspartic protease from the malaria parasite Plasmodium falciparum) is a target for anti-malarial drug design, but its three-dimensional structure is not yet known. This work predicted the structure of the enzyme, and the conformation of bound substrate, using a combination of homology modelling, automated docking and molecular dynamics/reaction free energy profile simulations. The only amino acid residue involved directly in the reaction in the predicted mechanism is a catalytic aspartate, with a histidine residue providing stabilization. The calculated reaction rate agreed well with experimental kinetic data for a hexapeptide substrate derived from human haemoglobin.
6. Modelling with quantum chemical (electronic structure) methods
Quantum chemical methods (e.g. ab initio molecular orbital or density functional theory calculations) can currently be used practically to study reactions in non-periodic, molecular systems containing of the order of tens of atoms. Small ‘cluster’ models of around this size can represent key features of an enzyme reaction and can identify probable mechanisms. The active site of an enzyme is a relatively small region, often within a cleft or crevice in the protein, where the substrate(s) (and cofactor(s) in cases where they are involved) bind. It contains the residues that are directly involved in the chemical reaction and the residues involved in binding. The substrates are typically bound at the active site by multiple weak interactions, such as hydrogen bonds, electrostatic and van der Waals forces. Clusters of small molecules can be used to represent important functional groups (e.g. key amino acid side chains involved in catalysis and (parts of) the substrate or cofactors) with their positions typically taken from a representative X-ray crystal structure of an enzyme complex. For example, acetate can represent an aspartate side chain, imidazole can represent histidine, etc. Calculations on models of this type can examine interactions between groups at the active site and can provide useful models of transition states and reaction intermediates. They can also be used to test the accuracy of different levels of calculations (e.g. comparing the results of semi-empirical with ab initio molecular orbital calculations or different levels of ab initio treatment). Applications of QM modelling in drug design have recently been reviewed by Raha et al. (2007).
The approach of modelling small clusters has proved especially useful in studying reaction mechanisms of metalloenzymes. In many metalloenzymes, all the important chemical steps take place at the metal centre (one or a small number of metal ions bound at one site). Typically, the metal ion(s) may also hold its ligands in place. This gives the technical advantage of limiting the requirement for restraints or constraints to maintain the correct active site structure in calculations. Reliable calculations on metalloenzymes have been made possible by methods based on density functional theory. Popular functionals, such as the widely used B3LYP hybrid functional, give good results for many reactions without requiring excessive amounts of computer time, memory or disk space, for clusters of quite large size. The work of Siegbahn and collaborators (Himo & Siegbahn 2003) on many enzymes provides an excellent example of the mechanistic insight that calculations on small clusters can give.
In a cluster model containing various small molecules representing important functional groups, it may be possible to optimize the geometries of complexes representing the reactants, transition state, intermediates and products of steps in the reaction. This can often be sufficient to discriminate between alternative possible mechanisms (Harvey et al. 2006), as the energy difference between alternative mechanisms is often very large, larger than the probable effects of the environment on the relative energies. For discriminating between alternative proposed mechanisms, a mechanism can be excluded if the calculated barriers for it are significantly higher than the experimentally derived activation energy, within the limits of accuracy of the computational method. A small model, though, might lack some important functional groups, and careful consideration should be given as to which groups to include, balancing computational feasibility against the desire for a larger, more extensive model. Also, perhaps counterintuitively, a larger cluster model is not always a better model: a larger model will involve greater conformational complexity (conformational changes distant from the reaction centre might artificially affect relative energies along the reaction path) and including unshielded charged groups could also have unrealistically large effects on reaction energies. Environmental effects, such as solvation, can be included approximately in calculations on small cluster models, e.g. by the use of continuum solvation models, but these cannot fully represent the heterogeneous electrostatic environment in an enzyme (Shurki & Warshel 2003). An important technical and practical aspect of cluster/supermolecule calculations is that it can often be difficult to optimize the geometry of the model (e.g. to locate a transition-state structure), while at the same time maintaining the correct orientations of the groups in the protein.
More approximate quantum chemical methods (such as the semi-empirical molecular orbital techniques AM1 and PM3) can model larger molecular systems (containing of the order of hundreds of atoms). However, semi-empirical methods are well known to be inaccurate for many applications (e.g. they can sometimes be subject to very large errors in calculated reaction energies). They also cannot straightforwardly be used for some types of system (e.g. for many transition metals). Techniques (such as ‘linear-scaling’ methods) have been developed, which allow semi-empirical electronic structure calculations on whole proteins (Van der Vaart et al. 2000; Khandogin et al. 2003; Khandogin & York 2004). Considerable steps are also being made in improving the scaling properties of higher level quantum chemical methods, which will allow their application to larger systems (Claeyssens et al. 2006; Mata et al. 2008).
Typical enzyme–substrate complexes, particularly when modelled using an explicit representation of surrounding solvent, will contain at least thousands of atoms, and perhaps many more. This places them currently beyond even semi-empirical quantum chemical methods for modelling reactions. An equally important consideration in modelling a reaction is that the calculation of (single point) energies is not enough: important points (such as transition-state structures) and preferably entire reaction pathways should be optimized. Extensive conformational sampling may be required to generate a representative ensemble of structures. These are in themselves significant challenges for large molecules. One should also consider the environment of the enzyme: aqueous solution (but some enzymes operate in concentrated solutions, e.g. high concentrations of other proteins, or in acidic or basic conditions), membranes or in protein or nucleic acid complexes. Protein internal motions are highly complex: many conformational substates can exist and a single structure may not be truly representative (Zhang et al. 2003). To carry out extensive conformational sampling (e.g. to calculate free energy profiles; Gao & Truhlar 2002), a dynamics simulation method must be capable of calculating trajectories of at least many picoseconds length. One useful approach can be to use MM molecular dynamics simulations (which can run to relatively long, nanosecond time scales) to generate multiple models for mechanism calculations, thus ensuring wide sampling of possible enzyme configurations (Lodola et al. 2007). If multiple different crystal structures of the same enzyme are available, these can be used as different starting models to examine the effects of structural variation on the reaction. For modelling on the large scale (e.g. for large models or to incorporate conformational variability), a QM/MM approach can be useful, as discussed in §7.
7. Combined QM/MM methods
Combined QM/MM methods are increasingly important in the modelling of biological systems, particularly in the growing field of computational enzymology, i.e. the computational modelling of enzyme-catalysed reaction mechanisms (Mulholland 2005, 2008). In essence, the QM/MM approach is simple: a small part of the system is treated quantum mechanically, while the rest is treated using MM (see §4). In a study of an enzyme reaction mechanism, the QM region would typically be the active site and include the reacting groups of the enzyme, substrate and any cofactors; the MM region would consist of the large non-reactive part of the system (figure 2). The QM treatment (by an electronic structure method, e.g. at the ab initio or semi-empirical molecular orbital, or density functional theory level) allows the modelling of chemical bond breaking and making, and the electronic rearrangements and polarization. Different types of treatment of the interaction between the QM and MM regions can be employed. For applications to biological macromolecules, such as proteins, which are polar, it is probably important to include the polarization of the QM region by the MM environment. The combination of the versatility and range of applicability of a QM electronic structure method with the efficiency and speed of the MM force field allows reactions in large systems to be studied. As noted in §4, modern MM methods deal well with protein structure and interactions, so we can ensure that these are treated accurately in the QM/MM approach. With lower levels of QM theory (e.g. semi-empirical molecular orbital or approximate density functional methods), QM/MM molecular dynamics simulations are feasible.
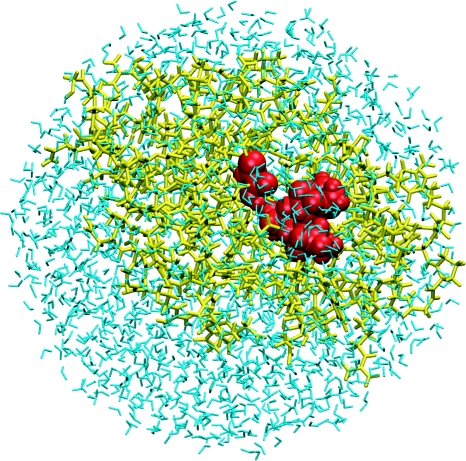
Figure 2.
QM/MM methods are a good approach for modelling enzyme-catalysed reactions. This figure shows the general set-up for a QM/MM simulation of an enzyme. The substrate, catalytic residues and any cofactors (red) are treated by a QM method, which can treat bond making and breaking processes. The surrounding protein (yellow) and solvent (blue) are treated by a standard empirical MM force field. The QM and MM regions interact, so that the enzyme and solvent environment of the reaction are taken into account.
A QM/MM method was first applied to an enzyme-catalysed reaction by Warshel & Levitt (1976) in their seminal study of the reaction mechanism of hen egg white lysozyme. Simple QM/MM methods, for example, involving basic QM treatments of π-electrons with an MM description of the σ-bonded framework, were developed for conjugated biomolecules such as retinal (Warshel & Karplus 1972; Warshel 1976). Interest in QM/MM methods has grown rapidly in recent years. It is becoming apparent that QM/MM calculations can provide useful insight into enzyme-catalysed reactions (Garcia-Viloca et al. 2004; Mulholland 2005; Senn & Thiel 2007; Mulholland 2008). An example is the identification of catalytic functions for the active site residues (such as a conserved proline in two flavin-dependent monooxygenases; Ridder et al. 2000, 2003), investigating questions of mechanism (e.g. comparing and differentiating between alternative proposed mechanisms; van der Kamp et al. 2008), and suggesting and testing catalytic principles (such as the possible contribution of conformational effects and transition-state stabilization in chorismate mutase; Lyne et al. 1995; Ranaghan et al. 2003, 2004; Marti et al. 2004; Ranaghan & Mulholland 2004; Claeyssens et al. 2005; Guimaraes et al. 2005). Even for this apparently simple enzyme reaction (a Claisen rearrangement), there are lively current debates on the origin of catalysis. Modelling has been central in formulating and testing proposed mechanisms and hypotheses (figure 3).
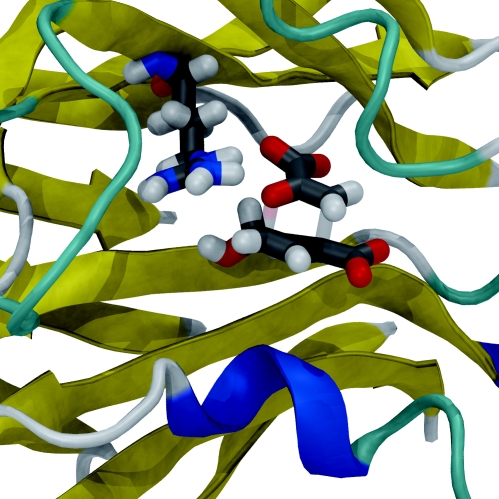
Figure 3.
QM/MM modelling of the reaction in the enzyme chorismate mutase (Lyne et al. 1995; Ranaghan et al. 2003; Claeyssens et al. 2006). The transition state for the conversion of chorismate to prephenate, bound in the active site of the enzyme, is shown. Chorismate mutase catalyses the reaction by electrostatic stabilization of the transition state, in particular by a charged arginine residue (also shown) close to the substrate (Strajbl et al. 2003a; Claeyssens et al. 2005; Guimaraes et al. 2005).
Many different QM/MM implementations are available in a number of widely used programs. QM/MM calculations can be carried out at various different levels of QM electronic structure calculation: ab initio (Mulholland et al. 2000; Woodcock et al. 2003) or semi-empirical molecular orbital (Field et al. 1990), density functional (Lyne et al. 1999) or approximate density functional (Cui et al. 2001) levels. It is now possible to carry out QM/MM calculations using very high levels of electronic structure theory (e.g. coupled cluster theory with single and double excitations). Such methods offer the potential of highly accurate results, approaching ‘chemical accuracy’ (within 1 kcal mol−1), at least for the QM part of the calculation (Claeyssens et al. 2006; Mulholland 2007).
Transition-state structures can be optimized in QM/MM calculations (Prat-Resina et al. 2004; Marti & Moliner 2005). Also free energy differences, such as activation free energies, can be calculated, as can quantum effects such as tunnelling and zero-point corrections. More approximate, less computer-intensive QM/MM methods (such as semi-empirical or self-consistent charge density functional tight-binding (SCC-DFTB) QM/MM) have an important role as they allow more extensive simulations to be performed (e.g. molecular dynamics; Ridder et al. (2002) or Monte Carlo simulations for extensive conformational sampling, and calculation of reaction pathways and Hessians). Specifically, parametrized semi-empirical methods can give improved accuracy for a particular reaction (Gonzalez-Lafont et al. 1991; Bowman et al. 2007). In addition to parametrizing the semi-empirical (QM) model, the Lennard-Jones (MM) parameters for QM/MM interactions can be optimized, which may be necessary to ensure a proper balance of the interactions between the QM and MM regions (Martin et al. 2002). The high-level QM/MM calculations (e.g. ab initio or density functional level QM) are needed for some systems and have an important role in testing more approximate (e.g. semi-empirical QM/MM) methods, but are highly demanding of computational resources.
Current developments include the use of QM/MM methods in calculations of relative free energies (e.g. either directly (Riccardi et al. 2005) or indirectly via a reference potential (Muller & Warshel 1995; Wood et al. 1999; Strajbl et al. 2002, 2003a; Ming et al. 2004; Rod & Ryde 2005; Rosta et al. 2006; Woods et al. 2008)), for example, to calculate relative binding affinities, or molecular ‘docking’ and ‘scoring’ for ligands in proteins (Raha & Merz 2004). QM/MM methods provide several advantages over MM methods in studies of small molecule ligands bound to proteins, including potentially a better physical description of the ligand (e.g. by including electronic polarization), and avoiding the need for time-consuming development of MM parameters for the ligand. Also, MM methods may be inadequate for some types of pharmaceutically important target proteins, particularly metalloproteins (such as cytochrome P450 enzymes involved in drug metabolism; Bathelt et al. 2005). For such biomolecular systems, the use of QM/MM approaches for the predictions of relative binding affinity or binding mode may be significantly better. With increasing computer power, and continuing methodological development, QM/MM methods will certainly become ever more important in practical applications such as drug design, and related areas such as the prediction of drug metabolism and toxicity (Mulholland 2005).
8. Ab initio (Car–Parrinello) molecular dynamics simulations
An increasingly important technique in biomolecular simulations (Carloni et al. 2002) is the ab initio molecular dynamics technique first proposed by Car and Parrinello about 20 years ago (Car & Parrinello 1985; Remler & Madden 1990). The scheme combines molecular dynamics simulation and density functional theory: it integrates fictitious wave function coefficient dynamics with classical molecular dynamics in a single extended Lagrangian. Crucially, the electronic wave functions are included as dynamical variables. Initially, a converged wave function is determined, and the orbitals subsequently evolve simultaneously with the changes in nuclear position. The orbital parameters are included as variables with fictitious masses in the dynamics, analogous to the nuclear positions and masses. The nuclear forces are not exactly correct in dynamics, as the electronic wave function is not converged in the orbital parameter space, but this error is controlled by an appropriate choice of dynamic parameters (e.g. the fictitious masses). Constraints are applied to the system to ensure that the orbitals remain orthonormal. Applications of Car–Parrinello molecular dynamics simulations to biomolecular systems have recently been reviewed (Dal Peraro et al. 2007). One relevant application of these techniques examined the catalytic site of galactose oxidase and a biomimetic catalyst (Röthlisberger et al. 2000). A recent QM/MM Car–Parrinello molecular dynamics study examined the protonation state of residues in the KcsA potassium channel (Bucher et al. 2007).
Despite the development of highly efficient codes and algorithms, ab initio molecular dynamics simulations are extremely computationally expensive, requiring very large amounts of supercomputing time. They provide an advantage over molecular dynamics employing empirical force fields in that the electronic structure methods are able to describe bond breaking and forming reactions and therefore Car–Parrinello methods can, in principle, allow the direct simulation of chemical reactions. Similarly, they overcome other limitations of MM force fields: for example, electronic polarization effects are included naturally. The major practical limitations are the size of the systems that can be simulated, and the time scale of feasible dynamics simulations. For this reason, combined QM/MM approaches are also attractive for ab initio molecular dynamics simulations. For example, Parrinello and co-workers have developed a scheme for Car–Parrinello molecular dynamics simulations with a QM/MM method, with the CPMD and EGO programs (Eichinger et al. 1999). Using these interfaced programs, efficient and consistent QM/MM Car–Parrinello simulations of large systems can be performed, including the steric and electrostatic effects of the protein and its solvent environment explicitly.
9. Conclusions
Biomolecular simulation is a rapidly developing area that is contributing increasingly to biology. The whole field of biomolecular modelling is very large, and so it has not been possible to describe all its aspects here. One notable important area of application is structure-aided drug design. Developments in structure-based virtual screening have been reviewed, for example, by McInnes (2007); ligand docking methods and scoring functions have been specifically covered by others (Krömer 2007; Rajamani & Good 2007). Applications of ligand docking in drug design and the current developments of inclusion of protein flexibility have been described by Cavasotto & Orry (2007) and Joseph-McCarthy et al. (2007). Others have discussed the field of calculations of protein–ligand binding free energies, including covering scoring functions for ranking binding affinities in such complexes (Raha & Merz 2005) and new physics-based methods (Huang et al. 2006). One exciting developing area in which molecular modelling plays a vital part is protein design (Baker 2006; Lippow & Tidor 2007): developments here, relying on practical and reliable modelling methods, promise a route to new catalysts and components for biologically inspired nanotechnology and molecular medicine. It is also worth pointing out some theoretical developments, for example, in the calculation of free energies by non-equilibrium approaches, such as through the application of the Jarzynski relation to calculate free energies from steered molecular dynamics simulations (Crespo et al. 2005; Roitberg 2005; Bastug & Kuyucak 2007). Altogether, the field of biomolecular modelling and simulation is thriving and growing, and its importance looks certain to increase in the future.
Acknowledgments
A.J.M. thanks his co-workers on the work described here in which he has been involved. He also thanks BBSRC (with K.E.S.), EPSRC (with C.J.W.), the European Commission and IBM (with M.W.v.d.K.) for their support. The authors thank Ulla Pentikäinen for supplying the data for figure 1.
Footnotes
One contribution of 9 to a Theme Supplement ‘Biomolecular simulation’.
References
- Allinger N.L., Rahman M., Lii J.H. A molecular mechanics force-field (MM3) for alcohols and ethers. J. Am. Chem. Soc. 1990;112:8293–8307. doi: 10.1021/ja00179a012. [DOI] [Google Scholar]
- Anisimov V.M., Lamoureux G., Vorobyov I.V., Huang N., Roux B., MacKerell A.D. Determination of electrostatic parameters for a polarizable force field based on the classical Drude oscillator. J. Chem. Theory Comput. 2005;1:153–168. doi: 10.1021/ct049930p. [DOI] [PubMed] [Google Scholar]
- Ayton G.S., Voth G.A. Mesoscopic lateral diffusion in lipid bilayers. Biophys. J. 2004;87:3299–3311. doi: 10.1529/biophysj.104.047811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A. Biomolecular applications of Poisson–Boltzmann methods. In: Lipkowitz K.B., Larter R., Cundari T.R., editors. Reviews in computational chemistry. Wiley; Hoboken, NJ: 2005a. pp. 349–379. [Google Scholar]
- Baker N.A. Improving implicit solvent simulations: a Poisson-centric view. Curr. Opin. Struct. Biol. 2005b;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baker D. Prediction and design of macromolecular structures and interactions. Phil. Trans. R. Soc. B. 2006;361:459–463. doi: 10.1098/rstb.2005.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.M., Grant G.H. The structure of liquid benzene. J. Chem. Theory Comput. 2006;2:947–955. doi: 10.1021/ct060024h. [DOI] [PubMed] [Google Scholar]
- Baker C.M., Grant G.H. Modeling aromatic liquids: toluene, phenol, and pyridine. J. Chem. Theory Comput. 2007;3:530–548. doi: 10.1021/ct600218f. [DOI] [PubMed] [Google Scholar]
- Bas D.C., Rogers D.M., Jensen J.H. Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins: Struct. Funct. Bioinform. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- Bastug T., Kuyucak S. Application of Jarzynski's equality in simple versus complex systems. Chem. Phys. Lett. 2007;436:383–387. doi: 10.1016/j.cplett.2007.01.078. [DOI] [Google Scholar]
- Bathelt C.M., Zurek J., Mulholland A.J., Harvey J.N. Electronic structure of compound I in human isoforms of cytochrome P450 from QM/MM modeling. J. Am. Chem. Soc. 2005;127:12 900–12 908. doi: 10.1021/ja0520924. [DOI] [PubMed] [Google Scholar]
- Beckstein O., Biggin P.C., Bond P., Bright J.N., Domene C., Grottesi A., Holyoake J., Sansom M.S.P. Ion channel gating: insights via molecular simulations. FEBS Lett. 2003;555:85–90. doi: 10.1016/S0014-5793(03)01151-7. [DOI] [PubMed] [Google Scholar]
- Bentzien J., Muller R.P., Florian J., Warshel A. Hybrid ab initio quantum mechanics molecular mechanics calculations of free energy surfaces for enzymatic reactions: the nucleophilic attack in subtilisin. J. Phys. Chem. B. 1998;102:2293–2301. doi: 10.1021/jp973480y. [DOI] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R.B., Clarke J., Karplus M. What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J. Mol. Biol. 2005;349:185–203. doi: 10.1016/j.jmb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Bjelic S., Åqvist J. Computational prediction of structure, substrate binding mode, mechanism, and rate for a malaria protease with a novel type of active site. Biochemistry. 2004;43:14 521–14 528. doi: 10.1021/bi048252q. [DOI] [PubMed] [Google Scholar]
- Bowman A.L., Ridder L., Rietjens I.M.C.M., Vervoort J., Mulholland A.J. Molecular determinants of xenobiotic metabolism: QM/MM simulation of the conversion of 1-chloro-2,4-dinitrobenzene catalyzed by M1-1 glutathione S-transferase. Biochemistry. 2007;46:6353–6363. doi: 10.1021/bi0622827. [DOI] [PubMed] [Google Scholar]
- Brannigan G., Lin L.C.L., Brown F.L.H. Implicit solvent simulation models for biomembranes. Eur. Biophys. J. Biophys. Lett. 2006;35:104–124. doi: 10.1007/s00249-005-0013-y. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Brown S.P., Muchmore S.W. Rapid estimation of relative protein–ligand binding affinities using a high-throughput version of MM-PBSA. J. Chem. Inf. Model. 2007;47:1493–1503. doi: 10.1021/ci700041j. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D. Molecular dynamics applied to X-ray structure refinement. Acc. Chem. Res. 2002;35:404–412. doi: 10.1021/ar010034r. [DOI] [PubMed] [Google Scholar]
- Bucher D., Guidoni L., Röthlisberger U. The protonation state of the Glu-71/Asp-80 residues in the KcsA potassium channel: a first-principles QM/MM molecular dynamics study. Biophys. J. 2007;93:2315–2324. doi: 10.1529/biophysj.106.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Bouguet-Bonnet S., Pastor R.W., MacKerell A.D. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Car R., Parrinello M. Unified approach for molecular-dynamics and density-functional theory. Phys. Rev. Lett. 1985;55:2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- Carloni P., Röthlisberger U., Parrinello M. The role and perspective of ab initio molecular dynamics in the study of biological systems. Acc. Chem. Res. 2002;35:455–464. doi: 10.1021/ar010018u. [DOI] [PubMed] [Google Scholar]
- Carlsson J., Ander M., Nervall M., Åqvist J. Continuum solvation models in the linear interaction energy method. J. Phys. Chem. B. 2006;110:12 034–12 041. doi: 10.1021/jp056929t. [DOI] [PubMed] [Google Scholar]
- Case D.A. Molecular dynamics and NMR spin relaxation in proteins. Acc. Chem. Res. 2002;35:325–331. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- Case D.A., et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C.N., Orry A.J.W. Ligand docking and structure-based virtual screening in drug discovery. Curr. Top. Med. Chem. 2007;7:1006–1014. doi: 10.2174/156802607780906753. [DOI] [PubMed] [Google Scholar]
- Cembran A., Gao J. Potential energy functions for an intramolecular proton transfer reaction in the ground and excited state. Theor. Chem. Acc. 2007;118:211–218. doi: 10.1007/s00214-007-0272-z. [DOI] [Google Scholar]
- Charlier L., Nespoulous C., Fiorucci S., Antonczaka S., Golebiowski J. Binding free energy prediction in strongly hydrophobic biomolecular systems. Phys. Chem. Chem. Phys. 2007;9:5761–5771. doi: 10.1039/b710186d. [DOI] [PubMed] [Google Scholar]
- Cheatham T.E. Simulation and modeling of nucleic acid structure, dynamics and interactions. Curr. Opin. Struct. Biol. 2004;14:360–367. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cheatham, T. E. 2005 Molecular modeling and atomistic simulation of nucleic acids. In Annual reports in computational chemistry, vol. 1 (ed. C. Simmerling), pp. 75–90. Oxford, UK: Elsevier.
- Cheatham T.E., Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- Chen J.H., Won H.S., Im W.P., Dyson H.J., Brooks C.L. Generation of native-like protein structures from limited NMR data, modern force fields and advanced conformational sampling. J. Biomol. NMR. 2005;31:59–64. doi: 10.1007/s10858-004-6056-z. [DOI] [PubMed] [Google Scholar]
- Chen J.H., Im W.P., Brooks C.L. Balancing solvation and intramolecular interactions: toward a consistent generalized Born force field. J. Am. Chem. Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.N., Wu Y.J., Voth G.A. Proton transport behavior through the influenza a M2 channel: insights from molecular simulation. Biophys. J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M., Van Gunsteren W.F. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: a review. J. Comput. Chem. 2008;29:157–166. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- Chu J.W., Ayton G.S., Izvekov S., Voth G.A. Emerging methods for multiscale simulation of biomolecular systems. Mol. Phys. 2007;105:167–175. doi: 10.1080/00268970701256696. [DOI] [Google Scholar]
- Claeyssens, F., Ranaghan, K. E. Manby, F. R., Harvey, J. N. & Mulholland, A. J. 2005 Multiple high-level QM/MM reaction paths demonstrate transition-state stabilization in chorismate mutase: correlation of barrier height with transition-state stabilization. Chem. Commun. 5068–5070. ( 10.1039/b508181e) [DOI] [PubMed]
- Claeyssens F., et al. High-accuracy computation of reaction barriers in enzymes. Angew. Chem. Int. Ed. 2006;45:6856–6859. doi: 10.1002/anie.200602711. [DOI] [PubMed] [Google Scholar]
- Cornell W.D., et al. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. doi: 10.1021/ja00124a002. [DOI] [Google Scholar]
- Crespo A., Marti M.A., Estrin D.A., Roitberg A.E. Multiple-steering QM–MM calculation of the free energy profile in chorismate mutase. J. Am. Chem. Soc. 2005;127:6940–6941. doi: 10.1021/ja0452830. [DOI] [PubMed] [Google Scholar]
- Cui Q., Elstner M., Kaxiras E., Frauenheim T., Karplus M. A QM/MM implementation of the self-consistent charge density functional tight binding (SCC-DFTB) method. J. Phys. Chem. B. 2001;105:569–585. doi: 10.1021/jp0029109. [DOI] [Google Scholar]
- Daggett V. Protein folding-simulation. Chem. Rev. 2006;106:1898–1916. doi: 10.1021/cr0404242. [DOI] [PubMed] [Google Scholar]
- Daggett V., Fersht A. The present view of the mechanism of protein folding. Nat. Rev. Mol. Cell Biol. 2003;4:497–502. doi: 10.1038/nrm1126. [DOI] [PubMed] [Google Scholar]
- Dal Peraro M., Ruggerone P., Raugei S., Gervasi F.L., Carloni P. Investigating biological systems using first principles Car–Parrinello molecular dynamics simulations. Curr. Opin. Struct. Biol. 2007;17:149–156. doi: 10.1016/j.sbi.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Dedmon M.M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., Dobson C.M. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- Deng Y.Q., Roux B. Computation of binding free energy with molecular dynamics and grand canonical Monte Carlo simulations. J. Chem. Phys. 2008;128:115 103. doi: 10.1063/1.2842080. [DOI] [PubMed] [Google Scholar]
- Dittrich M., Hayashi S., Schulten K. ATP hydrolysis in the βTP and βDP catalytic sites of F1-ATPase. Biophys. J. 2004;87:2954–2967. doi: 10.1529/biophysj.104.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Eichinger M., Tavan P., Hutter J., Parrinello M. A hybrid method for solutes in complex solvents: density functional theory combined with empirical force fields. J. Chem. Phys. 1999;110:10 452–10 467. doi: 10.1063/1.479049. [DOI] [Google Scholar]
- Elber R. Long-timescale simulation methods. Curr. Opin. Struct. Biol. 2005;15:151–156. doi: 10.1016/j.sbi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Elcock A.H. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. Plos Comput. Biol. 2006;2:824–841. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English N.J. Calculation of binding affinities of HIV-1 RT and β-secretase inhibitors using the linear interaction energy method with explicit and continuum solvation approaches. J. Mol. Model. 2007;13:1081–1097. doi: 10.1007/s00894-007-0229-0. [DOI] [PubMed] [Google Scholar]
- Feller S.E., Yin D.X., Pastor R.W., MacKerell A.D. Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys. J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M.J., Bash P.A., Karplus M. A combined quantum-mechanical and molecular mechanical potential for molecular-dynamics simulations. J. Comput. Chem. 1990;11:700–733. doi: 10.1002/jcc.540110605. [DOI] [Google Scholar]
- Florian J., Goodman M.F., Warshel A. Computer simulation of the chemical catalysis of DNA polymerases: discriminating between alternative nucleotide insertion mechanisms for T7 DNA polymerase. J. Am. Chem. Soc. 2003;125:8163–8177. doi: 10.1021/ja028997o. [DOI] [PubMed] [Google Scholar]
- Foloppe N., MacKerell A.D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. doi: 10.1002/(SICI)1096-987X(20000130)21:2%3C86::AID-JCC2%3E3.0.CO;2-G. [DOI] [Google Scholar]
- Fossi M., Oschkinat H., Nilges M., Ball L.J. Quantitative study of the effects of chemical shift tolerances and rates of SA cooling on structure calculation from automatically assigned NOE data. J. Magn. Reson. 2005;175:92–102. doi: 10.1016/j.jmr.2005.03.020. [DOI] [PubMed] [Google Scholar]
- French A.D., Johnson G.P., Kelterer A.M., Dowd M.K., Cramer C.J. QM/MM distortion energies in di- and oligosaccharides complexed with proteins. Int. J. Quantum Chem. 2001;84:416–425. doi: 10.1002/qua.1111. [DOI] [Google Scholar]
- Friesner R.A., Gunn J.R. Computational studies of protein folding. Annu. Rev. Biophys. Biomol. Struct. 1996;25:315–342. doi: 10.1146/annurev.biophys.25.1.315. [DOI] [PubMed] [Google Scholar]
- Gao J.L., Truhlar D.G. Quantum mechanical methods for enzyme kinetics. Annu. Rev. Phys. Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- Gao Y.Q., Yang L.J., Fan Y.B., Shao Q. Thermodynamics and kinetics simulations of multi-time-scale processes for complex systems. Int. Rev. Phys. Chem. 2008;27:201–227. doi: 10.1080/01442350801920334. [DOI] [Google Scholar]
- Garcia-Viloca M., Gao J., Karplus M., Truhlar D.G. How enzymes work: analysis by modern rate theory and computer simulations. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- Gilson M.K., Zhou H.-X. Calculation of protein–ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lafont A., Truong T.N., Truhlar D.G. Direct dynamics calculations with neglect of diatomic differential overlap molecular orbital theory with specific reaction parameters. J. Phys. Chem. 1991;95:4618–4627. doi: 10.1021/j100165a009. [DOI] [Google Scholar]
- Gresh N., Piquemal J.P., Krauss M. Representation of Zn(II) complexes in polarizable molecular mechanics. Further refinements of the electrostatic and short-range contributions. Comparisons with parallel ab initio computations. J. Comput. Chem. 2005;26:1113–1130. doi: 10.1002/jcc.20244. [DOI] [PubMed] [Google Scholar]
- Guimaraes C.R.W., Udier-Blagovic M., Tubert-Brohman I., Jorgensen W.L. Effects of Arg90 neutralization on the enzyme-catalyzed rearrangement of chorismate to prephenate. J. Chem. Theory Comput. 2005;1:617–625. doi: 10.1021/ct0500803. [DOI] [PubMed] [Google Scholar]
- Gumbart J., Wang Y., Aksimentiev A., Tajkhorshid E., Schulten K. Molecular dynamics simulations of proteins in lipid bilayers. Curr. Opin. Struct. Biol. 2005;15:423–431. doi: 10.1016/j.sbi.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler A.T., Huler E., Lifson S. Energy functions for peptides and proteins. 1. Derivation of a consistent force-field including hydrogen-bond from amide crystals. J. Am. Chem. Soc. 1974;96:5319–5327. doi: 10.1021/ja00824a004. [DOI] [PubMed] [Google Scholar]
- Hansmann U.H.E. Parallel tempering algorithm for conformational studies of biological molecules. Chem. Phys. Lett. 1997;281:140–150. doi: 10.1016/S0009-2614(97)01198-6. [DOI] [Google Scholar]
- Harder E., Kim B.C., Friesner R.A., Berne B.J. Efficient simulation method for polarizable protein force fields: application to the simulation of BPTI in liquid. J. Chem. Theory Comput. 2005;1:169–180. doi: 10.1021/ct049914s. [DOI] [PubMed] [Google Scholar]
- Harvey J.N., Aggarwal V.K., Bathelt C.M., Carreon-Macedo J.L., Gallagher T., Holzmann N., Mulholland A.J., Robiette R. QM and QM/MM studies of selectivity in organic and bioorganic chemistry. J. Phys. Org. Chem. 2006;19:608–615. doi: 10.1002/poc.1030. [DOI] [Google Scholar]
- Hemmingsen L., Madsen D.E., Esbensen A.L., Olsen L., Engelsen S.B. Evaluation of carbohydrate molecular mechanical force fields by quantum mechanical calculations. Carbohydr. Res. 2004;339:937–948. doi: 10.1016/j.carres.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Himo F., Siegbahn P.E.M. Quantum chemical studies of radical-containing enzymes. Chem. Rev. 2003;103:2421–2456. doi: 10.1021/cr020436s. [DOI] [PubMed] [Google Scholar]
- Hofinger S., Almeida B., Hansmann U.H.E. Parallel tempering molecular dynamics folding simulation of a signal peptide in explicit water. Proteins: Struct. Funct. Bioinform. 2007;68:662–669. doi: 10.1002/prot.21268. [DOI] [PubMed] [Google Scholar]
- Hong G.Y., Rosta E., Warshel A. Using the constrained DFT approach in generating diabatic surfaces and off diagonal empirical valence bond terms for modeling reactions in condensed phases. J. Phys. Chem. B. 2006;110:19 570–19 574. doi: 10.1021/jp0625199. [DOI] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinform. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Kalyanaraman C., Bernacki K., Jacobson M.P. Molecular mechanics methods for predicting protein–ligand binding. Phys. Chem. Chem. Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- Huang X.H., Hagen M., Kim B., Friesner R.A., Zhou R.H., Berne B.J. Replica exchange with solute tempering: efficiency in large scale systems. J. Phys. Chem. B. 2007;111:5405–5410. doi: 10.1021/jp068826w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W., Chen J.H., Brooks C.L. Peptide and protein folding and conformational equilibria: theoretical treatment of electrostatics and hydrogen bonding with implicit solvent models. Adv. Protein Chem. 2006;72:173–198. doi: 10.1016/S0065-3233(05)72007-6. [DOI] [PubMed] [Google Scholar]
- Imberty A., Perez S. Structure, conformation, and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem. Rev. 2000;100:4567–4588. doi: 10.1021/cr990343j. [DOI] [PubMed] [Google Scholar]
- Jang Y.H., Goddard W.A., Noyes K.T., Sowers L.C., Hwang S., Chung D.S. pKa values of guanine in water: density functional theory calculations combined with Poisson–Boltzmann continuum-solvation model. J. Phys. Chem. B. 2003;107:344–357. doi: 10.1021/jp020774x. [DOI] [Google Scholar]
- Jensen J.H., Li H., Robertson A.D., Molina P.A. Prediction and rationalization of protein pKa values using QM and QM/MM methods. J. Phys. Chem. A. 2005;109:6634–6643. doi: 10.1021/jp051922x. [DOI] [PubMed] [Google Scholar]
- Johansson A.C.V., Lindahl E. Position-resolved free energy of solvation for amino acids in lipid membranes from molecular dynamics simulations. Proteins: Struct. Funct. Bioinform. 2008;70:1332–1344. doi: 10.1002/prot.21629. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Tirado-Rives J. The OPLS potential functions for proteins—energy minimizations for crystals of cyclic-peptides and crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc. Natl Acad. Sci. USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11 225–11 236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- Joseph-McCarthy D., Baber J.C., Feyfant E., Thompson D.C., Humblet C. Lead optimization via high-throughput molecular docking. Curr. Opin. Drug Discov. Devel. 2007;10:264–274. [PubMed] [Google Scholar]
- Kamberaj H., van der Vaart A. Multiple scaling replica exchange for the conformational sampling of biomolecules in explicit water. J. Chem. Phys. 2007;127:234 102. doi: 10.1063/1.2806930. [DOI] [PubMed] [Google Scholar]
- Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. doi: 10.1021/jp003919d. [DOI] [Google Scholar]
- Kaminski G.A., Stern H.A., Berne B.J., Friesner R.A., Cao Y.X.X., Murphy R.B., Zhou R.H., Halgren T.A. Development of a polarizable force field for proteins via ab initio quantum chemistry: first generation model and gas phase tests. J. Comput. Chem. 2002;23:1515–1531. doi: 10.1002/jcc.10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski G.A., Stern H.A., Berne B.J., Friesner R.A. Development of an accurate and robust polarizable molecular mechanics force field from ab initio quantum chemistry. J. Phys. Chem. A. 2004;108:621–627. doi: 10.1021/jp0301103. [DOI] [Google Scholar]
- Kamiya N., Yonezawa Y., Nakamura H., Higo J. Protein-inhibitor flexible docking by a multicanonical sampling: native complex structure with the lowest free energy and a free-energy barrier distinguishing the native complex from the others. Proteins: Struct. Funct. Bioinform. 2008;70:41–53. doi: 10.1002/prot.21409. [DOI] [PubMed] [Google Scholar]
- Karplus M., Kuriyan J. Molecular dynamics and protein function. Proc. Natl Acad. Sci. USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M., Gao Y.Q., Ma J.P., van der Vaart A., Yang W. Protein structural transitions and their functional role. Phil. Trans. R. Soc. A. 2005;363:331–355. doi: 10.1098/rsta.2004.1496. [DOI] [PubMed] [Google Scholar]
- Khandogin J., York D.M. Quantum descriptors for biological macromolecules from linear-scaling electronic structure methods. Proteins: Struct. Funct. Bioinform. 2004;56:724–737. doi: 10.1002/prot.20171. [DOI] [PubMed] [Google Scholar]
- Khandogin J., Musier-Forsyth K., York D.M. Insights into the regioselectivity and RNA-binding affinity of HIV-1 nucleocapsid protein from linear-scaling quantum methods. J. Mol. Biol. 2003;330:993–1004. doi: 10.1016/S0022-2836(03)00658-2. [DOI] [PubMed] [Google Scholar]
- Kim B.C., Young T., Harder E., Friesner R.A., Berne B.J. Structure and dynamics of the solvation of bovine pancreatic trypsin inhibitor in explicit water: a comparative study of the effects of solvent and protein polarizability. J. Phys. Chem. B. 2005;109:16 529–16 538. doi: 10.1021/jp051569v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnear B.S., Jarrold M.F., Hansmann U.H.E. All-atom generalized-ensemble simulations of small proteins. J. Mol. Graph. Model. 2004;22:397–403. doi: 10.1016/j.jmgm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt A., Schuurmann G. Cosmo—a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 1993;2:799–805. doi: 10.1039/P29930000799. [DOI] [Google Scholar]
- Koehl P. Electrostatics calculations: latest methodological advances. Curr. Opin. Struct. Biol. 2006;16:142–151. doi: 10.1016/j.sbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Kouwijzer M., Grootenhuis P.D.J. Parametrization and applications of CHEAT95, an extended atom force-field for hydrated (oligo) saccharides. J. Phys. Chem. 1995;99:13 426–13 436. doi: 10.1021/j100036a017. [DOI] [Google Scholar]
- Krömer R.T. Structure-based drug design: docking and scoring. Curr. Protein Pept. Sci. 2007;8:312–328. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- Kuttel M., Brady J.W., Naidoo K.J. Carbohydrate solution simulations: producing a force field with experimentally consistent primary alcohol rotational frequencies and populations. J. Comput. Chem. 2002;23:1236–1243. doi: 10.1002/jcc.10119. [DOI] [PubMed] [Google Scholar]
- Leontiadou H., Mark A.E., Marrink S.J. Ion transport across transmembrane pores. Biophys. J. 2007;92:4209–4215. doi: 10.1529/biophysj.106.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer-simulation of protein folding. Nature. 1975;253:694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- Lim, D., Jenson, J., Repasky, M. P. & Jorgensen, W. L. 1999 In Transition state modeling for catalysis (eds D. G. Truhlar & K. Morokuma). ACS Symposium Series 721, pp. 74–85. Washington, DC: Oxford University Press.
- Lindorff-Larsen K., Kristjansdottir S., Teilum K., Fieber W., Dobson C.M., Poulsen F.M., Vendruscolo M. Determination of an ensemble of structures representing the denatured state of the bovine acyl-coenzyme a binding protein. J. Am. Chem. Soc. 2004;126:3291–3299. doi: 10.1021/ja039250g. [DOI] [PubMed] [Google Scholar]
- Lippow S.M., Tidor B. Progress in computational protein design. Curr. Opin. Biotechnol. 2007;18:305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.B., Warshel A. The catalytic effect of dihydrofolate reductase and its mutants is determined by reorganization energies. Biochemistry. 2007;46:6011–6025. doi: 10.1021/bi700201w. [DOI] [PubMed] [Google Scholar]
- Lodola A., Mor M., Zurek J., Tarzia G., Piomelli D., Harvey J.N., Mulholland A.J. Conformational effects in enzyme catalysis: reaction via a high energy conformation in fatty acid amide hydrolase. Biophys. J. 2007;92:L20–L22. doi: 10.1529/biophysj.106.098434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan I., Head-Gordon T. An analytical electrostatic model for salt screened interactions between multiple proteins. J. Chem. Theory Comput. 2006;2:541–555. doi: 10.1021/ct050263p. [DOI] [PubMed] [Google Scholar]
- Lyne P.D., Hodoscek M., Karplus M. A hybrid QM–MM potential employing Hartree–Fock or density functional methods in the quantum region. J. Phys. Chem. A. 1999;103:3462–3471. doi: 10.1021/jp982115j. [DOI] [Google Scholar]
- Lyne P.D., Mulholland A.J., Richards W.G. Insights into chorismate mutase catalysis from a combined QM/MM simulation of the enzyme reaction. J. Am. Chem. Soc. 1995;117:11 345–11 350. doi: 10.1021/ja00150a037. [DOI] [Google Scholar]
- Mackerell A.D. Empirical force fields for biological macromolecules: overview and issues. J. Comput. Chem. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- MacKerell, A. D. 2005 Empirical force fields for proteins: current status and future directions. In Annual reports in computational chemistry, vol. 1 (ed. C. Simmerling), pp. 91–111. Oxford, UK: Elsevier.
- MacKerell A.D., Banavali N.K. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000;21:105–120. doi: 10.1002/(SICI)1096-987X(20000130)21:2%3C105::AID-JCC3%3E3.0.CO;2-P. [DOI] [Google Scholar]
- MacKerell A.D., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Feig M., Brooks C.L. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004a;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Feig M., Brooks C.L. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004b;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- Marti S., Moliner V. Improving the QM/MM description of chemical processes: a dual level strategy to explore the potential energy surface in very large systems. J. Chem. Theory Comput. 2005;1:1008–1016. doi: 10.1021/ct0501396. [DOI] [PubMed] [Google Scholar]
- Marti S., Roca M., Andres J., Moliner V., Silla E., Tunon I., Bertran J. Theoretical insights in enzyme catalysis. Chem. Soc. Rev. 2004;33:98–107. doi: 10.1039/b301875j. [DOI] [PubMed] [Google Scholar]
- Martin M.E., Aguilar M.A., Chalmet S., Ruiz-Lopez M.F. An iterative procedure to determine Lennard-Jones parameters for their use in quantum mechanics/molecular mechanics liquid state simulations. Chem. Phys. 2002;284:607–614. doi: 10.1016/S0301-0104(02)00785-1. [DOI] [Google Scholar]
- Mata R.A., Werner H.J., Thiel S., Thiel W. Toward accurate barriers for enzymatic reactions: QM/MM case study on p-hydroxybenzoate hydroxylase. J. Chem. Phys. 2008;128:025 104. doi: 10.1063/1.2823055. [DOI] [PubMed] [Google Scholar]
- Mayor U., et al. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- McCammon J.A., Gelin B.R., Karplus M. Dynamics of folded proteins. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- McGuffee S.R., Elcock A.H. Atomically detailed simulations of concentrated protein solutions: the effects of salt, pH, point mutations, and protein concentration in simulations of 1000-molecule systems. J. Am. Chem. Soc. 2006;128:12 098–12 110. doi: 10.1021/ja0614058. [DOI] [PubMed] [Google Scholar]
- McInnes C. Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 2007;11:494–502. doi: 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- McNamara J.P., Muslim A.M., Abdel-Aal H., Wang H., Mohr M., Hillier I.H., Bryce R.A. Towards a quantum mechanical force field for carbohydrates: a reparametrized semi-empirical MO approach. Chem. Phys. Lett. 2004;394:429–436. doi: 10.1016/j.cplett.2004.07.037. [DOI] [Google Scholar]
- Ming Y., Lai G.L., Tong C.H., Wood R.H., Doren D.J. Free energy perturbation study of water dimer dissociation kinetics. J. Chem. Phys. 2004;121:773–777. doi: 10.1063/1.1756574. [DOI] [PubMed] [Google Scholar]
- Moitessier N., Englebienne P., Lee D., Lawandi J., Corbeil C.R. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br. J. Pharmacol. 2008;153:S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y.G., Yang Y., Xu W.X. Hybrid Hamiltonian replica exchange molecular dynamics simulation method employing the Poisson–Boltzmann model. J. Chem. Phys. 2007;127:084 119. doi: 10.1063/1.2772264. [DOI] [PubMed] [Google Scholar]
- Mulholland A.J. Modelling enzyme reaction mechanisms, specificity and catalysis. Drug Discov. Today. 2005;10:1393–1402. doi: 10.1016/S1359-6446(05)03611-1. [DOI] [PubMed] [Google Scholar]
- Mulholland A.J. Chemical accuracy in QM/MM calculations on enzyme-catalysed reactions. Chem. Central J. 2007;1:1–5. doi: 10.1186/1752-153X-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland A.J. Computational enzymology: modelling the mechanisms of biological catalysts. Biochem. Soc. Trans. 2008;36:22–26. doi: 10.1042/BST0360022. [DOI] [PubMed] [Google Scholar]
- Mulholland A.J., Lyne P.D., Karplus M. Ab initio QM/MM study of the citrate synthase mechanism. A low-barrier hydrogen bond is not involved. J. Am. Chem. Soc. 2000;122:534–535. doi: 10.1021/ja992874v. [DOI] [Google Scholar]
- Muller R.P., Warshel A. Ab initio calculations of free-energy barriers for chemical-reactions in solution. J. Phys. Chem. 1995;99:17 516–17 524. doi: 10.1021/j100049a009. [DOI] [PubMed] [Google Scholar]
- Neria E., Fischer S., Karplus M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 1996;105:1902–1921. doi: 10.1063/1.472061. [DOI] [Google Scholar]
- Nielsen J.E., McCammon J.A. Calculating pKa values in enzyme active sites. Protein Sci. 2003;12:1894–1901. doi: 10.1110/ps.03114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.O., Lopez C.F., Srinivas G., Klein M.L. Coarse grain models and the computer simulation of soft materials. J. Phys. Condens. Matter. 2004;16:R481–R512. doi: 10.1088/0953-8984/16/15/R03. [DOI] [Google Scholar]
- Nymeyer H. How efficient is replica exchange molecular dynamics? An analytic approach. J. Chem. Theory Comput. 2008;4:626–636. doi: 10.1021/ct7003337. [DOI] [PubMed] [Google Scholar]
- Okamoto Y. Generalized-ensemble algorithms: enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J. Mol. Graph. Model. 2004;22:425–439. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Okur A., Strockbine B., Hornak V., Simmerling C. Using PC clusters to evaluate the transferability of molecular mechanics force fields for proteins. J. Comput. Chem. 2003;24:21–31. doi: 10.1002/jcc.10184. [DOI] [PubMed] [Google Scholar]
- Olsson M.H.M., Sharma P.K., Warshel A. Simulating redox coupled proton transfer in cytochrome c oxidase: looking for the proton bottleneck. FEBS Lett. 2005;579:2026–2034. doi: 10.1016/j.febslet.2005.02.051. [DOI] [PubMed] [Google Scholar]
- Pang J.Y., Pu J.Z., Gao J.L., Truhlar D.G., Allemann R.K. Hydride transfer reaction catalyzed by hyperthermophilic dihydrofolate reductase is dominated by quantum mechanical tunneling and is promoted by both inter- and intramonomeric correlated motions. J. Am. Chem. Soc. 2006;128:8015–8023. doi: 10.1021/ja061585l. [DOI] [PubMed] [Google Scholar]
- Park C., Carlson M.J., Goddard W.A. Solvent effects on the secondary structures of proteins. J. Phys. Chem. A. 2000;104:2498–2503. doi: 10.1021/jp9911189. [DOI] [Google Scholar]
- Patel S., Mackerell A.D., Brooks C.L. CHARMM fluctuating charge force field for proteins: II. Protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J. Comput. Chem. 2004;25:1504–1514. doi: 10.1002/jcc.20077. [DOI] [PubMed] [Google Scholar]
- Patriksson A., van der Spoel D. A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- Pentikäinen U., Pentikäinen O.T., Mulholland A.J. Cooperative symmetric to asymmetric conformational transition of the apo-form of scavenger decapping enzyme revealed by simulations. Proteins: Struct. Funct. Bioinform. 2008;70:498–508. doi: 10.1002/prot.21540. [DOI] [PubMed] [Google Scholar]
- Perez A., Marchan I., Svozil D., Sponer J., Cheatham T.E., Laughton C.A., Orozco M. Refinenement of the Amber force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.C., et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinini E., Ceccarelli M., Affinito F., Brunetti R., Jacoboni C. Biased molecular simulations for free-energy mapping: a comparison on the KcsA channel as a test case. J. Chem. Theory Comput. 2008;4:173–183. doi: 10.1021/ct7001896. [DOI] [PubMed] [Google Scholar]
- Ponder J.W., Case D.A. Force fields for protein simulations. Adv. Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- Ponder J.W., Richards F.M. An efficient newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 1987;8:1016–1024. doi: 10.1002/jcc.540080710. [DOI] [Google Scholar]
- Prat-Resina X., Bofill J.M., Gonzalez-Lafont A., Lluch J.M. Geometry optimization and transition state search in enzymes: different options in the microiterative method. Int. J. Quantum Chem. 2004;98:367–377. doi: 10.1002/qua.20072. [DOI] [Google Scholar]
- Raha K., Merz K.M. A quantum mechanics-based scoring function: study of zinc ion-mediated ligand binding. J. Am. Chem. Soc. 2004;126:1020–1021. doi: 10.1021/ja038496i. [DOI] [PubMed] [Google Scholar]
- Raha, K. & Merz, K. M. 2005 Calculating binding free energy in protein–ligand interaction. In Annual reports in computational chemistry, vol. 1 (ed. C. Simmerling), pp. 113–130. Oxford, UK: Elsevier.
- Raha K., Peters M.B., Wang B., Yu N., WollaCott A.M., Westerhoff L.M., Merz K.M. The role of quantum mechanics in structure-based drug design. Drug Discov. Today. 2007;12:725–731. doi: 10.1016/j.drudis.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Rajamani R., Good A.C. Ranking poses in structure-based lead discovery and optimization: current trends in scoring function development. Curr. Opin. Drug Discov. Devel. 2007;10:308–315. [PubMed] [Google Scholar]
- Ranaghan, K. E. & Mulholland, A. J. 2004 Conformational effects in enzyme catalysis: QM/MM free energy calculation of the ‘NAC’ contribution in chorismate mutase. Chem. Commun. 1238–1239. [DOI] [PubMed]
- Ranaghan K.E., Ridder L., Szefczyk B., Sokalski W.A., Hermann J.C., Mulholland A.J. Insights into enzyme catalysis from QM/MM modelling: transition state stabilization in chorismate mutase. Mol. Phys. 2003;101:2695–2714. doi: 10.1080/00268970310001593286. [DOI] [Google Scholar]
- Ranaghan K.E., Ridder L., Szefczyk B., Sokalski W.A., Hermann J.C., Mulholland A.J. Transition state stabilization and substrate strain in enzyme catalysis: ab initio QM/MM modelling of the chorismate mutase reaction. Org. Biomol. Chem. 2004;2:968–980. doi: 10.1039/b313759g. [DOI] [PubMed] [Google Scholar]
- Remler D.K., Madden P.A. Molecular-dynamics without effective potentials via the Car–Parrinello approach. Mol. Phys. 1990;70:921–966. doi: 10.1080/00268979000101451. [DOI] [Google Scholar]
- Ren P.Y., Ponder J.W. Polarizable atomic multipole water model for molecular mechanics simulation. J. Phys. Chem. B. 2003;107:5933–5947. doi: 10.1021/jp027815+. [DOI] [Google Scholar]
- Riccardi D., Schaefer P., Cui Q. pKa calculations in solution and proteins with QM/MM free energy perturbation simulations: a quantitative test of QM/MM protocols. J. Phys. Chem. B. 2005;109:17 715–17 733. doi: 10.1021/jp0517192. [DOI] [PubMed] [Google Scholar]
- Ridder L., Mulholland A.J., Rietjens I.M.C.M., Vervoort J. A quantum mechanical/molecular mechanical study of the hydroxylation of phenol and halogenated derivatives by phenol hydroxylase. J. Am. Chem. Soc. 2000;122:8728–8738. doi: 10.1021/ja0007814. [DOI] [Google Scholar]
- Ridder L., Rietjens I.M.C.M., Vervoort J., Mulholland A.J. Quantum mechanical/molecular mechanical free energy simulations of the glutathione S-transferase (M1-1) reaction with phenanthrene 9,10-oxide. J. Am. Chem. Soc. 2002;124:9926–9936. doi: 10.1021/ja0256360. [DOI] [PubMed] [Google Scholar]
- Ridder L., Harvey J.N., Rietjens I.M.C.M., Vervoort J., Mulholland A.J. Ab initio QM/MM modeling of the hydroxylation step in p-hydroxybenzoate hydroxylase. J. Phys. Chem. B. 2003;107:2118–2126. doi: 10.1021/jp026213n. [DOI] [Google Scholar]
- Ritchie D.W. Recent progress and future directions in protein–protein docking. Curr. Protein Pept. Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- Rod T.H., Ryde U. Accurate QM/MM free energy calculations of enzyme reactions: methylation by catechol O-methyltransferase. J. Chem. Theory Comput. 2005;1:1240–1251. doi: 10.1021/ct0501102. [DOI] [PubMed] [Google Scholar]
- Roitberg, A. E. 2005 Non-equilibrium approaches to free energy calculations. In Annual reports in computational chemistry, vol. 1 (ed. C. Simmerling), pp. 103–112. Oxford, UK: Elsevier.
- Rosta E., Klahn M., Warshel A. Towards accurate ab initio QM/MM calculations of free-energy profiles of enzymatic reactions. J. Phys. Chem. B. 2006;110:2934–2941. doi: 10.1021/jp057109j. [DOI] [PubMed] [Google Scholar]
- Röthlisberger U., Carloni P., Doclo K., Parrinello M. A comparative study of galactose oxidase and active site analogs based on QM/MM Car Parrinello simulations. J. Biol. Inorg. Chem. 2000;5:236–250. doi: 10.1007/s007750050368. [DOI] [PubMed] [Google Scholar]
- Roux B. Computational studies of the gramicidin channel. Acc. Chem. Res. 2002;35:366–375. doi: 10.1021/ar010028v. [DOI] [PubMed] [Google Scholar]
- Sansom M.S.P., Bond P.J., Deol S.S., Grottesi A., Haider S., Sands Z.A. Molecular simulations and lipid–protein interactions: potassium channels and other membrane proteins. Biochem. Soc. Trans. 2005;33:916–920. doi: 10.1042/BST20050916. [DOI] [PubMed] [Google Scholar]
- Sansom M.S.P., Scott K.A., Bond P.J. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem. Soc. Trans. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- Schuler L.D., Daura X., van Gunsteren W.F. An improved Gromos96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 2001;22:1205–1218. doi: 10.1002/jcc.1078. [DOI] [Google Scholar]
- Scott W.R.P., et al. The Gromos biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. doi: 10.1021/jp984217f. [DOI] [Google Scholar]
- Senderowitz H., Parish C., Still W.C. Carbohydrates: united atom Amber* parameterization of pyranoses and simulations yielding anomeric free energies. J. Am. Chem. Soc. 1996;118:2078–2086. doi: 10.1021/ja9529652. [DOI] [Google Scholar]
- Senn H.M., Thiel W. QM/MM studies of enzymes. Curr. Opin. Chem. Biol. 2007;11:182–187. doi: 10.1016/j.cbpa.2007.01.684. [DOI] [PubMed] [Google Scholar]
- Shurki A., Warshel A. Structure/function correlations of proteins using MM, QM/MM, and related approaches: methods, concepts, pitfalls, and current progress. Adv. Protein Chem. 2003;66:249–313. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- Siebert H.C., et al. A new combined computational and NMR-spectroscopical strategy for the identification of additional conformational constraints of the bound ligand in an aprotic solvent. Chembiochem. 2000;1:181–195. doi: 10.1002/1439-7633(20001002)1:3%3C181::AID-CBIC181%3E3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Sindhikara D., Meng Y.L., Roitberg A.E. Exchange frequency in replica exchange molecular dynamics. J. Chem. Phys. 2008;128:024 103. doi: 10.1063/1.2816560. [DOI] [PubMed] [Google Scholar]
- Stanton C.L., Houk K.N. Benchmarking pKa prediction methods for residues in proteins. J. Chem. Theory Comput. 2008;4:951–966. doi: 10.1021/ct8000014. [DOI] [PubMed] [Google Scholar]
- Stortz C.A., Cerezo A.S. MM3 potential energy surfaces of the 2-linked glucosyl trisaccharides α-kojitriose and β-sophorotriose. Carbohydr. Res. 2003;338:1679–1689. doi: 10.1016/S0008-6215(03)00265-9. [DOI] [PubMed] [Google Scholar]
- Strajbl M., Hong G.Y., Warshel A. Ab initio QM/MM simulation with proper sampling: “first principle” calculations of the free energy of the autodissociation of water in aqueous solution. J. Phys. Chem. B. 2002;106:13 333–13 343. doi: 10.1021/jp021625h. [DOI] [Google Scholar]
- Strajbl M., Shurki A., Kato M., Warshel A. Apparent NAC effect in chorismate mutase reflects electrostatic transition state stabilization. J. Am. Chem. Soc. 2003a;125:10 228–10 237. doi: 10.1021/ja0356481. [DOI] [PubMed] [Google Scholar]
- Strajbl M., Shurki A., Warshel A. Converting conformational changes to electrostatic energy in molecular motors: the energetics of ATP synthase. Proc. Natl Acad. Sci. USA. 2003b;100:14 834–14 839. doi: 10.1073/pnas.2436328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y., Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. doi: 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- Taft C.A., da Silva V.B., de Paula da Silva C.H.T. Current topics in computer-aided drug design. J. Pharm. Sci. 2008;97:1089–1098. doi: 10.1002/jps.21293. [DOI] [PubMed] [Google Scholar]
- Tai K. Conformational sampling for the impatient. Biophys. Chem. 2004;107:213–220. doi: 10.1016/j.bpc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Tomasi J. Thirty years of continuum solvation chemistry: a review, and prospects for the near future. Theor. Chem. Acc. 2004;112:184–203. doi: 10.1007/s00214-004-0582-3. [DOI] [Google Scholar]
- Torrie G.M., Valleau J.P. Non-physical sampling distributions in Monte–Carlo free-energy estimation—umbrella sampling. J. Comput. Phys. 1977;23:187–199. doi: 10.1016/0021-9991(77)90121-8. [DOI] [Google Scholar]
- Tozzini V. Coarse-grained models for proteins. Curr. Opin. Struct. Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Truhlar D.G. Valence bond theory for chemical dynamics. J. Comput. Chem. 2007;28:73–86. doi: 10.1002/jcc.20529. [DOI] [PubMed] [Google Scholar]
- van der Kamp, M. W., Perruccio, F. & Mulholland, A. J. 2008 High-level QM/MM modelling predicts an arginine as the acid in the condensation reaction catalysed by citrate synthase. Chem. Commun 1874–1876. ( 10.1039/B800496J) [DOI] [PubMed]
- Van der Vaart A., Gogonea V., Dixon S.L., Merz K.M. Linear scaling molecular orbital calculations of biological systems using the semiempirical divide and conquer method. J. Comput. Chem. 2000;21:1494–1504. doi: 10.1002/1096-987X(200012)21:16%3C1494::AID-JCC6%3E3.0.CO;2-4. [DOI] [Google Scholar]
- Van Gunsteren W.F., et al. Biomolecular modeling: goals, problems, perspectives. Angew. Chem. Int. Ed. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- Varnai P., Warshel A. Computer simulation studies of the catalytic mechanism of human aldose reductase. J. Am. Chem. Soc. 2000;122:3849–3860. doi: 10.1021/ja994246j. [DOI] [Google Scholar]
- Venturoli M., Sperotto M.M., Kranenburg M., Smit B. Mesoscopic models of biological membranes. Phys. Lett. 2006;437:1–54. [Google Scholar]
- Villa J., Warshel A. Energetics and dynamics of enzymatic reactions. J. Phys. Chem. B. 2001;105:7887–7907. doi: 10.1021/jp011048h. [DOI] [Google Scholar]
- Von Dreele R.B., Stephens P.W., Smith G.D., Blessing R.H. The first protein crystal structure determined from high-resolution X-ray powder diffraction data: a variant of T3R3 human insulin-zinc complex produced by grinding. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:1549–1553. doi: 10.1107/S0907444900013901. [DOI] [PubMed] [Google Scholar]
- Vorobyov I.V., Anisimov V.M., MacKerell A.D. Polarizable empirical force field for alkanes based on the classical Drude oscillator model. J. Phys. Chem. B. 2005;109:18 988–18 999. doi: 10.1021/jp053182y. [DOI] [PubMed] [Google Scholar]
- Wang Z.X., Zhang W., Wu C., Lei H.X., Cieplak P., Duan Y. Strike a balance: optimization of backbone torsion parameters of Amber polarizable force field for simulations of proteins and peptides. J. Comput. Chem. 2006;27:781–790. doi: 10.1002/jcc.20386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warme P.K., Momany F.A., Rumball S.V., Tuttle R.W., Scheraga H.A. Computation of structures of homologous proteins—α-lactalbumin from lysozyme. Biochemistry. 1974;13:768–782. doi: 10.1021/bi00701a020. [DOI] [PubMed] [Google Scholar]
- Warshel A. Bicycle-pedal model for 1st step in vision process. Nature. 1976;260:679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- Warshel A. Wiley; New York, NY: 1997. Computer modeling of chemical reactions in enzymes and solutions. [Google Scholar]
- Warshel A. Molecular dynamics simulations of biological reactions. Acc. Chem. Res. 2002;35:385–395. doi: 10.1021/ar010033z. [DOI] [PubMed] [Google Scholar]
- Warshel A. Computer simulations of enzyme catalysis: methods, progress, and insights. Annu. Rev. Biophys. Biomol. Struct. 2003;32:425–443. doi: 10.1146/annurev.biophys.32.110601.141807. [DOI] [PubMed] [Google Scholar]
- Warshel A., Karplus M. Calculation of ground and excited-state potential surfaces of conjugated molecules. 1. Formulation and parametrization. J. Am. Chem. Soc. 1972;94:5612–5625. doi: 10.1021/ja00771a014. [DOI] [Google Scholar]
- Warshel A., Levitt M. Theoretical studies of enzymic reactions—dielectric, electrostatic and steric stabilization of carbonium-ion in reaction of lysozyme. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Warshel A., Levitt M., Lifson S. Consistent force field for calculation of vibrational spectra and conformations of some amides and lactam rings. J. Mol. Spectrosc. 1970;33:84–99. doi: 10.1016/0022-2852(70)90054-8. [DOI] [Google Scholar]
- Warshel A., Sharma P.K., Kato M., Parson W.W. Modeling electrostatic effects in proteins. Biochim. Biophys. Acta Proteins Proteomics. 2006a;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Warshel A., Sharma P.K., Kato M., Xiang Y., Liu H.B., Olsson M.H.M. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006b;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- Warshel A., Kato M., Pisliakov A.V. Polarizable force fields: history, test cases, and prospects. J. Chem. Theory Comput. 2007;3:2034–2045. doi: 10.1021/ct700127w. [DOI] [PubMed] [Google Scholar]
- Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P. A new force-field for molecular mechanical simulations of nucleic-acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. doi: 10.1021/ja00315a051. [DOI] [Google Scholar]
- Wood R.H., Yezdimer E.M., Sakane S., Barriocanal J.A., Doren D.J. Free energies of solvation with quantum mechanical interaction energies from classical mechanical simulations. J. Chem. Phys. 1999;110:1329–1337. doi: 10.1063/1.478009. [DOI] [Google Scholar]
- Woodcock H.L., Hodoscek M., Sherwood P., Lee Y.S., Schaefer H.F., Brooks B.R. Exploring the quantum mechanical/molecular mechanical replica path method: a pathway optimization of the chorismate to prephenate Claisen rearrangement catalyzed by chorismate mutase. Theor. Chem. Acc. 2003;109:140–148. [Google Scholar]
- Woods, C. J. & Mulholland, A. J. 2008 Multiscale modelling of biological systems. In RSC specialist periodical reports: chemical modelling and theory, vol. 5 (ed. A. Hinchliffe), pp. 1–38. Cambridge, UK: Royal Society of Chemistry.
- Woods R.J., Dwek R.A., Edge C.J., Fraser-Reid B. Molecular mechanical and molecular dynamical simulations of glycoproteins and oligosaccharides. 1. GLYCAM-93 parameter development. J. Phys. Chem. 1995;99:3832–3846. doi: 10.1021/j100011a061. [DOI] [Google Scholar]
- Woods C.J., et al. Grid computing and biomolecular simulation. Phil. Trans. R. Soc. A. 2005;363:2017–2035. doi: 10.1098/rsta.2005.1626. [DOI] [PubMed] [Google Scholar]
- Woods C.J., Manby F.R., Mulholland A.J. An efficient method for the calculation of quantum mechanics/molecular mechanics free energies. J. Chem. Phys. 2008;128:014 109. doi: 10.1063/1.2805379. [DOI] [PubMed] [Google Scholar]
- Xu Z.T., Luo H.H., Tieleman D.P. Modifying the OPLS-AA force field to improve hydration free energies for several amino acid side chains using new atomic charges and an off-plane charge model for aromatic residues. J. Comput. Chem. 2007;28:689–697. doi: 10.1002/jcc.20560. [DOI] [PubMed] [Google Scholar]
- Yu Z.Y., Jacobson M.P., Friesner R.A. What role do surfaces play in GB models? A new-generation of surface-generalized Born model based on a novel Gaussian surface for biomolecules. J. Comput. Chem. 2006;27:72–89. doi: 10.1002/jcc.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.K., Kua J., McCammon J.A. Influence of structural fluctuation on enzyme reaction energy barriers in combined quantum mechanical/molecular mechanical studies. J. Phys. Chem. B. 2003;107:4459–4463. doi: 10.1021/jp022525e. [DOI] [Google Scholar]